Introduction
Colorectal cancer (CRC) is the third leading cause
of cancer-associated mortality worldwide (1). In China, CRC ranks fifth among the major
causes of mortality (2). In recent
years, increasing attention has been paid to the diagnosis,
treatment and prognosis of CRC, however, CRC remains a challenge.
The 5-year survival rate of patients with CRC remains low. Numerous
factors account for the increasing incidence of CRC, including
environmental pollution, an unhealthy diet and food safety
problems. The development of CRC is a complex procedure involving
multiple stages and factors; however, the underlying mechanism is
not fully understood (3).
MicroRNAs (miRNAs) are endogenous, small non-coding
RNAs, 22–25 nucleotides in length, which are widely present in
eukaryotes (4). Previous studies
revealed that miRNAs have essential functions in the development of
cancer, which supports their roles as potential therapeutic targets
and prognostic indicators for cancer (5–7). miRNA
(miR)-455-5p, located on chromosome 6, has been reported to have
low expression levels in numerous cancer types, including medullary
thyroid carcinoma (8), melanoma
(9) and gastric cancer (10), compared with normal tissues. However,
there is a limited number of studies that have investigated the
expression levels of miR-455-5p and its potential role in CRC.
Whether miR-455-5p may affect metastasis and its regulatory
mechanism in CRC remains unclear.
In the present study, the in vivo expression
profile of miR-455-5p was studied in CRC, in order to gain insights
into the underlying mechanisms of human CRC, and to assess the
biological function of miR-455-5p in CRC. The expression levels of
miR-455-5p in colorectal cancer tissues and paracancerous tissues
were detected and the differences were evaluated. Using
bromodeoxyuridine (BrdU) cell proliferation, apoptosis and
migration assays, the biological characteristics of miR-455-5p were
determined and its target gene identified.
Materials and methods
Patients and samples
Carcinoma tissues and adjacent tissue specimens were
randomly collected from 40 patients with CRC (21 cases of rectal
cancer, 19 cases of colon cancer), who underwent surgery at the
anorectal branch of Ningbo First Hospital (Ningbo, China) between
March 2016 and July 2016. CRC specimens and the normal specimens
were collected and immediately placed into the RNA preservation
solution, and stored at −80°C.
The inclusion criteria of patients included the
clinical and pathological diagnosis of colorectal cancer with
stable vital signs. Patients with a history of chemotherapy or
other malignancies prior to surgery were excluded. Patients whose
family had a history of similar neoplastic disease were also
excluded. Tumors were classified according to the Tumor Node
Metastasis classification staging system (11). All post-operative pathology results
were reviewed by two pathologists. Informed written consent (no.
S2016-055-10) was provided by all patients and the study was
approved by the Ningbo First Hospital Ethics Committee. All the
procedures were performed in accordance with the ethical standards
laid down in the 1964 Declaration of Helsinki and its later
amendments.
Cell culture
The colorectal cancer cell lines (SW-480, HT-29 and
HCT-116) were provided by Shanghai Nuobai Pharmaceutical Co., Ltd.
(Shanghai, China). Cells were cultured in DMEM (GE Healthcare Life
Sciences, Logan, UT, USA) containing 10% fetal bovine serum (FBS;
BioWest, Nuaillé, France) and 1% penicillin/streptomycin (P/S)
solution at 37°C in 5% CO2 and 95% air.
RNA extraction and quantitative
analysis of miR-455-5p and phosphoinositide-3-kinase regulatory
subunit 1 (PIK3R1) using reverse transcriptase-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or transfected
cells using TRIzol® reagent (Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
Reverse transcription was performed with the following parameters:
25°C for 5 min, followed by 50°C for 60 min and 70°C for 15 min.
The expression of miR-455-5p in CRC and adjacent healthy tissue,
and PIK3R1 in transfected CRC cell lines, was measured using
RT-qPCR using a SYBR-Green PCR system (Tiangen Biotech Co., Ltd.,
Beijing, China). The thermocycling conditions were as follows: 95°C
for 2 min, followed by 40 cycles of 95°C for 10 sec, 60°C for 30
sec and 70°C for 45 sec. All the primer sequences are listed in
Table I. Relative quantitative
analysis was performed using the 2−ΔΔCq method (12).
 | Table I.Primer sequences used for miR-455-5p
and PIK3R1 expression analysis. |
Table I.
Primer sequences used for miR-455-5p
and PIK3R1 expression analysis.
| Author, year | Name of primer | Sequence
(5′→3′) | (Refs.) |
|---|
| – | miR-455-5p-RT |
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCGATGTAG | – |
| – | miR-455-5p-F |
ACACTCCAGCTGGGTATGTGCCTTTGGACT | – |
| – | miR-455-5p-R |
TGGTGTCGTGGAGTCG | – |
| Ai et al,
2010 | U6-RT |
CGCTTCACGAATTTGCGTGTCAT |
(15) |
| Ai et al,
2010 | U6-F |
GCTTCGGCAGCACATATACTAAAAT |
(15) |
| Ai et al,
2010 | U6-R |
CGCTTCACGAATTTGCGTGTCAT |
(15) |
| Cao et al,
2013 | Actin-F |
GCTCAGGAGGAGCAATGATCTTG |
(16) |
| Cao et al,
2013 | Actin-R |
GTACGCCAACACAGTGCTGTC |
(16) |
| – | PIK3R1-F |
ACCACTACCGGAATGAATCTCT | – |
| – | PIK3R1-R |
GGGATGTGCGGGTATATTCTTC | – |
Cell transfection of miR-455-5p
mimics
The three CRC cell lines (SW480, HT-29 and HCT-116)
were cultured in DMEM and transfected with miR-455-5p mimics (miRNA
mimics, forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) or non-specific control (NC) miRNA
mimic (forward, 5′-UUCUCCGAACGUGUCACGUTT-3′ and reverse,
5′-ACGUGACACGUUCGGAGAATT-3′) using Hieff Trans™ Liposomal
Transfection Reagent (Yeasen Biotechnology Co., Ltd., Shanghai,
China), according to the manufacturer's protocol. In brief, cells
were grown in 12-well plates until 80–90% confluence was reached.
miRNA mimics or NC mimic (20 pmol) were mixed with 2 µl
transfection reagent in 50 µl Opti-MEM (Invitrogen; Thermo Fisher
Scientific, Inc.). Following incubation at room temperature for 20
min, the mixture was added to the cells. After 6 h, the medium was
changed to fresh medium supplemented with 10% FBS and P/S.
BrdU cell proliferation assays
Following transfection with miR-455-5p mimics or NC
mimic for 48 h, the BrdU assay kit (Boster Biological Technology,
Pleasanton, USA) was used to evaluate the proliferative ability of
the cells, according to the manufacturer's protocol. Fluorescence
microscopy was used to observe the fluorescence ratio
(magnification, ×10).
Apoptosis assays
A total of 48 h following transfection, cell medium
was replaced with DMEM supplemented with 2% FBS. A total of 24 h
later, the cells were resuspended in 1X Annexin Binding Buffer
(Nanjing KeyGen Biotech Co., Ltd., Nanjing, China). A total of 5 µl
of 7-Aminoactinomycin D (7-AAD) viability Dye and 1 µl of Annexin
conjugated to phycoerythrin (PE) solution (Nanjing KeyGen Biotech
Co., Ltd.) were added to 100 µl of cell suspension solution. The
tubes were maintained on ice and incubated for 15 min in the dark.
Subsequently, 400 µl of ice-cold 1X Binding Buffer was added into
the tubes followed by flow cytometry analysis on a BD FACS Calibur
Flow Cytometry System using BD FACStation™ 6.1 software (BD
Biosciences, Franklin Lakes, NJ, USA), according to the
manufacturer's protocols. The cells with Annexin V-PE+/7-AAD- were
calculated to represent apoptotic cells.
Wound healing migration assays
Wound healing cell migration assays were carried out
as described previously (13).
Briefly, 48 h following transfection, HT-29 cells (8×105
cells/well) were seeded into a 24-well plate and grown until ~100%
confluent. An uninterrupted scratch was made with a sterile P200
pipette tip across the monolayer. Following washing with PBS, cells
were cultured in DMEM without FBS for 0, 6, 12 and 24 h.
Alterations in the area of the scratch were observed with an
inverted microscope (magnification, ×10) at different time points
(0, 6, 12 and 24 h).
Bioinformatics analysis
To predict the target genes of miR-455-5p, the
following computational programs were used: TargetScan (http://www.targetscan.org); miRanda (http://www.microrna.org); PICTAR (http://www.pictar.mdc-berlin.de); and MICROCOSM
Targets (http://www.ebi.ac.uk/enright-srv/microcosm).
Western blotting
Western blotting was performed as previously
described (14). At 48 h following
transfection, cells were harvested and lysed, and the supernatants
were collected by centrifugation at 12,000 × g for 15 min at 4°C.
The protein concentration was measured using the bicinchoninic acid
protein assay kit (Thermo Fisher Scientific, Inc.), according to
the manufacturer's protocols. The protein was separated by SDS-PAGE
on a 12.5% gel and transferred on to a polyvinylidene difluoride
membrane. The membrane was blocked and subsequently incubated
overnight at 4°C with anti-PIK3R1 (dilution, 1:1,000; cat. no.
ab191606; Abcam, Cambridge, UK). A goat anti-rabbit HRP-conjugated
antibody (dilution, 1:5,000; cat. no. ab97080, Abcam) was used as
the secondary antibody. The membrane was developed using an
enhanced chemiluminescence system (GE Healthcare Bio-Sciences,
Pittsburgh, PA, USA). Protein expression was quantified by
Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville,
MD, USA).
Statistical analysis
SPSS 18.0 (SPSS, Inc., Chicago, IL, USA) statistical
software was used for all statistical analyses. The
Kolmogorov-Smirnov test was used to test the normality of
continuous variables. Data were counted and are presented as the
number of cases (n, %). The statistical inferences of intergroup
differences were evaluated using the χ2 test. Data
conforming to a normal distribution were presented as the mean ±
standard error of the mean, and one-way analysis of variance
followed by the least significant difference post-hoc test was used
for multiple comparisons between groups. Data not conforming to a
normal distribution are presented as the median with interquartile
range, and non-parametric tests, including Kruskal-Wallis tests
with Nemenyi post hoc test, and Mann-Whitney tests, were used for
multiple comparisons between groups. P<0.05 was considered to
indicate a statistically significant difference.
Results
Patient characteristics
A total of 40 patients (21 cases of rectal cancer
and 19 cases of colon cancer) were included in the present study.
These comprised 24 males and 16 females, aged 43–85 years with an
average age of 65±11 years. A total of 28 patients were ≥60 years
old and 12 patients were <60 years old. Tumors were classified
as stage I (n=6), stage II (n=17), stage III (n=13) or stage IV
(n=4). There were 33 cases of moderate differentiation and four
cases of poor differentiation. Patients with no previous history of
malignancies had not received radiotherapy, chemotherapy or
immunotherapy prior to surgery.
Expression of miR-455-5p is
downregulated in CRC tissues
The expression levels of miR-455-5p were measured
using RT-qPCR in the 40 CRC tissues and adjacent non-tumor tissues.
The results indicated that the relative expression levels of
miR-455-5p in CRC tissues were 0.59±0.34, while the relative
expression in adjacent normal tissues was 2.4±1.60. Compared with
the normal group, the expression levels of miR-455-5p in the CRC
group were decreased by 75% (P<0.01; Fig. 1).
Association between miR-455-5p
expression levels and clinicopathological characteristics
Following the observation that miR-455-5p expression
levels were significantly decreased in colorectal tumor tissues,
the association between miR-455-5p expression levels and the
characteristics of CRC patients was investigated. The expression of
miR-455-5p was reported to be negatively associated with the
differentiation of colorectal tumors (P<0.05; Table II). According to the results of the
present study (Table II), the
expression level of miR-455-5p was not associated with the
following indicators (P>0.05): Sex, age, tumor location,
pathological type, tumor stage and lymph node metastasis of
patients with CRC.
 | Table II.Correlation between the level of
miR-455-5p and clinical pathological characteristics of patients
with colorectal cancer. |
Table II.
Correlation between the level of
miR-455-5p and clinical pathological characteristics of patients
with colorectal cancer.
| Clinical
characteristic | No. patients | Expression of
miR-455-5p |
Z/χ2 | P-value |
|---|
| Sex |
|
Male | 24 | 0.879 (0.401,
1.222) | −0.939 | 0.392a |
|
Female | 16 | 0.594 (0.226,
1.016) |
|
|
| Age, years |
|
≥60 | 28 | 0.662 (0.328,
1.102) | −1.107 | 0.348a |
|
<60 | 12 | 1.002 (0.446,
1.635) |
|
|
| Location |
|
LSCC | 6 | 0.934 (0.492,
1.516) | 2.875 | 0.238b |
|
RSCC | 13 | 0.736 (0.441,
1.813) |
|
|
|
Rectum | 21 | 0.599 (0.263,
1.015) |
|
|
| Type |
|
Protuberant | 13 | 0.550 (0.314,
0.770) | 2.100 | 0.350b |
|
Ulcerative | 21 | 0.953 (0.361,
1.548) |
|
|
|
Invasive | 6 | 0.700 (0.399,
1.792) |
|
|
|
Differentiation |
|
Poorly-differentiated | 7 | 0.477 (0.290,
1.721) | 8.837 | 0.012b |
|
Moderately-poorly-differentiated | 5 | 0.174 (0.171,
0.383) |
|
|
|
Moderately-differentiated | 28 | 0.863 (0.501,
1.222) |
|
|
|
Well-differentiated | 0 | 0 |
|
|
| TNM |
|
I/II | 17 | 0.588 (0.324,
1.194) | −0.328 | 0.743a |
|
III/IV | 23 | 0.818 (0.390,
1.285) |
|
|
| Lymph node
metastases |
| No | 26 | 0.647 (0.380,
1.203) | −0.128 | 0.898a |
|
Yes | 14 | 0.863 (0.297,
1.169) |
|
|
| No. of metastatic
lymph nodes |
|
1–3 | 9 | 0.908 (0.253,
1.239) | −0.200 | 0.841a |
|
>3 | 5 | 0.477 (0.307,
2.772) |
|
|
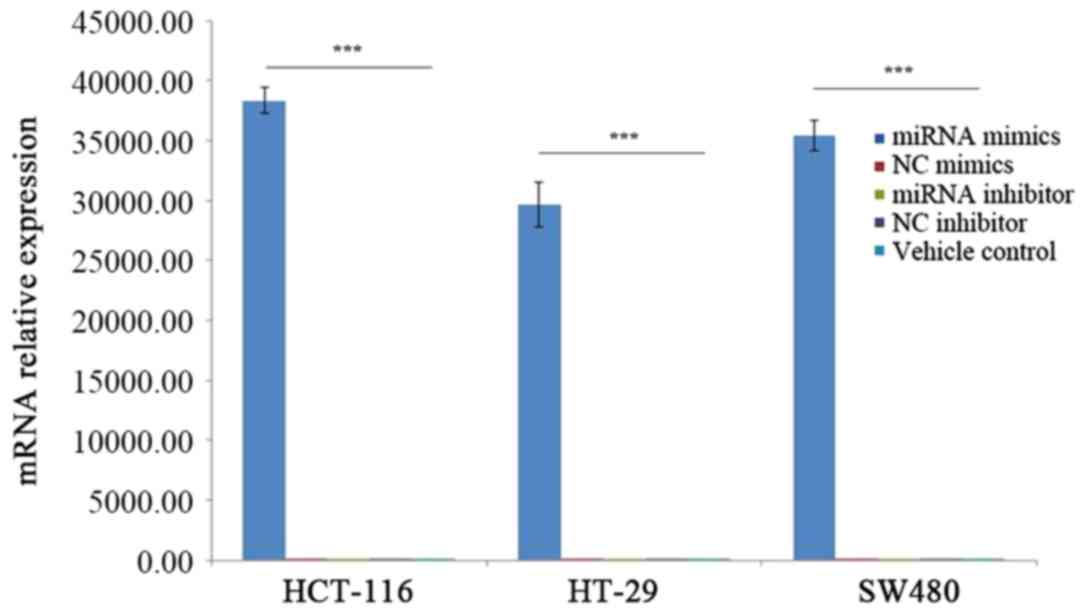
miR-455-5p mimics transfection of CRC
cells was successful
RT-qPCR experiments were performed to demonstrate
that the transfection with miR-455-5p mimics was successful. As
presented in Fig. 2, the relative
expression levels of mRNA in the miRNA mimics groups were 54,343-,
43,085- and 18,856-times higher compared with the NC mimics groups
in the HCT-116, HT-29 and SW480 cell lines. The mRNA expression
level of miR-455-5p was significantly upregulated in CRC cell lines
transfected with miR-455-5p mimics (P<0.001).
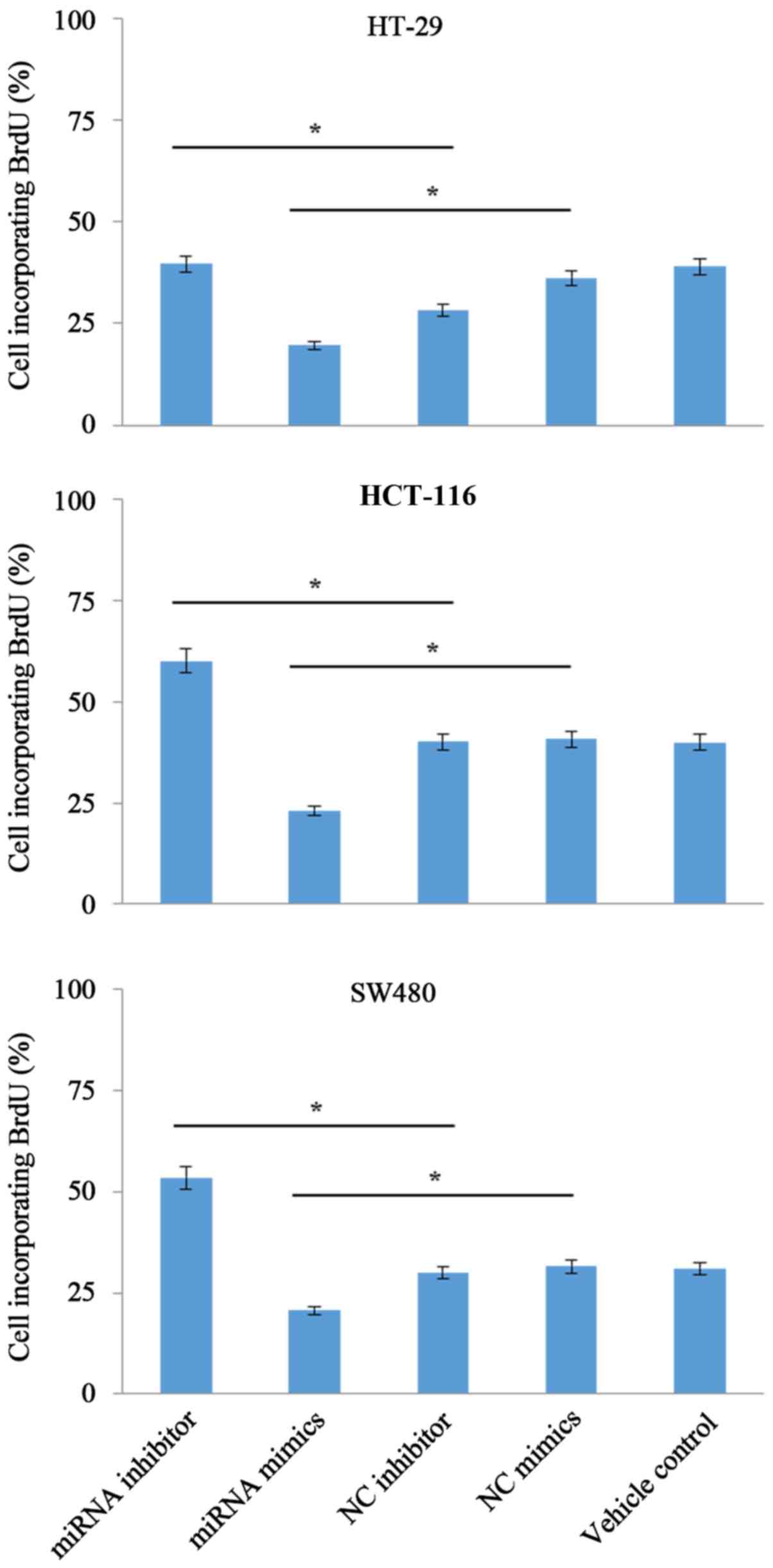
miR-455-5p suppresses the
proliferation of CRC cells
According to results from the present study
(Fig. 3), it was demonstrated that
the number of viable cells in the miRNA mimics group was
significantly reduced (P<0.05) compared with the NC group. In
addition, inhibition of endogenous miR-455-5p with miR-455-5p
inhibitors increased the cell proliferation by ~20% compared with
the control group (HT-29, 40% in miRNA inhibitor vs. 28% in NC
inhibitor; HCT-116, 60% in miRNA inhibitor vs. 40% in NC inhibitor;
SW480, 53% in miRNA inhibitor vs. 30% in NC inhibitor).
miR-455-5p promotes apoptosis in CRC
cells
As displayed in Fig.
4, transient transfection of miR-455-5p mimics in HCT-116 cells
markedly promoted apoptosis compared with the NC mimic group
(P<0.05; 48 and 39%, respectively). Apoptosis was significantly
inhibited in HCT-116 cells transfected with the miRNA inhibitor
compared with the NC inhibitor group (P<0.05). In addition,
similar patterns of apoptosis in an additional two CRC cell lines
(HT-29 and SW480) were observed.
miR-455-5p inhibits the migration of
CRC cells
The effect of miR-455-5p on CRC cell migration was
detected by wound healing cell migration assays using HT-29 cells.
As displayed in Fig. 5, the distance
of cell migration for the miRNA mimics-treated group was smaller
compared with that of the NC mimic group at a variety of different
time points (6, 12 and 24 h). Specifically, the relative mobility
percentages were as follows: i) 6 h, 4% in miRNA mimics vs. 8% in
NC mimics; ii) 12 h, 6% in miRNA mimics vs. 14% in NC mimics; and
iii) 24 h, 9% in miRNA mimics vs. 12% in NC mimics. By contrast,
depletion of miR-455-5p with an miRNA inhibitor enhanced the cell
migratory ability (Fig. 5). The
comparisons of relative mobility between the two groups were as
follows: i) 6 h, 8% in miRNA inhibitor vs. 3% in NC inhibitor; ii)
12 h, 13% in miRNA inhibitor vs. 9% in NC inhibitor; and iii) 24 h,
15% in miRNA inhibitor vs. 14% in NC inhibitor. These results
indicated that miR-455-5p may inhibit the migration of HT-29
cells.
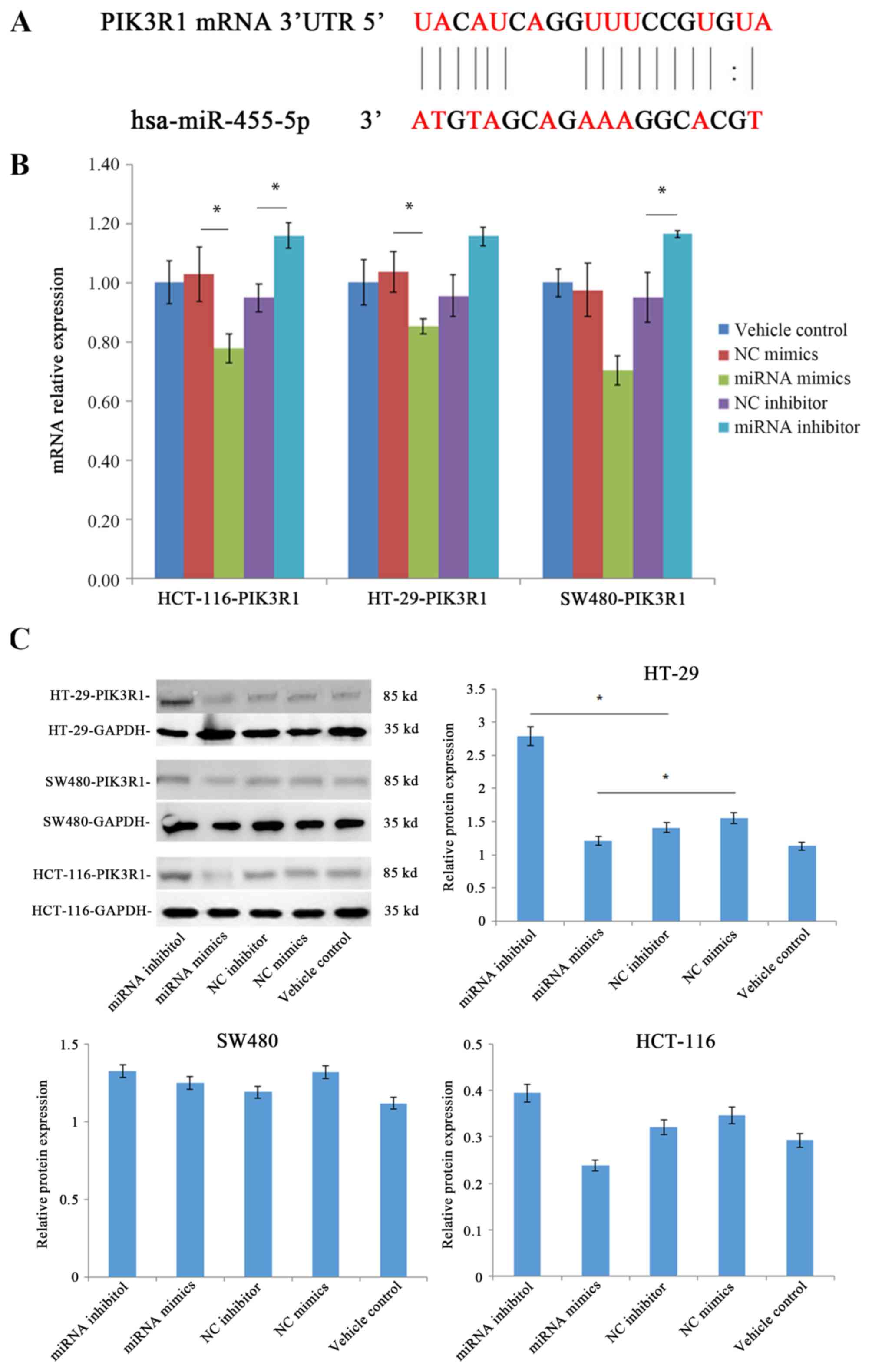
miR-455-5p negatively regulates the
expression levels of the target gene PIK3R1
The above results prompted an investigation into the
molecular mechanism underlying the aforementioned differences in
the behavior of CRC cells treated with miR-455-5p. The binding
region of miR-455-5p in the target gene PIK3R1 mRNA 3′-untranslated
region was predicted using target gene prediction databases
(Fig. 6A).
To validate these predictions, the effect of
miR-455-5p on the expression levels of PIK3R1 was determined using
RT-qPCR and western blotting experiments. As displayed in Fig. 6B, the expression levels of mRNA in the
miRNA mimics group were 25, 19 and 32% lower in HCT-116, HT-29 and
SW480 cell lines, respectively, compared with the NC mimics group.
By contrast, the mRNA expression levels in the miRNA inhibitor
group were 21, 20 and 21% higher compared with the NC inhibitor
group in HCT-116, HT-29 and SW480 cell lines, respectively. The
mRNA level of PIK3R1 was significantly reduced by the expression of
miR-455-5p in CRC cells (P<0.05). Furthermore, the levels of
protein expression of PIK3R1 were also observed to be downregulated
by miR-455-5p (Fig. 6C). Western blot
quantification reported that the protein expression levels in the
mimics group (1.21) were lower compared with the NC mimics group
(1.55) in the HT-29 cell lines, while the inhibitor groups
exhibited the opposite trend. The results of the western blotting
were consistent with the RT-qPCR results (Fig. 6B). Together, these results indicated
that miR-455-5p may function as a CRC suppressor by negatively
regulating the expression levels of the oncogene PIK3R1.
Discussion
The present study demonstrated that miR-455-5p may
suppress the proliferation and migration of CRC cells and
downregulate PIK3R1 in CRC. Ex vivo research revealed that
the expression level of miR-455-5p was significantly downregulated
in human CRC. Further in vitro studies suggested that
miR-455-5p prevented the development of CRC, possibly by
downregulating the oncogene PIK3R1.
miRNAs are widely distributed in viruses, plants and
higher mammals (17). Since the
discovery of the first miRNA gene (lin-4) in 1993 (4,18), the
function of miRNAs has been widely investigated (19–22). Given
their essential role, even a small alteration in the expression
levels of miRNAs may lead to notable differences in the levels of
downstream target mRNAs. As a member of the miR-455 family,
miR-455-5p has been reported to have differential expression levels
in a variety of carcinomas. For instance, Shoshan et al
(9) reported that miR-455-5p was
overexpressed in melanoma and downregulated the tumor suppressor
gene cytoplasmic polyadenylation element binding protein 1, which
consequently promoted the proliferation and metastasis of cancer
cells. Similarly, Sand et al (23) demonstrated that miR-455-5p was
significantly upregulated in basal cell carcinoma. By contrast, the
downregulation of miR-455-5p was also reported in other cancer
types, including esophageal, gastric and thymic epithelial cancer
(10,24,25). Yang
et al (26) reported that
mir-455-5p is upregulated in colon cancer and functions as an
oncogene, promoting the development of colon cancer. This
contradicts the results from the present study, and this may be due
to the fact that Yang et al (26) only studied 10 cases of colon cancer
tissue, while rectal cancer tissues were not included. Furthermore,
only one cell line was studied. The number of tissue specimens and
cells was lower compared with the present study, and the results
were not representative. There are limited studies on the
expression levels of miR-455-5p in human CRC tissues, thus the
association remains unclear.
CRC is known to be associated with abnormalities in
a large collection of genes, which may be reflected by the
identification of numerous molecular markers (27–31). In
the present study, it was reported that there was significantly low
expression levels of miR-455-5p in CRC, which correlated with tumor
differentiation, suggesting that miR-455-5p may have tumor
suppressing potential in CRC. However, given the relatively limited
tissue samples in the present study, an extensive study with a
larger sample size is required in the future.
To investigate the potential role of miR-455-5p in
CRC, the effects of up-and downregulation of miR-455-5p on cell
proliferation, migration and apoptosis were assessed. The results
of the present study reported that the upregulation of miR-455-5p
may lead to the inhibition of cell proliferation and migration, in
addition to promoting apoptosis in CRC cells. Together, these
findings support the tumor suppressor role of miR-455-5p in CRC.
Furthermore, expression levels of miR-455-5p in HCT-116 cells
regulated apoptosis positively. It was demonstrated that apoptosis
was not significantly altered in HT-29 and SW-480 cells treated
with the miRNA inhibitor and miRNA mimics, respectively. Further
investigation is required in order to address this
inconsistency.
To gain insights into the underlying molecular
mechanism of CRC, the potential target gene of miR-455-5p was
investigated using various online miRNA target gene predictors. As
a result, the established oncogene PIK3R1 was identified as the
downstream target which serves a key role in activating the
PI3K-AKT signaling pathway (32,33). Upon
activation, AKT is translocated to the nucleus where it
phosphorylates a variety of downstream target proteins, including
mechanistic target of rapamycin, X-linked inhibitor of apoptosis
protein and E3 ubiquitin-protein ligase (34). According to recent studies, the
PI3K/AKT signaling pathway has been reported to contribute to the
pathogenesis of various cancer types, including breast cancer,
ovarian cancer and colon cancer (35). In the present study, it was
demonstrated that the expression of PIK3R1 at the mRNA and protein
level was downregulated by miR-455-5p, which strongly confirmed the
bioinformatics prediction that PIK3R1 may be a functional target
for miR-455-5p. Notably, it was reported that miR-455 may
positively regulate PIK3R1 gene expression in kidney carcinoma
cells, suggesting an association between these two molecules
(36). Huang et al (37) reported that miR-4686-5p is
downregulated in hepatocellular carcinoma, and suppresses tumor
growth by directly targeting PIK3R1. In future studies, the
association between miR-455-5p and PIK3R1 will be investigated,
along with the intermediate role of the PIK pathway in the tumor
suppressive function of miR-455-5p in CRC (38).
In conclusion, it was demonstrated that the
expression of miR-455-5p was significantly downregulated in human
CRC tissues. In addition, miR-455-5p was reported to serve an
inhibitory role in cell proliferation and migration, while
promoting cell apoptosis in numerous different CRC cell lines. It
was demonstrated that the well-established oncogene PIK3R1 was
predicted to serve an essentially intermediate role in the
molecular mechanism underlying the anti-cancer role of miR-455-5p.
This was validated by assessing the mRNA and protein expression
levels. The results of the present study may facilitate future
efforts to elucidate the exact role of miR-455-5p in CRC, and thus
hold potential for the treatment of CRC by allowing for the
identification of novel gene therapy targets.
Acknowledgements
Not applicable.
Funding
This study was funded by the Ningbo Science and
Technology Project (grant no. 2010C50033).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Author's contributions
JW and YG contributed to the conception and design
of the research. JW performed the histological examination of the
colorectal cancer, collected the data, and was a major contributor
in writing the manuscript. YL analyzed and interpreted the patient
data regarding the colorectal cancer. LZ contributed to the
experimental analysis of the data; KK and YZ participated in the
statistical processing of data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Informed written consent was provided by all
patients and the study was approved by the Ningbo First Hospital
Ethics Committee (approval no., S2016-055-10). All the procedures
were performed in accordance with the ethical standards laid down
in the 1964 Declaration of Helsinki and its later amendments.
Patient consent for publication
Informed written consent was provided by all
patients for the publication of the present study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y, Shi J, Huang H, Ren J, Li N and
Dai M: Burden of colorectal cancer in China. Zhonghua Liu Xing Bing
Xue Za Zhi. 36:709–714. 2015.(In Chinese). PubMed/NCBI
|
|
3
|
Brenner H, Stock C and Hoffmeister M:
Colorectal cancer screening: The time to act is now. BMC Med.
13:2622015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lee RC, Feinbaum RL and Ambros V: The
C. elegans heterochronic gene lin-4 encodes small RNAs with
antisense complementarity to lin-14. Cell. 75:843–854. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zoni E, van der Pluijm G, Gray PC and
Kruithof-de Julio M: Epithelial plasticity in cancer: Unmasking a
microRNA network for TGF-beta-, notch-, and Wnt-mediated EMT. J
Oncol. 2015:1989672015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kohlhapp FJ, Mitra AK, Lengyel E and Peter
ME: MicroRNAs as mediators and communicators between cancer cells
and the tumor microenvironment. Oncogene. 34:5857–5868. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yi R, Li Y, Wang FL, Miao G, Qi RM and
Zhao YY: MicroRNAs as diagnostic and prognostic biomarkers in
colorectal cancer. World J Gastrointest Oncol. 8:330–340. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hudson J, Duncavage E, Tamburrino A,
Salerno P, Xi L, Raffeld M, Moley J and Chernock RD: Overexpression
of miR-10a and miR-375 and downregulation of YAP1 in medullary
thyroid carcinoma. Exp Mol Pathol. 95:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shoshan E, Mobley AK, Braeuer RR, Kamiya
T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al:
Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma
growth and metastasis. Nat Cell Biol. 17:311–321. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu J, Zhang J, Li Y, Wang L, Sui B and
Dai D: MiR-455-5p acts as a novel tumor suppressor in gastric
cancer by down-regulating RAB18. Gene. 592:308–315. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hari DM, Leung AM, Lee JH, Sim MS, Vuong
B, Chiu CG and Bilchik AJ: AJCC Cancer Staging Manual 7th edition
criteria for colon cancer: Do the complex modifications improve
prognostic assessment? J Am Coll Surg. 217:181–190. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu S, Xie F, Wang H, Liu Z, Liu X, Sun L
and Niu Z: Ubenimex inhibits cell proliferation, migration and
invasion in renal cell carcinoma: The effect is
autophagy-associated. Oncol Rep. 33:1372–1380. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hirano S: Western blot analysis. Methods
Mol Biol. 926:87–97. 2014. View Article : Google Scholar
|
|
15
|
Ai J, Zhang R, Li Y, Pu J, Lu Y, Jiao J,
Li K, Yu B, Li Z, Wang R, et al: Circulating microRNA-1 as a
potential novel biomarker for acute myocardial infarction. Biochem
Biophys Res Commun. 391:73–77. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cao J, Cai J, Huang D, Han Q, Yang Q, Li
T, Ding H and Wang Z: miR-335 represents an invasion suppressor
gene in ovarian cancer by targeting Bcl-w. Oncol Rep. 30:701–706.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Niwa R and Slack FJ: The evolution of
animal microRNA function. Curr Opin Genet Dev. 17:145–150. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans.
Cell. 75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Feinbaum R and Ambros V: The timing of
lin-4 RNA accumulation controls the timing of postembryonic
developmental events in Caenorhabditis elegans. Dev Biol.
210:87–95. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moss EG, Lee RC and Ambros V: The cold
shock domain protein LIN-28 controls developmental timing in C.
elegans and is regulated by the lin-4 RNA. Cell. 88:637–646.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rodriguez A, Vigorito E, Clare S, Warren
MV, Couttet P, Soond DR, van Dongen S, Grocock RJ, Das PP, Miska
EA, et al: Requirement of bic/microRNA-155 for normal immune
function. Science. 316:608–611. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sand M, Skrygan M, Sand D, Georgas D, Hahn
SA, Gambichler T, Altmeyer P and Bechara FG: Expression of
microRNAs in basal cell carcinoma. Br J Dermatol. 167:847–855.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hummel R, Wang T, Watson DI, Michael MZ,
Van der Hoek M Haier J and Hussey DJ: Chemotherapy-induced
modification of microRNA expression in esophageal cancer. Oncol
Rep. 26:1011–1017. 2011.PubMed/NCBI
|
|
25
|
Bellissimo T, Russo E, Ganci F, Vico C,
Sacconi A, Longo F, Vitolo D, Anile M, Disio D, Marino M, et al:
Circulating miR-21-5p and miR-148a-3p as emerging non-invasive
biomarkers in thymic epithelial tumors. Cancer Biol Ther. 17:79–82.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang Q, Hou C, Huang D, Zhuang C, Jiang W,
Geng Z, Wang X and Hu L: miR-455-5p functions as a potential
oncogene by targeting galectin-9 in colon cancer. Oncol Lett.
13:1958–1964. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Peeters M and Price T: Biologic therapies
in the metastatic colorectal cancer treatment continuum-applying
current evidence to clinical practice. Cancer Treat Rev.
38:397–406. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Morillas JD, Castells A, Oriol I, Pastor
A, Pérez-Segura P, Echevarría JM, Caballero B, González-Navarro A,
Bandrés F, Brullet E, et al: The Alliance for the prevention of
colorectal cancer in Spain. A civil commitment to society.
Gastroenterol Hepatol. 35:109–128. 2012.(In Spanish).
|
|
29
|
He K, Jin K, Wang H and Teng L:
Anti-angiogenic therapy for colorectal cancer: On the way to
getting better! Hepatogastroenterology. 59:1113–1117.
2012.PubMed/NCBI
|
|
30
|
Ng F, Ganeshan B, Kozarski R, Miles KA and
Goh V: Assessment of primary colorectal cancer heterogeneity by
using whole-tumor texture analysis: Contrast-enhanced CT texture as
a biomarker of 5-year survival. Radiology. 266:177–184. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Troiani T, Martinelli E, Orditura M, De
Vita F, Ciardiello F and Morgillo F: Beyond bevacizumab: New
anti-VEGF strategies in colorectal cancer. Expert opin Investig
Drugs. 21:949–959. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Cizkova M, Vacher S, Meseure D, Trassard
M, Susini A, Mlcuchova D, Callens C, Rouleau E, Spyratos F,
Lidereau R and Bièche I: PIK3R1 underexpression is an independent
prognostic marker in breast cancer. BMC Cancer. 13:5452013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee SR, Kim SJ, Park Y, Sung HJ, Choi CW
and Kim BS: Bortezomib and melphalan as a conditioning regimen for
autologous stem cell transplantation in multiple myeloma. Korean J
Hema. 45:183–187. 2010. View Article : Google Scholar
|
|
34
|
Franke TF: PI3K/Akt: Getting it right
matters. Oncogene. 27:6473–6488. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bellacosa A, de Feo D, Godwin AK, Bell DW,
Cheng JQ, Altomare DA, Wan M, Dubeau L, Scambia G, Masciullo V, et
al: Molecular alterations of the AKT2 oncogene in ovarian and
breast carcinomas. Int J Cancer. 64:280–285. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie J, Chen Y, Meng F, Shu T, Liu Y, Zhang
L and Zhang ZX: Study on the relationship between the RASSF10 gene
and the biological behavior of hepatocellular carcinoma cells. Eur
Rev Med Pharmacol Sci. 21:3576–3580. 2017.PubMed/NCBI
|
|
37
|
Huang XP, Hou J, Shen XY, Huang CY, Zhang
XH, Xie YA and Luo XL: MicroRNA-486-5p, which is downregulated in
hepatocellular carcinoma, suppresses tumor growth by targeting
PIK3R1. FEBS J. 282:579–594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kaur J and Sanyal SN: PI3-kinase/Wnt
association mediates COX-2/PGE(2) pathway to inhibit apoptosis in
early stages of colon carcinogenesis: Chemoprevention by
diclofenac. Tumour Biol. 31:623–631. 2010. View Article : Google Scholar : PubMed/NCBI
|