Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
most common malignant tumor in the region of the head and neck, and
the diagnosis of more than half of patients with HNSCC is
accompanied by lymph node metastasis (1). Previous studies have indicated that
lymph node metastasis is an important prognostic factor for
patients with HNSCC, particularly for locally advanced cases
(2,3).
Therefore, it is important to identify the mechanism of lymph node
metastasis in patients with HNSCC, which may contribute to the
prevention, diagnosis and treatment of HNSCC.
C-C chemokine receptor type 7 (CCR7) is an important
chemokine that is expressed on the surface of a number of tumor
cells and belongs to the C-C subgroup of chemokines (4). CCR7 is expressed in numerous malignant
tumors, including leukemia, lymphomas, lymphoproliferative
syndromes and certain epithelial solid tumors (5,6). It also
has effects on tumor migration and lymph node dissemination through
combined action with CC ligand (CCL)21, and CCL19 (5,6). Previous
studies have demonstrated that the expression of CCR7 is associated
with the invasion and metastasis of tumors through various
signaling pathways in HNSCC (7–9).
MicroRNAs (miRNAs/miRs) belong to a group of short
non-coding RNAs existing in eukaryotic organisms, which are usually
comprised of 19–25 nucleotides (10,11). The
mapping association between miRNAs and mRNAs is not one-to-one,
thus the same miRNAs can perform a different role in different
types of cancer through identifying specific sequences combined
with mRNA, and serve as important post-transcriptional regulators
of gene expression (12).
hsa-miR-125a-5p, located at 19q13.41, is a member of
the mature miR-125a family (13).
Numerous studies (14–16) in different tumors have investigated
the role of hsa-miR-125a-5p. However, there are few studies on the
role between the expression of hsa-miR-125a-5p and CCR7 in HNSCC,
particularly in cell lines with distinct metastatic, and invasive
properties. The present study explored the role of hsa-miR-125a-5p
in HNSCC, and the association between hsa-miR-125a-5p and CCR7 in
HNSCC PCI-37B cells. In in vitro studies, overexpression of
hsa-miR-125a-5p was identified to upregulate the expression of CCR7
and serves an oncogenic role in HNSCC.
Materials and methods
Cell lines and cell culture
The PCI-37B cell line has been well characterized as
a HNSCC cell line with evident ability of metastasis and invasion,
and expresses a high level of CCR7 mRNA (7). The PCI-37B cell line was provided by the
University of Pittsburgh Cancer Institute (Pittsburgh, PA, USA).
The cells were maintained in Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
containing 10% fetal bovine serum (FBS; Gibco, USA), 100 U/ml
penicillin and 100 µg/ml streptomycin (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2 atmosphere. The
cells were trypsinized with 0.25% trypsin (Beyotime Institute of
Biotechnology, Haimen, China) when 80% confluent.
Patients and samples
A total of 15 patients (11 males and 4 females; age
range, 47–72) with HNSCC who underwent surgery at the Department of
Oromaxillofacial-Head and Neck Surgery, School of Stomatology,
China Medical University (Shenyang, China) between August 2014 and
September 2014 were recruited for the present study. No patients
have received chemotherapy or radiotherapy prior to surgery.
Written informed consent was obtained from all patients prior to
collection of the samples. Subsequent to tumor resection, tumor
tissue and matched normal tissue adjacent to the tumor were
immediately collected into a collecting pipe. Tissues were then
conserved in liquid nitrogen within a minute until use in
experiments. The diagnosis of the patients was confirmed by the
pathological results of intraoperative and postoperative
examination according to the benchmark of the national
comprehensive cancer network (17).
The research was approved by the Ethics Committee of the China
Medical University.
Plasmid construction
To create miR-125a-5p overexpression (miR-125a-5p+)
and negative control (miR-125a-5p-) plasmids, GV214 was used as the
vector (Shanghai GenePharma Co., Ltd., Shanghai, China). The
objective sequence (5′-UCCCUGAGACCCUUUAACCUGUGA-3′) and negative
control sequence (5′-TTCTCCGAACGTGTCACGT-3′) were inserted into the
vector separately. The constructed plasmid was then tested by
sequencing following polymerase chain reaction (PCR) with
PrimerSTAR HS DNA Polymerase (Takara Biotechnology Co., Ltd.,
Dalian, China) with the primers (Thermo Fisher Scientific, Inc.):
P1, 5′-GTATGAGACCACTCGGATCCGGTCTTTCTGTCTCTGG-3′; and P2,
5′-AGCGGTTTAAACTTAAGCTTAAAAAATCAGTTGGTGGTC-3′, following
thermocycling conditions: 98°C for 5 min, 30 cycles of 98°C for 10
sec, 55°C for 10 sec and 72°C for 20 sec, and then 72°C for 5
min.
Transfection of miRNA-125a-5p
A total of 3×105 PCI-37B cells per well
were maintained in 6-wells plate for 24 h in an incubator with 5%
CO2 at 37°C. The cells were then transfected with 2 µg
of the two aforementioned plasmids (miR-125a-5p+, miR-125a-5p-)
using 8 µl Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
The cells were cultured for 48 h at 37°C in 5% CO2 prior
to subsequent experiments.
Total RNA isolation and fluorescence
reverse transcription-quantitative (RT-q) PCR
A high purity total RNA rapid extraction kit (BioTek
China, Beijing, China) was used to isolate the total RNA of the
cell lines, according to the manufacturer's protocol. The cell
suspensions were centrifuged at 4°C and 16,000 × g for 10 min. The
concentrations of samples were tested by microplate reader. The
corresponding cDNA was generated using RT subsequent to total RNA
of the 37B, 37B miR-125a-5p+ and 37B miR-125a-5p-, and 15 pairs of
tissue samples being obtained.
The method of qPCR was applied to analyze the
expression of miRNA125a-5p and the reference gene U6 in samples.
Relative quantification mode was adopted to process the data with
triplicate measurements and followed the formulation of the
following thermocycling conditions: 95°C for 10 min, 40 cycles of
95°C for 10 sec, 60°C for 20 sec and 72°C for 30 sec.
The oligonucleotide primers for qPCR were as
follows: miR-125a-5p forward, 5′-CCGTCCCTGAGACCCTTTAAC-3′ and
reverse, 5′-GTGCAGGGTCCGAGGTATTC-3′; U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The results of qPCR were quantified using the 2−ΔΔCq
method (18). Reverse transcriptase
(BioTek, Beijing, China), SYBR-Green Master mix (Solarbio Science
& Technology Co., Ltd., Beijing, China), ultraviolet
spectrophotometer NanoDrop™ 2000 (Thermo Fisher
Scientific, Inc.) and fluorescence qPCR thermal cycler Exicycler 96
(Bioneer Corporation, Daejeon, Korea) were the main instruments,
and reagents used in the aforementioned experiments.
Cell Counting Kit-8 (CCK-8), Transwell
and wound healing assay for cell proliferation, invasion and
metastasis
The three groups of cells (37B, 37B miR-125a-5p+,
37B miR-125a-5p-) were prepared on 96-well plates at a density of
~3,000 cells/well. At a particular time point (0, 24, 48, 72 and 96
h), 10 µl CCK-8 (Beyotime, China) was added to the corresponding
wells in each group of cells. The cells were then maintained in the
incubator for 1 h at 37°C in a 5% CO2 atmosphere.
Optical density was measured at 4.90 nm with the microplate reader
(BioTek, Beijing, China), and the data was analyzed to detect cell
proliferation.
The cell invasion assay was performed using a
Transwell chamber (Corning, NY, USA) and Matrigel (BD, Franklin
Lakes, USA). A transwell chamber was put into the 24-well plate and
the transwell chamber was covered with the Matrigel. DMEM (800 µl)
with 20% FBS was added to the lower chamber and 200 µl cell
suspension was added to the upper chamber. Cells were seeded at a
density of 2×104 cells/well. The cells were cultured for
24 h at 37°C in an atmosphere containing 5% CO2. Then
the chamber was washed with PBS twice (HyClone; GE Healthcare Life
Sciences, Logan, UT, USA), and filtered cells were fixed with
polyoxymethylene (Sinopharm Chemical Reagent Co., Ltd., Shanghai,
China) for 20 min at room temperature followed by 0.5% crystal
violet (Amresco, LLC, Solon, OH, USA) staining for 5 min at room
temperature. An inverted microscope (magnification, ×200;
Motic-AE31; Motic Incorporation, Ltd., Causeway Bay, Hong Kong) was
used to count the cells that had migrated to the lower microporous
membrane. A total of five fields were selected in each sample to
count the number of cells, and the mean was calculated.
The medium of the cells (37B, 37B miR-125a-5p +, 37B
miR-125a-5p-) was replaced with serum-free medium and 1 µg/ml of
mitomycin C (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was
added for 1 h prior to the experiment. Cell scratches were made
using a 200 µl pipette tip in each group, and the cell debris was
washed away using serum-free medium. Images of the cells were
captured at 0, 6, 12 and 24 h using an inverted microscope
(magnification, ×100; Motic-AE31; Motic Incorporation, Ltd.) camera
to note the location of the cells. The distance of migration was
calculated for each group.
Protein extraction and western
blotting
Before the protein extraction, mirnaviewer
(http://cbio.mskcc.org/cgi-bin/mirnaviewer/mirnaviewer.pl)
was used to predict the target protein of the hsa-miR-125a-5p. A
total protein extraction kit (cat. no. WLA019; Wanleibio, Shanghai,
China) and a bicinchoninic acid protein concentration assay kit
(cat. no. WLA004; Wanleibio, Co., Ltd.) were used to extract total
protein, and determine the protein concentration according to the
manufacturer's protocol. In the western blot analysis assays, 40 µg
of protein were size-fractionated through a SDS-PAGE gel (5%
concentration gel and 10% separation gel) and transferred onto
polyvinylidene fluorides membranes (EMD Millipore, Billerica, MA,
USA). The membranes were blocked in 5% (M/V) non-fat dry milk (room
temperature, shaken for 1 h) and then incubated with primary
antibodies (4°C overnight, 37°C 45 min for secondary antibodies).
The anti-CCR7 antibody (dilution, 1:500; cat. no. BYK-1305R;
Shanghai Bioye Biotechnology, Shanghai, China) and internal
reference antibody anti-β-actin (dilution, 1:1,000; cat. no.
WL0002; Wanleibio Co., Ltd.) were used as the primary antibodies.
Goat anti-rabbit IgG-horseradish peroxidase (1:5,000 diluted) (cat.
no. WLA023; Wanleibio Co., Ltd.) was used as the secondary
antibody. The protein expression of CCR7 was observed and analyzed
using a Gel Imaging System (cat. no. WD-9413B; Beijing Liuyi,
Biological Technology Co., Ltd., Beijing, China). Gel-Pro-Analyzer
software (version 6.0; Media Cybernetics, Inc., Rockville, MD, USA)
was used to analyze the optical density of the target strip
subsequent to being scanned with the gel image processing
system.
Survival analysis
The clinical data and hsa-miR-125a-5p expression
profile of HNSCC were downloaded from the Cancer Genome Atlas
database (https://cancergenome.nih.gov/). A total of 397
patients with hsa-miR-125a-5p expression data and clinical data
were derived from two high throughput sequencing platforms: BCGSC
IlluminaGA miRNASeq and BCGSC IlluminaHiSeq miRNASeq (https://wiki.nci.nih.gov/display/TCGA/miRNASeq; final
data download: July 20, 2014). The 397 cases of microRNA and
clinical follow-up data were used for analysis. The staging of
disease is according to the American Joint Committee on Cancer TNM
Staging Classification for the Lip and Oral Cavity (17). Patients in stage I and II were divided
into the low-grade group, patients in stage III and IV are divided
into the high-grade group. The threshold of microRNA expression was
analyzed by receiver operating characteristic curve. The cut-off
value was determined as the highest true positive rate together
with the lowest false positive rate. The patients were then divided
into two groups (high expression and low expression) according to
the threshold. A log rank test was used to obtain significant
P-values of the Kaplan-Meier overall survival curve. The last
contact time was kept as censored data.
Statistical analysis
The data of all assays are expressed as the mean ±
standard deviation from at least three independent experiments.
Differences between two groups were analyzed using the paired
Student's t-test. Differences among multiple groups were analyzed
using one-way analysis of variance followed by the
Student-Newman-Keuls test for post hoc analysis. The association
between the expression of microRNA and tumor stage/survival time
was analyzed by the survival data matrix. P<0.05 was considered
to indicate a statistically significant difference. Statistical
analyses were performed using SPSS version 22 (IBM Corp., Armonk,
NY, USA).
Results
hsa-miR-125a-5p expression is
increased in cancer tissue compared with adjacent normal
tissue
hsa-miR-125a-5p expression of cancer tissue and
adjacent normal tissue in 15 patients with HNSCC were determined by
fluorescence qPCR. The 2−ΔΔCq of the first cancer tissue
sample was taken as a reference to obtain the relative expression
levels of hsa-miR-125a-5p in each group of samples (Table I). In 10/15 groups of samples,
hsa-miR-125a-5p expression of normal adjacent tissue was higher
compared with the amount in the cancer tissue. In the other five
groups of samples, there was little difference in the expression of
hsa-miR-125a-5p, with a difference of less than 0.1. The expression
data of hsa-miR-125a-5p was analyzed using an single-sample t-test,
which revealed that the expression of hsa-miR-125a-5p in cancer
tissue was significantly lower compared with the corresponding
adjacent normal tissues (P=0.038).
 | Table I.Details of clinical tissue samples of
patients and miR-125a-5p relative expression. |
Table I.
Details of clinical tissue samples of
patients and miR-125a-5p relative expression.
|
| miR-125a-5p
expressiona |
|
|
|
|
|
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Sample number | Cancer tissue | Adjacent normal
tissue | Age, years | Sex | Tumor location | Differentiation
degree | Reactive hyperplasia
of lymph nodes | TNM |
|---|
| 1 | 1.00 | 0.55 | 59 | Male | Buccal | Medium | Positive | T2N0M0 |
| 2 | 0.49 | 0.92 | 54 | Male | Tongue and soft
palate | High | Positive | T2N0M0 |
| 3 | 0.62 | 0.88 | 64 | Female | Buccal | Medium | Positive | T2N0M0 |
| 4 | 0.53 | 4.18 | 50 | Male | Tongue | Medium | Positive | T2N1M0 |
| 5 | 5.67 | 9.17 | 66 | Female | Buccal | Medium | Positive | T4N1M0 |
| 6 | 0.68 | 0.61 | 54 | Female | Tongue | Medium | Positive | T2N0M0 |
| 7 | 2.43 | 4.89 | 51 | Male | Mouth floor | Poor | Positive | T2N0M2 |
| 8 | 0.70 | 0.54 | 47 | Male | Soft palate | Medium | Positive | T1NOM0 |
| 9 | 3.27 | 6.39 | 68 | Male | Tongue | Medium | Positive | T2N0M0 |
| 10 | 0.89 | 1.12 | 61 | Male | Gingival | Medium | Positive | T1N2bM0 |
| 11 | 4.46 | 5.05 | 72 | Male | Tongue | Poor | Positive | T2N0M0 |
| 12 | 0.52 | 0.45 | 63 | Male | Tongue | Poor | Positive | T2N2cM0 |
| 13 | 0.61 | 0.67 | 61 | Female | Tongue | Poor | Positive | T2N0M0 |
| 14 | 0.31 | 0.36 | 58 | Male | Tongue | Medium | Positive | T2N0M0 |
| 15 | 4.81 | 3.99 | 50 | Male | Tongue | Medium | Positive | T2N0M0 |
Overexpression of hsa-miR-125a-5p
upregulates CCR7 protein in PCI-37B
Fluorescence qPCR was used to examine the expression
of has-miR-125a-5p in the three groups of PCI-37B cells (37B, 37B
miR-125a-5p+, 37B miR-125a-5p). The relative 2−ΔΔCq
means of the 37B miR-125a-5p+ and 37B miR-125a-5p-groups were 2.33,
and 0.94 respectively (F=520.81; P<0.01; Fig. 1A). The results demonstrated that
miR-125a-5p+ cell transfection was significantly higher compared
with the blank control and negative transfection group in the
expression of hsa-miR-125a-5p. Therefore, it was evident that cell
transfection had achieved the intended purpose, and the further
functional experiments and western blot analysis could be
performed.
CCR7 protein expression of the three groups of
PCI-37B cells (37B, 37B miR-125a-5p+, 37B miR-125a-5p-) was
detected by western blotting. The results of western
electrophoretic bands and gray analysis revealed that the CCR7
protein expression of 37B miR-125a-5p+ was significantly higher
compared with the non-transfected 37B cells and 37B
miR-125a-5p-cells (F=113.87; P<0.01; Fig. 1B). Therefore, it was hypothesized that
upregulating the expression levels of hsa-miR-125a-5p can increase
the protein expression of CCR7 correspondingly. In other words,
there is a positive regulatory association between them.
Overexpression of hsa-miR-125a-5p
enhances proliferation, migration and invasion of PCI-37B
As presented in Fig.
2A, a CCK-8 assay was used to examine the changes in absorbance
of the three groups at different time points. The number of live
cells in the 37B miR-125a-5p+ group was significantly higher
compared with the other two groups at 24, 48, 72 and 96 h
(P<0.05). However, no significant difference in the number of
live cells was observed between the 37B cell miR-125a-5p-group and
the 37B group at each time point.
 | Figure 2.Results of CCK-8 and scratch assays
for testing cell proliferation and migration. (A) OD (450 nm)
values (mean of five wells) of three groups were detected at five
time points (0, 24, 48, 72 and 96 h) by CCK-8 assay. Cell migration
was measured by cell scratch assay for the three groups at four
time points (0, 6, 12 and 24 h). miR-125a-5p+ vs. PCI-37B,
*P<0.05. The (B) cell migration rate was calculated from (C) the
mean width of the scratch in the cell scratch image at each time
point. miR-125a-5p+ vs. PCI-37B at 6, 12 and 24 h, **P<0.01
separately. Light microscopy with original magnification, ×100.
37B, blank control group; miR-125a-5p+, positive transfection;
miR-125a-5p-, negative transfection group; CCK-8, Cell Counting
Kit-8; OD, optical density; miR, microRNA. |
Migration results of cell scratch assay for the
three groups at each time point (0, 6, 12 and 24 h) are presented
in Fig. 2B and C. The cell migration
rate of the 37B cell miR-125a-5p+ group at 6, 12 and 24 h was
significantly higher compared with the non-transfected group
(P<0.05). However, no significant difference was observed in the
cell migration rate between the 37B miR-125a-5p-transfected and
non-transfected cells at all time points (P>0.05).
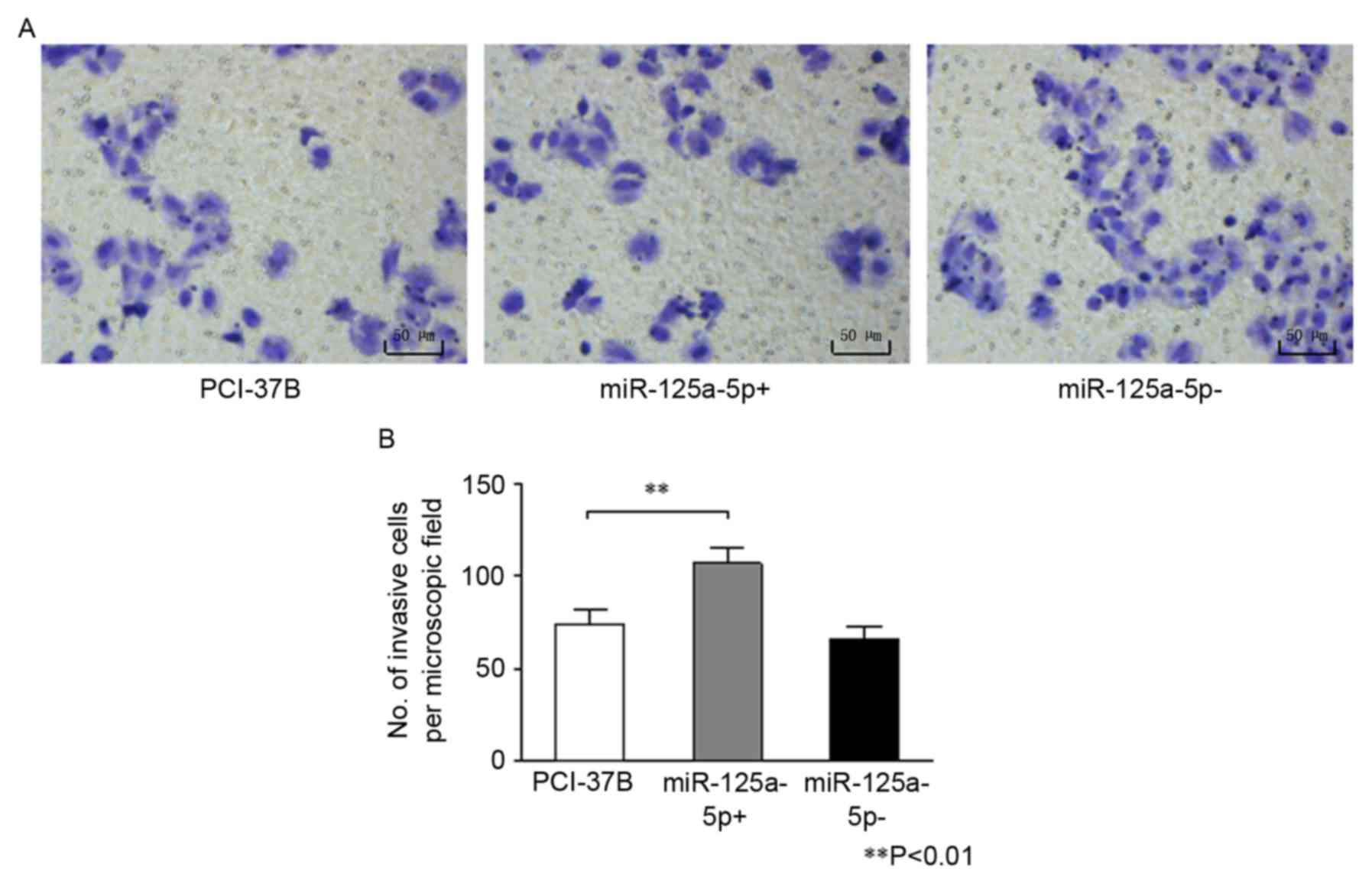
Results of the Transwell chamber assay are presented
in Fig. 3. The number of invasive
cells was significantly greater in the 37B miR-125a-5p+ group
compared with the 37B group (P<0.01). However, no difference was
observed in the number of invasive cells between the miR-125a-5p-
and the 37B group (P=0.135).
The results of the three assays illustrated that
upregulating hsa-miR-125a-5p expression can enhance the ability of
proliferation, migration and invasion of 37B cells.
Patients with high hsa-miR-125a-5p
expression tend to have shorter survival times
Results of the t-test according to the expression
values of hsa-miR-125a-5p of patients with different clinical
grading show that the expression of hsa-miR-125a-5p in patients
with high grade was significantly lower compared with patients with
low grade (P=0.011; Fig. 4A). P-value
of Kaplan-Meier curves was obtained using the log rank test
(Fig. 4B). The results revealed that
patients in the low expression group of hsa-miR-125a-5p tended to
have longer survival times compared with patients in the high
expression group (P=0.045). Significant differences existed in the
survival time between two groups.
Discussion
In recent years, numerous studies (12,19) have
investigated the association between miRNA expression and tumor
formation, diagnosis, treatment or prognosis. The molecular
mechanism of tumor formation is affected by the interaction and
regulation between proteins, and cytokines, and the existence of
complex regulating association between miRNAs and the target
protein (20,21). In studies investigating the
association between has-miR-125a-5p expression and tumors,
differences have been observed between different types of tumors,
as well as between studies on the same tumor type.
A previous study demonstrated that patients with
HNSCC have a lower hsa-miR-125a-5p expression in saliva compared
with those without HNSCC (14). In
the present study, patients with HNSCC belonging to low tumor stage
(stage I and II) had a higher hsa-miR-125a-5p expression compared
with patients with high stage (stage III and IV). This indicated
that hsa-miR-125a-5p is associated the clinical stage of HNSCC.
Although patients with a low tumor stage had a higher expression of
hsa-miR-125a-5p, patients with a higher expression tended to have a
shorter survival time in the survival analysis. No significant
association was observed between the expression and the different
levels of lymph node metastasis. The present results, to a certain
extent, indicated that hsa-miR-125a-5p can be used as a potential
factor in the prognosis of patients with HNSCC. A previous study in
gastric cancer considered that hsa-miR-125a-5p is an independent
prognostic factor by analyzing the corresponding expression of
clinical samples with tumor size, invasion, metastasis and survival
time (15). Therefore, an evident
association between hsa-miR-125a-5p expression and the prognosis of
HNSCC requires more efforts to be identified.
Studies have demonstrated that there is a lower
expression of hsa-miR-125a-5p in patients with oral squamous cell
carcinoma (OSCC) compared with those without OSCC in saliva
(16). In the present study, the
expression of hsa-miR-125a-5p was lower in cancer tissues compared
with that of adjacent normal tissues in patients with OSCC. The
results of these studies support the conclusion that the expression
of hsa-miR-125a-5p can be used as a biomarker for the diagnosis of
OSCC. The expression of hsa-miR-125a-5p was also detected in HNSCC
cancer and adjacent tissues. A higher expression was detected in
cancer tissues compared with adjacent normal tissues (22,23).
Although OSCC is an important component of HNSCC, there are a
number differences between them, at least regarding the expression
of hsa-miR-125a-5p.
CCR7 protein and hsa-miR-125a-5p have a high target
score in the predict software of mirnaviewer. There may be a direct
regulatory effect between hsa-miR-125a-5p and CCR7 protein, which
is consistent with the results of another study predicted using
miRBase gene prediction software (24).
Previous studies investigating the role of
miR-125a-5p in tumors have reported controversial results regarding
the oncogenic and tumor suppressive properties of miR-125a-5p
(19,25). A previous study demonstrated that
suppressing the expression of hsa-miR-125a-5p can significantly
reduce the cholangiocarcinoma cell viability in the HuH28 cell line
(19). Hsa-miR-125a-5p performs an
important role in promoting tumor formation. This is consistent
with the role of hsa-miR-125a-5p in HNSCC PCI-37B cells in the
present study. The hsa-miR-125a-5p expression of OSCC cell lines
was detected in a previous study. It was concluded that
upregulation of hsa-miR-125a-5p expression can reduce the
expression of estrogen-related receptor-α protein, and inhibit the
proliferation and invasion of tumor cells (25). The reasons for this difference can be
primarily summarized as two points. On the one hand, miRNAs
regulate proteins through a complex network role: A miRNA can
regulate multiple mRNAs of proteins, and there are also multiple
miRNAs binding sites on the same mRNA of protein (20). By contrast, the regulatory function of
miRNAs primarily depends on the different expression of miRNA in
cells from different tissues and on the tissue specificity
(21). The HNSCC PCI-37B cell line
was used in the present study, which is a HNSCC cell line with
clear invasion and metastasis, and high expression of CCR7 protein.
In our previous studies, a more extensive study was performed on
the downstream signal pathway of CCR7, and it was confirmed that
CCR7 can regulate the proliferation, invasion and metastasis of
PCI-37B cells by mitogen-activated protein kinases, Janus kinase
2/signal transducer, and activator of transcription 3 and matrix
metalloproteinase-9 (8,26,27). Based
on the experimental results of cell function in the present study,
it was concluded that the upregulation of hsa-miR-125a-5p can
increase the expression of CCR7, and thus perform a significant
role in promoting proliferation, migration and invasion of
HNSCC.
Intracellular microRNAs can inhibit the expression
of target genes and perform a corresponding role in the regulation
through binding the target gene 3′ non encoding region (28,29).
However, in the present study, the expression of target protein
CCR7 was significantly upregulated. There are two primary
mechanisms that could result in this phenomenon. The first one is
that there is a number of endogenous microRNA binding sites on the
CCR7 protein gene, and exogenous microRNAs can increase the protein
expression by competitively binding to endogenous binding sites
(30). The other is that microRNAs
can be combined with the target protein 5′ non-translation regions,
and thereby increase the expression of protein (31). However, further studies are required
to determine the clear mechanism.
In conclusion, the present study demonstrated that
the expression of hsa-miR-125a-5p miRNAs in OSCC tissue samples can
be used as a potential biomarker for diagnosis of OSCC. The
expression of miRNAs in the HNSCC cell line, and its regulatory
role between CCR7 and hsa-miR-125a-5p was also studied. It was
observed that hsa-miR-125a-5p could promote the proliferation,
migration and invasion of HNSCC cells by upregulating CCR7, which
may or may not be regulated directly. The present results indicated
that hsa-miR-125a-5p has a significant role in promoting cancer in
HNSCC, which may provide a basis for the treatment of HNSCC in
molecular targeted therapy. Additional studies are required to
ascertain the exact molecular mechanism of hsa-miR-125a-5p
upregulating CCR7 and the role of hsa-miR-125a-5p in other HNSCC
cell lines or in vivo.
Acknowledgements
The authors would like to thank the University of
Pittsburgh Cancer Institute for providing the PCI-37B cell
line.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372877), the
National Young Scholars Science Foundation of China (grant no.
81102058), the Public Welfare Fund Project for Science of Liaoning
Province (grant no. 2011002001), the Excellent Talent Fund Project
of Higher Education of Liaoning Province (grant no. LJQ2014087) and
the Doctoral Scientific Research Foundation of Liaoning Province
(grant no. 201501002).
Availability of data and materials
The datasets and materials used or analyzed during
the present study are available from the corresponding author on
reasonable request.
Authors' contributions
SJ was a major contributor to clinical specimen
collection, basic experiments, cell function tests, data analysis
and in writing the manuscript. ML and SW participated in data
analysis. CS and FL are responsible for methodology and project
administration. HW and ZL were involved in collecting the clinical
specimens and analysising the experimental data. PP participated in
methodology. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from all
patients prior to collection of the samples. The research was
approved by the Ethics Committee of the China Medical
University.
Consent for publication
All patients provided written informed consent for
the publication of any associated data and accompanying images.
Competing interests
All authors declare that they have no conflict of
interest.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dünne AA, Müller HH, Eisele DW, Kessel K,
Moll R and Werner JA: Meta-analysis of the prognostic significance
of perinodal spread in head and neck squamous cell carcinomas
(HNSCC) patients. Eur J Cancer. 42:1863–1868. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xing Y, Zhang J, Lin H, Gold KA, Sturgis
EM, Garden AS, Lee JJ and William WN Jr: Relation between the level
of lymph node metastasis and survival in locally advanced head and
neck squamous cell carcinoma. Cancer. 122:534–545. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bachelerie F, Ben-Baruch A, Burkhardt AM,
Combadiere C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH,
Locati M, Luster AD, et al: International union of basic and
clinical pharmacology. [corrected]. LXXXIX. Update on the extended
family of chemokine receptors and introducing a new nomenclature
for atypical chemokine receptors. Pharmacol Rev. 66:1–79. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang J, Wang S, Zhao G and Sun B: Effect
of chemokine receptors CCR7 on disseminated behavior of human T
cell lymphoma: Clinical and experimental study. J Exp Clin Cancer
Res. 30:512011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Förster R, Schubel A, Breitfeld D, Kremmer
E, Renner-Müller I, Wolf E and Lipp M: CCR7 coordinates the primary
immune response by establishing functional microenvironments in
secondary lymphoid organs. Cell 1999.99: 23–33. J Immunol.
196:5–15. 2016.PubMed/NCBI
|
|
7
|
Wang J, Xi L, Hunt JL, Gooding W,
Whiteside TL, Chen Z, Godfrey TE and Ferris RL: Expression pattern
of chemokine receptor 6 (CCR6) and CCR7 in squamous cell carcinoma
of the head and neck identifies a novel metastatic phenotype.
Cancer Res. 64:1861–1866. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu FY, Safdar J, Li ZN, Fang QG, Zhang X,
Xu ZF and Sun CF: CCR7 regulates cell migration and invasion
through MAPKs in metastatic squamous cell carcinoma of head and
neck. Int J Oncol. 45:2502–2510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao ZJ, Liu FY, Li P, Ding X, Zong ZH and
Sun CF: CCL19-induced chemokine receptor 7 activates the
phosphoinositide-3 kinase-mediated invasive pathway through Cdc42
in metastatic squamous cell carcinoma of the head and neck. Oncol
Rep. 25:729–737. 2011.PubMed/NCBI
|
|
10
|
Chekulaeva M and Filipowicz W: Mechanisms
of miRNA-mediated post-transcriptional regulation in animal cells.
Curr Opin Cell Biol. 21:452–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V and Lee RC: Identification of
microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods
Mol Biol. 265:131–158. 2004.PubMed/NCBI
|
|
12
|
Volinia S, Galasso M, Sana ME, Wise TF,
Palatini J, Huebner K and Croce CM: Breast cancer signatures for
invasiveness and prognosis defined by deep sequencing of microRNA.
Proc Natl Acad Sci USA. 109:3024–3029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Olena AF and Patton JG: Genomic
organization of microRNAs. J Cell Physiol. 222:540–545.
2010.PubMed/NCBI
|
|
14
|
Park NJ, Zhou H, Elashoff D, Henson BS,
Kastratovic DA, Abemayor E and Wong DT: Salivary microRNA:
Discovery, characterization, and clinical utility for oral cancer
detection. Clin Cancer Res. 15:5473–5477. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Janiszewska J, Szaumkessel M and Szyfter
K: MicroRNAs are important players in head and neck carcinoma: A
review. Crit Rev Oncol Hematol. 88:716–728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
National Comprehensive Cancer Network, .
Head and Neck Cancer. (Version 1.2017). https://www.nccn.org/professionals/physician_gls/#siteFebruary
1–2017
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Okamoto K, Miyoshi K and Murawaki Y:
miR-29b, miR-205 and miR-221 enhance chemosensitivity to
gemcitabine in HuH28 human cholangiocarcinoma cells. PloS One.
8:e776232013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Volinia S, Galasso M, Costinean S,
Tagliavini L, Gamberoni G, Drusco A, Marchesini J, Mascellani N,
Sana ME, Jarour Abu R, et al: Reprogramming of miRNA networks in
cancer and leukemia. Genome Res. 20:589–599. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang Y, Ridzon D, Wong L and Chen C:
Characterization of microRNA expression profiles in normal human
tissues. BMC Genomics. 8:1662007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ramdas L, Giri U, Ashorn CL, Coombes KR,
El-Naggar A, Ang KK and Story MD: MiRNA expression profiles in head
and neck squamous cell carcinoma and adjacent normal tissue. Head
Neck. 31:642–654. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O'Sullivan B,
Waldron J, et al: Comprehensive MicroRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jiang L, Huang Q, Zhang S, Zhang Q, Chang
J, Qiu X and Wang E: Hsa-miR-125a-3p and hsa-miR-125a-5p are
downregulated in non-small cell lung cancer and have inverse
effects on invasion and migration of lung cancer cells. BMC Cancer.
10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tiwari A, Shivananda S, Gopinath KS and
Kumar A: MicroRNA-125a reduces proliferation and invasion of oral
squamous cell carcinoma cells by targeting estrogen-related
receptor alpha: Implications for cancer therapeutics. J Biol Chem.
289:32276–32290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu FY, Safdar J, Li ZN, Fang QG, Zhang X,
Xu ZF and Sun CF: CCR7 regulates cell migration and invasion
through JAK2/STAT3 in metastatic squamous cell carcinoma of the
head and neck. Biomed Res Int. 2014:4153752014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guo N, Liu F, Yang L, Huang J, Ding X and
Sun C: Chemokine receptor 7 enhances cell chemotaxis and migration
of metastatic squamous cell carcinoma of head and neck through
activation of matrix metalloproteinase-9. Oncol Rep. 32:794–800.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brennecke J, Stark A, Russell RB and Cohen
SM: Principles of microRNA-target recognition. PLoS Biol.
3:e852005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yuan J, Xiao G, Peng G, Liu D, Wang Z,
Liao Y, Liu Q, Wu M and Yuan X: MiRNA-125a-5p inhibits glioblastoma
cell proliferation and promotes cell differentiation by targeting
TAZ. Biochem Biophys Res Commun. 457:171–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Khan AA, Betel D, Miller ML, Sander C,
Leslie CS and Marks DS: Transfection of small RNAs globally
perturbs gene regulation by endogenous microRNAs. Nat Biotechnol.
27:549–555. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ørom UA, Nielsen FC and Lund AH:
MicroRNA-10a binds the 5′UTR of ribosomal protein mRNAs and
enhances their translation. Mol Cell. 30:460–471. 2008. View Article : Google Scholar : PubMed/NCBI
|