Introduction
Colorectal cancer (CRC) is a type of malignant tumor
derived from the colonic epithelial mucosa. Carcinogenesis of CRC
involves abnormal expression of genes associated with
proliferation, apoptosis, metastasis and angiogenesis (1). To date, the molecular mechanisms
underlying CRC oncogenesis are not fully understood. The
pathogenesis and development of CRC is a multi-factor, multi-step
process, in which gene mutations and abnormal expression may serve
important roles (2). The involvement
of DNA epigenetic modifications (3),
non-coding RNAs including microRNAs (miRNAs/miRs) (4), long non-coding RNA (lncRNA) (5), circular RNA (cirRNA) (6) and chromatin remodeling (7) in the development of CRC are receiving
increasing attention.
miRNAs are a class of non-coding, single-stranded
RNAs with a length of 18–25 nucleotides. They affect gene
expression by binding to specific sites at the 3′-untranslated
region of target mRNAs. miRNAs are involved in tumor development
and are potential biomarkers in cancer diagnostics and treatment,
including CRC (8,9). Accumulating evidence has demonstrated
that aberrantly expressed miRNAs acted as oncogenes or tumor
suppressor genes in CRC (4,10,11).
miR-32 is an intronic miRNA located within intron 14
of transmembrane protein 245 gene (TMEM245). Our previous studies
(12,13) revealed that miR-32 was upregulated in
CRC tissues and that high miR-32 levels were significantly
associated with lymph node and distant metastasis. Additionally,
patients with high miR-32 expression had a poor overall survival.
Furthermore, overexpression of miR-32 led to increased
proliferation, migration and invasion, and reduced apoptosis of CRC
cells via inhibition of phosphatase and tensin homolog (PTEN).
However, the mechanism underlying the upregulation of miR-32
remains unknown. The aim of the current study was to elucidate the
mechanisms involved in the upregulation of miR-32 in CRC by
analyzing the promoter of the miR-32 gene and investigating the
proteins that bind to the promoter. The results obtained may assist
in the further investigation of transcriptional regulatory
mechanisms of miR-32 expression.
Materials and methods
DNA cloning and construction of
truncated promoter plasmids
To analyze the promoter region responsible for
expression of the miR-32 gene, serially truncated fragments of five
different lengths of the 5′-flanking region of host gene TMEM245
(ENST00000374586.7 from University of California Santa Cruz Genome
Browser) were amplified by polymerase chain reaction (PCR) using
different pairs of primers (Table
I). PCR was performed with 2×HIFITaq PCR StarMix (Genstar,
Beijing, China) and the conditions were as follows: Initial
denaturation at 94°C for 5 min; 35 cycles of 94°C for 30 sec, 60°C
for 30 sec and 72°C for 2 min; and final extension at 72°C for 10
min. These primers also introduced a KpnI site at the 5′end
and an XhoI site at the 3′end of the amplified fragments.
The PCR fragments were purified, digested with KpnI and
XhoI and cloned into a pGL3-basic vector (Promega
Corporation, Madison, WI, USA). Five successive truncated
constructs from the 5′-flanking region termed pGL3-1987 (−1987 to
−1 bp), pGL3-1648 (−1648 to −1 bp), pGL3-1088 (−1088 to −1 bp),
pGL3-606 (−606 to −1 bp), pGL3-320 (−320 to −1 bp) were generated.
All inserts were verified by DNA sequencing (Beijing Genomics
Institute, Beijing, China).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
|
| Sequence 5′ →
3′ |
|
|---|
|
|
|
|
|---|
| Primer | Forward | Reverse | Position (bp) |
|---|
| pGL3-1987 |
AAAGGTACCCAGCCTGTTCA ACATGGTGAA |
AAACTCGAGGTAATGGGAGTCGGGC TAGAAAC | −1987 to −1 |
| pGL3-1648 | AAAGGTACCCTCCCACC
GGGAGACTGC |
AAACTCGAGGTAATGGGAGTCGG GCTAGAAAC | −1648 to −1 |
| pGL3-1088 | AAAGGTACCCTTGCAAGG
TTTGAAGCAATCA |
AAACTCGAGGTAATGGGAGTCGG GCTAGAAAC | 1088 to −1 |
| pGL3-606 | AAAGGTACCCTTGCCTGT
GCCACTTGG |
AAACTCGAGGTAATGGGAGTCGG GCTAGAAAC | 606 to −1 |
| pGL3-320 | AAAGGTACCTTCTAGTATG
CAGCTTGGGTTTTAATATC |
AAACTCGAGGTAATGGGAGTCG GGCTAGAAAC | - 320 to −1 |
Cell transfection and dual luciferase
assay
The CRC cell line HCT-116 was obtained from the Cell
Bank of the Type Culture Collection of Chinese Academy of Sciences
(Shanghai, China). HCT-116 cells were plated in six-well plates and
cultured in RPMI-1640 medium (GE Healthcare Life Sciences, Logan,
UT, USA) supplemented with 10% fetal bovine serum (Gibco; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) and 1%
penicillin/streptomycin (GE Healthcare Life Sciences) at 37°C with
5% CO2. For each transfection, cells were incubated with
0.5 µg of each promoter reporter plasmid respectively, and 0.5 µg
of pRL-TK (Promega Corporation), which was used as an internal
control. The pGL3-basic vector was used as the negative control.
Transfection was performed using Lipofectamine® 2000
(Thermo Fisher Scientific, Inc.). Cells were harvested for
subsequent experimentation 48 h after transfection using the
dual-luciferase reporter assay system (Promega Corporation),
following the manufacturer's protocol. Firefly luciferase activity
was normalized to Renilla luciferase activity and the ratio of
Firefly/Renilla luciferase in each group reflected the promoter
activity. Each experiment was performed in triplicate.
DNA pull-down assay
The DNA pull-down was performed with a DNA pull-down
test kit (catalog no. KT401), according to the manufacturer's
protocol (Gzscbio, Guangzhou, China). Briefly, the sequences of
−320 to −1 bp 5′-flanking region were amplified by PCR and tagged
with biotin. The biotin-labeled promoter was bound with
streptavidin magnetic beads (Dynabeads™ M-280 Streptavidin; Thermo
Fisher Scientific, Inc.) at 4°C for 4 h. The non-biotinylated
promoter was used as the negative control. All protein extracted
from HCT-116 cells in the input group was used as the positive
control. The bound promoter was incubated with 1 mg protein
extracted from HCT-116 cells with gentle agitation at 4°C
overnight. The bound beads-promoter protein complexes were washed
with wash buffer, which was included in the kit, and separated by
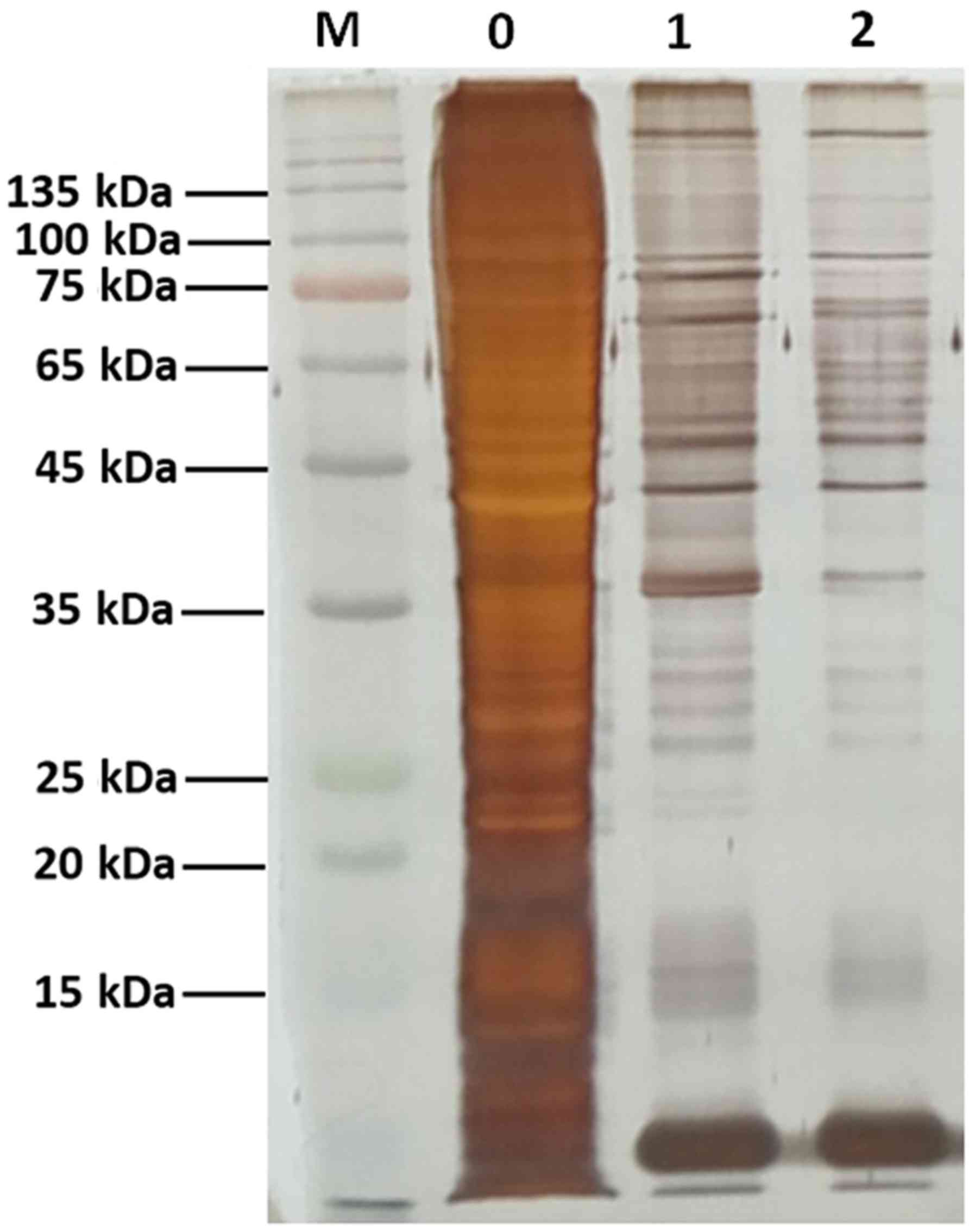
SDS-PAGE. Gel bands were visualized by silver staining.
Mass spectrometry (MS) and
bioinformatics analysis
The proteins were digested by incubating with 0.02
µg/µl trypsin at 37°C overnight. The resulting peptides were
extracted, purified and processed using a Q-Exactive mass
spectrometer (Thermo Fisher Scientific, Inc.). Proteins were
identified by comparing the MS data with the Uniprot human protein
sequence database (https://www.uniprot.org/taxonomy/9606) using the
Mascot search engine (http://www.matrixscience.com/; V2.3.02). Finally, the
protein identification results were verified by analyzing the
protein quality, matches of the secondary spectrum of the protein,
number of peptides matching the protein, protein abundance and
protein description between samples. The proteins exhibiting
differential binding to the biotin-labeled promoter and negative
control, identified by MS, were analyzed using Gene Ontology (GO)
functional enrichment (http://www.geneontology.org/), Kyoto Encyclopedia of
Genes and Genomes (KEGG) pathway analyses (https://www.kegg.jp/) and the AnimalTFDB database
(version 2; bioinfo.life.hust.edu.cn/AnimalTFDB) for possible
transcription factors (TFs). Proteins were considered significantly
enriched in GO terms and KEGG pathways when P<0.05. The MS and
bioinformatics analyses were performed by Sagene Biotech Co., Ltd.
(Guangzhou, China).
Statistical analysis
The experimental data were analyzed using one-way
analysis of variance and were presented as the mean ± standard
deviation from three independent experiments using SPSS software
(version 19.0; IBM Corp., Armonk, NY, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Functional analysis of the promoter of
the miR-32 gene
To investigate the mechanisms involved in the
expression of miR-32, the 5′-flanking region of the host gene
TMEM245 was dissected into a series of deletion fragments termed
pGL3-1987 (−1987 to −1 bp), pGL3-1648 (−1648 to −1 bp), pGL3-1088
(−1088 to −1 bp), pGL3-606 (−606 to −1 bp) and pGL3-320 (−320 to −1
bp) (Fig. 1A). A dual luciferase
reporter assay was performed to further detect the transcriptional
activity of the fragments. Compared with the pGL3-basic vector, the
luciferase activity in pGL3-320, pGL3-606, pGL3-1088, pGL3-1648 and
pGL3-1987 was significantly increased in HCT-116 cells (P<0.05;
Fig. 1B). The fragment −320 to −1 bp
exhibited the most increased activity, indicating the presence of
potential positive regulatory elements, which enhance miR-32
transcription in this region. However, a decrease in
transcriptional activity in the pGL3-606 group compared with the
pGL3-320 group (P<0.05) was observed (Fig. 1B), suggesting the presence of
repressive regulatory elements in the region between position −606
and −320 bp.
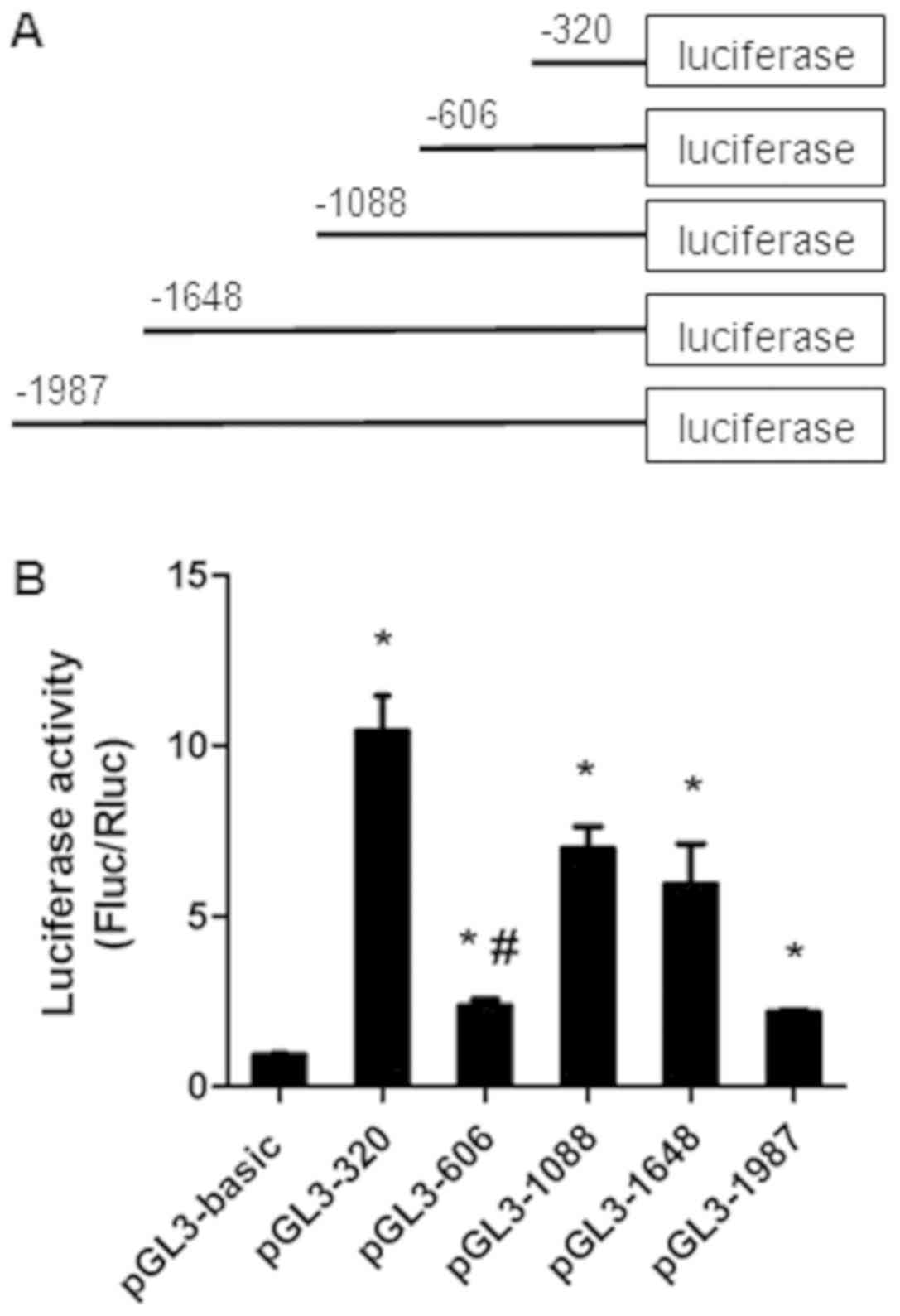 | Figure 1.The luciferase activity of the
truncated promoter of miR-32. (A) Schematics of each truncated
promoter plasmids. (B) Dual luciferase reporter assays of miR-32
gene promoter constructs. Various recombinant vectors, including
pGL3-1987, pGL3-1648, pGL3-1088, pGL3-606, pGL3-320 and pRL-TK,
were co-transfected into HCT-116 cells. pRL-TK and pGL3-basic were
used as internal and negative controls, respectively. Relative
luciferase activity was determined by the ratio of Fluc/Rluc
activity. Data presented as the mean ± standard deviation of three
independent experiments. *P<0.05, compared with the pGL3-basic
group. #P<0.05, compared with pGL3-320. Fluc, firefly
luciferase; Rluc, Renilla luciferase; miR, microRNA. |
Identification of promoter-binding
proteins
The DNA pull-down assay was used to identify
putative interacting proteins binding to the −320 to −1 bp fragment
using streptavidin magnetic beads coated with the biotin-labeled
promoter. The proteins were subsequently analyzed by SDS-PAGE and
silver staining (Fig. 2).
Identification of the differentially binding proteins in the two
groups by MS revealed that the binding factors of miR-32 promoter
included 403 proteins. These proteins were further analyzed by
bioinformatics tools.
GO, KEGG pathway and transcription
factor analysis
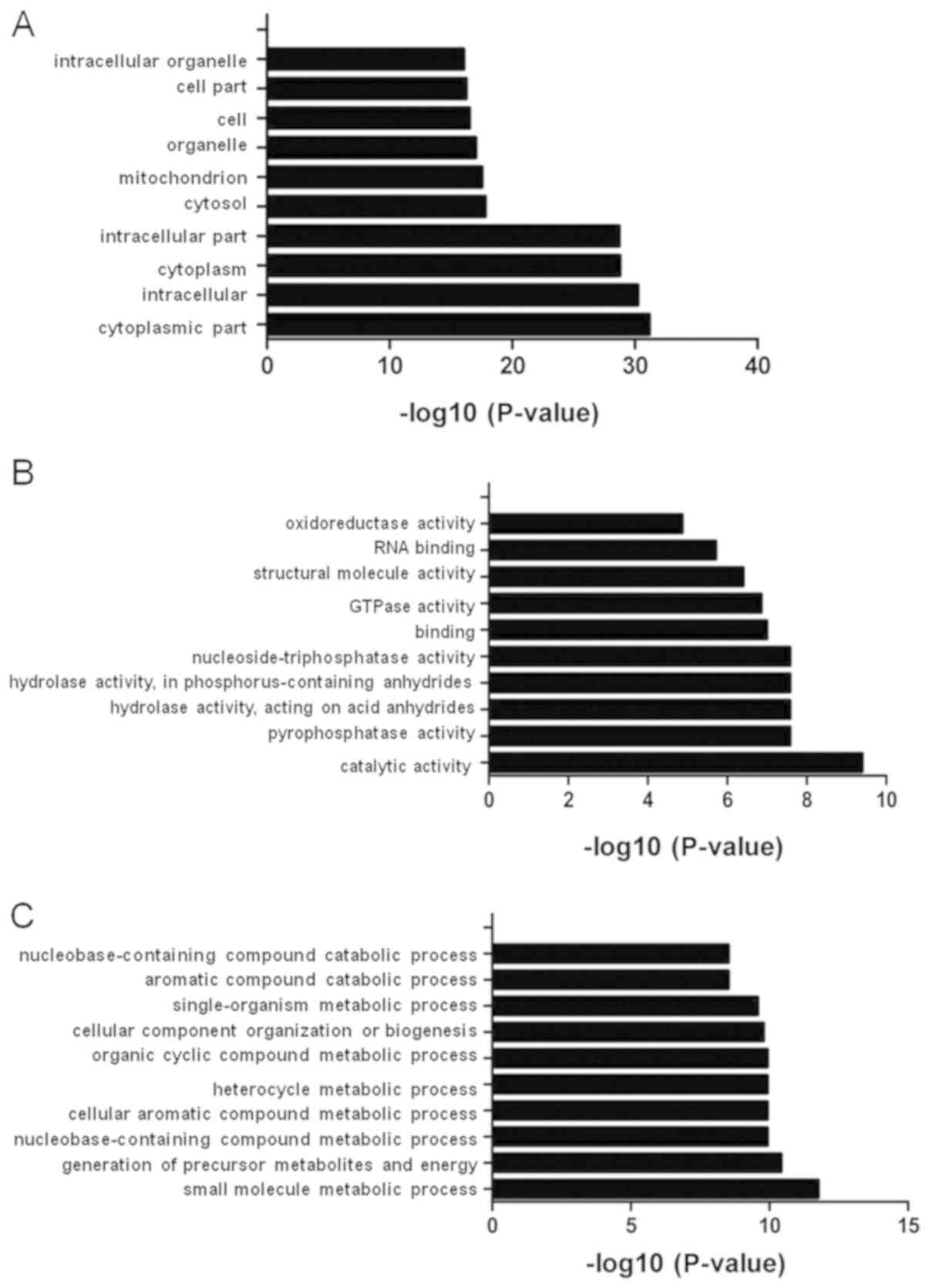
The 403 binding proteins were analyzed using GO
enrichment analysis. The GO analysis classified the proteins into
the following three functional categories: Biological process,
cellular component and molecular function (Fig. 3). Under cellular component, the top
ten GO terms were ‘cytoplasmic part’, ‘intracellular’, ‘cytoplasm’,
‘intracellular part’, ‘cytosol’, ‘mitochondrion’, ‘organelle’,
‘cell’, ‘cell part’ and ‘intracellular organelle’ (Fig. 3A). Under molecular function, the top
ten GO terms were ‘catalytic activity’, ‘pyrophosphatase activity’,
‘hydrolase activity’, ‘acting on acid anhydrides’, ‘hydrolase
activity’, ‘acting on acid anhydrides’, ‘in phosphorus-containing
anhydrides’, ‘nucleoside-triphosphatase activity’, ‘binding’,
‘GTPase activity’, ‘structural molecule activity’, ‘RNA binding’
and ‘oxidoreductase activity’ (Fig.
3B). Under biological process, the top ten GO terms were ‘small
molecule metabolic process’, ‘generation of precursor metabolites
and energy’, ‘nucleobase-containing compound metabolic process’,
‘cellular aromatic compound metabolic process’, ‘heterocycle
metabolic process’, ‘organic cyclic compound metabolic process’,
‘cellular component’, ‘organization or biogenesis’,
‘single-organism metabolic process’, ‘aromatic compound catabolic
process’ and ‘nucleobase-containing compound catabolic process’
(Fig. 3C).
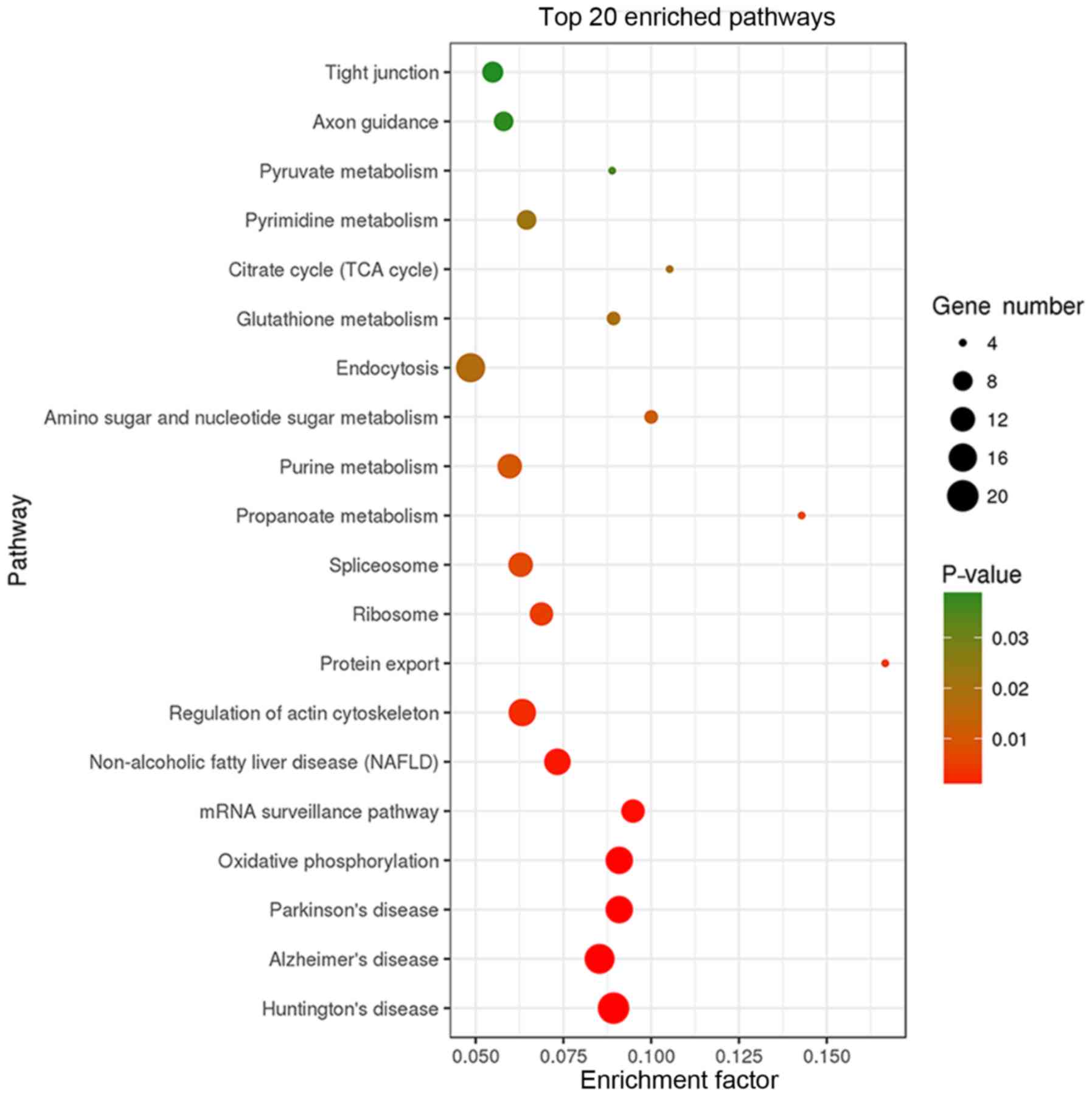
KEGG pathway analysis revealed that the 403 binding
proteins identified were involved in various cellular processes,
including ‘tight junction’, ‘oxidative phosphorylation’, ‘mRNA
surveillance’, ‘actin cytoskeleton regulation’, ‘protein export’,
and diseases, including ‘Huntington's disease’, ‘Alzheimer's
disease’, ‘Parkinson's disease’, ‘non-alcoholic fatty liver
disease’ (Fig. 4).
Possible transcription factors (TFs) involved were
predicted using the AnimalTFDB database. The analysis resulted in
the identification of 10 potential interacting TFs of the miR-32
promoter (Table II).
 | Table II.TFs potentially interacting with the
microRNA-32 promoter as detected by mass spectrometry. |
Table II.
TFs potentially interacting with the
microRNA-32 promoter as detected by mass spectrometry.
| Gene ID | TF symbol | Domain | TF name |
|---|
|
ENSG00000170365 | SMAD1 | MH1 | SMAD family member
1 |
|
ENSG00000061455 | PRDM6 | ZBTB | PR domain
containing 6 |
|
ENSG00000164916 | FOXK1 | Fork | Forkhead box
K1 |
|
ENSG00000165684 | SNAPC4 | MYB | Small nuclear RNA
activating complex, polypeptide 4 |
|
ENSG00000182359 | KBTBD3 | ZBTB | Kelch repeat and
BTB (POZ) domain containing 3 |
|
ENSG00000153048 | CARHSP1 | CSD | Calcium regulated
heat stable protein 1 |
|
ENSG00000167377 | ZNF23 | ZBTB | Zinc finger protein
23 |
|
ENSG00000121297 | TSHZ3 | ZBTB | Teashirt zinc
finger homeobox 3 |
|
ENSG00000115415 | STAT1 | STAT | Signal transducer
and activator of transcription 1 |
|
ENSG00000136535 | TBR1 | T-box | T-box, brain,
1 |
Discussion
In recent years, a number of studies reported the
potential role of miRNAs in different types of cancer (14–16). The
characterization of dysregulated miRNAs in CRC may help to improve
the understanding of carcinogenesis and develop treatments for the
disease. Previous studies have demonstrated that the overexpression
of miR-32 led to increased proliferation, migration, and invasion
and reduced apoptosis of CRC cells via inhibition of the
anti-oncogene PTEN (12,13). However, the regulation of miR-32
expression in CRC remains unknown. The aim of the current study was
to investigate the regulation of miR-32 expression.
The expression of miRNAs is regulated by regulatory
systems, including the promotors of their host genes, epigenetic
regulation and TFs (17–21). Dysregulation of miRNA expression in
different types of cancer may be due to an abnormal combination of
TFs acting on the promoter regions or due to epigenetic changes,
including aberrant DNA methylation and histone modification
(22–24). Various stimuli in the external
environment or signals at different stages of development may cause
different TFs to bind to transcriptional regulatory elements,
activating or inhibiting the transcription of miRNAs (25). Therefore, the identification of
proteins interacting with the promoter of miRNAs as well as
analysis of their function is important for investigating the
transcriptional regulation of miRNAs. Zhu et al (21) demonstrated that TF Kruppel like
factor 4 negatively regulated miR-106a expression by binding to the
promoter of miR-106a. A study by Kumar et al (26) revealed that the TF myocyte enhancer
factor-2 and hypermethylation and histone modifications may have
contributed to the downregulation of the miR-379/miR-656 cluster in
oligodendrogliomas, either acting independently or in synergy, in
oligodendroglioma. Nuclear factor-κB bound to the promoter of
miR-1275 and inhibited its transcription, in response to tumor
necrosis factor α (TNF-α) stimulation (27). Another study reported that
transforming growth factor β1 (TGFβ1) promoted the binding of
mothers against decapentaplegic homolog (SMAD)4 to the miR-155
promoter at a site located 454 bp from the transcription start
site, suggesting that miR-155 may be a transcriptional target of
the TGFβ1/SMAD4 pathway (28).
miRNAs are transcribed by RNA polymerase II to
generate the original transcript of miRNAs, called primary miRNAs
(pri-miRNAs) (29–31). Drosha, an enzyme in the polymerase
III family, processes the pri-miRNAs into a hairpin-like precursor
miRNA (pre-miRNA) (29–31). The pre-miRNA is exported into the
cytoplasm by exportin 5 and then cleaved by Dicer into 18–25
nucleotide double-stranded miRNAs, which are then unwound to
generate mature miRNAs (29–31). Half of the known mammalian miRNA
sequences are located in the introns of protein-coding host genes,
referred to as intronic miRNAs (32). Such intron-derived miRNAs are
commonly expressed coordinately and processed with their host gene
transcripts (33). miR-32 is an
intronic miRNA encoded by TMEM245, as described in the University
of California, Santa Cruz Genome Browser (genome.ucsc.edu). Several intronic miRNAs are
transcribed together with the host gene (18,34,35).
Human papillomavirus type 16 E6 may regulate miR-23b, an intronic
miRNA, indirectly through the methylation of its host gene TMEM245
(36). Lerner et al (37) demonstrated that deleted in
lymphocytic leukemia 2 (DLEU2) acts as a host gene of
miR-15a/miR-16-1, and the binding of the Myc to two alternative
DLEU2 promoters represses both the host gene transcription and
levels of mature miR-15a/miR-16-1. It is reported that the
transcript levels of TMEM245 and miR-32 are positively correlated
(38). Functional analysis of the
promoter of miR-32 is required to understand the molecular
mechanisms governing miR-32 gene expression. In the present study,
the truncation analysis and luciferase reporter assays demonstrated
that the cloned promoter fragment was capable of driving expression
of the luciferase gene in transfected HCT-116 cells. The core
promoter of miR-32 may be located within the −320 to −1 bp region
which exhibited the highest luciferase activity. The regions
spanning −606 to −320 bp potentially harbor negative regulatory
elements as a significant decrease in promoter activity was
observed. These data indicate that the miR-32 overexpression is due
to the complex interactions between different regulatory elements
and promoter.
A DNA pull-down assay in combination with MS was
performed to identify the proteins that bind to the miR-32 gene
promoter. In addition, bioinformatics analyses were performed to
characterize the binding proteins. The binding proteins were
involved in a variety of key biological processes, including
‘structural molecule activity’, ‘RNA binding’, ‘small molecule
metabolic process’ and ‘biogenesis’. This suggested that these
proteins may potentially serve a role in carcinogenesis. The KEGG
pathway analysis revealed that the 403 binding proteins identified
were involved in neuronal diseases. Yan et al (39) demonstrated that miR-32 promotes
neuroinflammation and neuropathic pain development through
regulation of dual-specificity phosphatase 5, and knockdown of
miR-32 suppressed mechanical allodynia and heat hyperalgesia and
decreased inflammatory cytokine [interleukin (IL)-1β, TNF-α and
IL-6] protein expression in rats following spinal nerve ligation.
Another study revealed that two single nucleotide polymorphisms in
the host gene TMEM245 were involved in genetic loci strongly
associated with schizophrenia (40).
Since miR-32 and its host gene TMEM245 may be involved in the
pathogenesis of nervous system-associated diseases, proteins
binding to the miR-32 promoter may also be involved in the
signaling pathways of nervous system diseases. The 403 binding
proteins identified in the current study are also involved in other
cellular processes, including ‘tight junction’, ‘oxidative
phosphorylation’, ‘mRNA surveillance’ and ‘actin cytoskeleton
regulation’. These pathways are also involved in the development of
tumors (41–44), including pancreatic, ovarian and
gastric cancer, which may be associated with pathogenesis of
colorectal cancer.
TFs are a group of proteins that can regulate RNA
transcription by binding to the promoter of the corresponding DNA
sequence (45). TF analysis revealed
10 potential interacting TFs, including SMAD1, signal transducer
and activator of transcription 1 (STAT1) and forkhead box K1
(Foxk1) among the binding proteins. Yang et al (46) revealed that SMAD1 promotes migration
of CRC cells by inducing Snail and ajuba LIM protein expression
simultaneously. The level of SMAD1 was significantly increased in
CRC tissues, and was confirmed as significant predictor for overall
survival (47). High STAT1 activity
was significantly associated with longer patient overall survival
in CRC (48). Wu et al
(49) demonstrated that higher
expression of Foxk1 could indicate a poor prognosis in patients
with CRC since Foxk1 induces epithelial-mesenchymal transition
(EMT) and promotes CRC cell invasion in vitro and in
vivo; knockdown of Foxk1 inhibited TGF-β1-induced EMT. These
transcription factors potentially serve a role in the pathogenesis
of CRC, and further investigation is required to identify, verify
and validate their involvement as binding proteins in miR-32
expression.
The current study demonstrated that the core
promoter region of the human miR-32 gene is located in the region
spanning −320 to −1 bp, and binding proteins, especially TFs, may
be involved in the transcriptional regulation of this gene. These
results provide insight into the mechanism of miR-32 gene
regulation. Due to the limitations of bioinformatics, which are
only based on bioinformatics predictions, it's not certain whether
these proteins can affect the expression of miR-32. Further
verifications, including gain-of-function and loss-of-function
studies, and a chromatin immunoprecipitation assay, are required to
clarify the function of possible binding proteins.
Acknowledgements
Not applicable.
Funding
This study was supported by Guangdong Natural
Science Foundation of China (grant no. 2017A030313546).
Availability of data and materials
The datasets used and/or analyzed in this study are
available from the corresponding author on reasonable request.
Authors' contributions
WW and YZ designed the experiments. WW, WT and SY
performed the experiments. WW and JQ performed statistical analysis
of the obtained data, and drafted the manuscript. YZ revised the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peddareddigari V, Wang D and Dubois RN:
The tumor microenvironment in colorectal carcinogenesis. Cancer
Microenviron. 3:149–166. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molnár B, Galamb O, Péterfia B, Wichmann
B, Csabai I, Bodor A, Kalmár A, Szigeti KA, Barták BK, Nagy ZB, et
al: Gene promoter and exon DNA methylation changes in colon cancer
development-mRNA expression and tumor mutation alterations. BMC
Cancer. 18:6952018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sakai E, Nakajima A and Kaneda A:
Accumulation of aberrant DNA methylation during colorectal cancer
development. World J Gastroenterol. 20:978–987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun W, Wang X, Li J, You C, Lu P, Feng H,
Kong Y, Zhang H, Liu Y, Jiao R, et al: MicroRNA-181a promotes
angiogenesis in colorectal cancer by targeting SRCIN1 to promote
the SRC/VEGF signaling pathway. Cell Death Dis. 9:4382018.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ,
Hsu CW, Chen WS and Wang JH: Linc00659, a long noncoding RNA, acts
as novel oncogene in regulating cancer cell growth in colorectal
cancer. Mol Cancer. 17:722018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Taborda MI, Ramírez S and Bernal G:
Circular RNAs in colorectal cancer: Possible roles in regulation of
cancer cells. World J Gastrointest Oncol. 9:62–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Pancione M, Remo A, Zanella C, Sabatino L,
Di Blasi A, Laudanna C, Astati L, Rocco M, Bifano D, Piacentini P,
et al: The chromatin remodelling component SMARCB1/INI1 influences
the metastatic behavior of colorectal cancer through a gene
signature mapping to chromosome 22. J Transl Med. 11:2972013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shirafkan N, Mansoori B, Mohammadi A,
Shomali N, Ghasbi M and Baradaran B: MicroRNAs as novel biomarkers
for colorectal cancer: New outlooks. Biomed Pharmacother.
97:1319–1330. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Toiyama Y, Takahashi M, Hur K, Nagasaka T,
Tanaka K, Inoue Y, Kusunoki M, Boland CR and Goel A: Serum miR-21
as a diagnostic and prognostic biomarker in colorectal cancer. J
Natl Cancer Inst. 105:849–859. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chang PY, Chen CC, Chang YS, Tsai WS, You
JF, Lin GP, Chen TW, Chen JS and Chan E: MicroRNA-223 and
microRNA-92a in stool and plasma samples act as complementary
biomarkers to increase colorectal cancer detection. Oncotarget.
7:10663–10675. 2016.PubMed/NCBI
|
|
11
|
Li J, Zou K, Yu L, Zhao W, Lu Y, Mao J,
Wang B, Wang L, Fan S, Song B and Li L: MicroRNA-140 inhibits the
epithelial-mesenchymal transition and metastasis in colorectal
cancer. Mol Ther Nucleic Acids. 10:426–437. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wu W, Yang J, Feng X, Wang H, Ye S, Yang
P, Tan W, Wei G and Zhou Y: MicroRNA-32 (miR-32) regulates
phosphatase and tensin homologue (PTEN) expression and promotes
growth, migration, and invasion in colorectal carcinoma cells. Mol
Cancer. 12:302013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu W, Yang P, Feng X, Wang H, Qiu Y, Tian
T, He Y, Yu C, Yang J, Ye S and Zhou Y: The relationship between
and clinical significance of MicroRNA-32 and phosphatase and tensin
homologue expression in colorectal cancer. Genes Chromosomes
Cancer. 52:1130–1140. 2013. View Article : Google Scholar
|
|
14
|
Porzycki P, Ciszkowicz E, Semik M and
Tyrka M: Combination of three miRNA (miR-141, miR-21, and miR-375)
as potential diagnostic tool for prostate cancer recognition. Int
Urol Nephrol. 50:1619–1626. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen Y, Min L, Ren C, Xu X, Yang J, Sun X,
Wang T, Wang F, Sun C and Zhang X: miRNA-148a serves as a
prognostic factor and suppresses migration and invasion through
Wnt1 in non-small cell lung cancer. PLoS One. 12:e01717512017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zeng Y, Wang KX, Xu H and Hong Y:
Integrative miRNA analysis identifies hsa-miR-3154, hsa-miR-7-3,
and hsa-miR-600 as potential prognostic biomarker for cervical
cancer. J Cell Biochem. 119:1558–1566. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Saito Y, Friedman JM, Chihara Y, Egger G,
Chuang JC and Liang G: Epigenetic therapy upregulates the tumor
suppressor microRNA-126 and its host gene EGFL7 in human cancer
cells. Biochem Biophys Res Commun. 379:726–731. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Baskerville S and Bartel DP: Microarray
profiling of microRNAs reveals frequent coexpression with
neighboring miRNAs and host genes. RNA. 11:241–247. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mekala JR, Naushad SM, Ponnusamy L,
Arivazhagan G, Sakthiprasad V and Pal-Bhadra M: Epigenetic
regulation of miR-200 as the potential strategy for the therapy
against triple-negative breast cancer. Gene. 641:248–258. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhu M, Zhang N, Lu X and He S: Negative
regulation of kruppel-like Factor 4 on microRNA-106a at upstream
transcriptional level and the role in gastric cancer metastasis.
Dig Dis Sci. 63:2604–2616. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kuang Q, Li J, You L, Shi C, Ji C, Guo X,
Xu M and Ni Y: Identification and characterization of NF-kappaB
binding sites in human miR-1908 promoter. Biomed Pharmacother.
74:158–163. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li Y, Xu Z, Li B, Zhang Z, Luo H, Wang Y,
Lu Z and Wu X: Epigenetic silencing of miRNA-9 is correlated with
promoter-proximal CpG island hypermethylation in gastric cancer in
vitro and in vivo. Int J Oncol. 45:2576–2586. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang W, Ji G, Xiao X, Chen X, Qin WW, Yang
F, Li YF, Fan LN, Xi WJ, Huo Y, et al: Epigenetically regulated
miR-145 suppresses colon cancer invasion and metastasis by
targeting LASP1. Oncotarget. 7:68674–68687. 2016.PubMed/NCBI
|
|
25
|
Tagne JB, Mohtar OR, Campbell JD,
Lakshminarayanan M, Huang J, Hinds AC, Lu J and Ramirez MI:
Transcription factor and microRNA interactions in lung cells: An
inhibitory link between NK2 homeobox 1, miR-200c and the
developmental and oncogenic factors Nfib and Myb. Respir Res.
16:222015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumar A, Nayak S, Pathak P, Purkait S,
Malgulawar PB, Sharma MC, Suri V, Mukhopadhyay A, Suri A and Sarkar
C: Identification of miR-379/miR-656 (C14MC) cluster downregulation
and associated epigenetic and transcription regulatory mechanism in
oligodendrogliomas. J Neurooncol. 139:23–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou YF, Fu ZY, Chen XH, Cui Y, Ji CB and
Guo XR: Tumor necrosis factor-α and interleukin-6 suppress
microRNA-1275 transcription in human adipocytes through nuclear
factor-κB. Mol Med Rep. 16:5965–5971. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhao H, Zhang J, Shao H, Liu J, Jin M,
Chen J and Huang Y: Transforming growth factor β1/smad4 signaling
affects osteoclast differentiation via regulation of miR-155
expression. Mol Cells. 40:211–221. 2017.PubMed/NCBI
|
|
29
|
Sand M: The pathway of miRNA maturation.
Methods Mol Biol. 1095:3–10. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J,
Lee J, Provost P, Rådmark O, Kim S and Kim VN: The nuclear RNase
III Drosha initiates microRNA processing. Nature. 425:415–419.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hammond SM: An overview of microRNAs. Adv
Drug Deliv Rev. 87:3–14. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kim YK and Kim VN: Processing of intronic
microRNAs. EMBO J. 26:775–783. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Zhang L and Sun T: Cohesive
regulation of neural progenitor development by microRNA miR-26, its
host gene ctdsp and target gene Emx2 in the mouse embryonic
cerebral cortex. Front Mol Neurosci. 11:442018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ma N, Wang X, Qiao Y, Li F, Hui Y, Zou C,
Jin J, Lv G, Peng Y, Wang L, et al: Coexpression of an intronic
microRNA and its host gene reveals a potential role for miR-483-5p
as an IGF2 partner. Mol Cell Endocrinol. 333:96–101. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yeung CL, Tsang TY, Yau PL and Kwok TT:
Human papillomavirus type 16 E6 suppresses microRNA-23b expression
in human cervical cancer cells through DNA methylation of the host
gene C9orf3. Oncotarget. 8:12158–12173. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lerner M, Harada M, Lovén J, Castro J,
Davis Z, Oscier D, Henriksson M, Sangfelt O, Grandér D and Corcoran
MM: DLEU2, frequently deleted in malignancy, functions as a
critical host gene of the cell cycle inhibitory microRNAs miR-15a
and miR-16-1. Exp Cell Res. 315:2941–2952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ambs S, Prueitt RL, Yi M, Hudson RS, Howe
TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, et al:
Genomic profiling of microRNA and messenger RNA reveals deregulated
microRNA expression in prostate cancer. Cancer Res. 68:6162–6170.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yan T, Zhang F, Sun C, Sun J, Wang Y, Xu
X, Shi J and Shi G: miR-32-5p-mediated Dusp5 downregulation
contributes to neuropathic pain. Biochem Biophys Res Commun.
495:506–511. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu C, Aragam N, Li X, Villla EC, Wang L,
Briones D, Petty L, Posada Y, Arana TB, Cruz G, et al: BCL9 and
C9orf5 are associated with negative symptoms in schizophrenia:
meta-analysis of two genome-wide association studies. PLoS One.
8:e516742013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kyuno D, Yamaguchi H, Ito T, Kono T,
Kimura Y, Imamura M, Konno T, Hirata K, Sawada N and Kojima T:
Targeting tight junctions during epithelial to mesenchymal
transition in human pancreatic cancer. World J Gastroenterol.
20:10813–10824. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Guo NL, Zhang JX, Wu JP and Xu YH:
Isoflurane promotes glucose metabolism through up-regulation of
miR-21 and suppresses mitochondrial oxidative phosphorylation in
ovarian cancer cells. Biosci Rep. 37(pii): BSR201708182017.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li L, Geng Y, Feng R, Zhu Q, Miao B, Cao J
and Fei S: The human RNA surveillance factor UPF1 modulates gastric
cancer progression by targeting long non-coding RNA MALAT1. Cell
Physiol Biochem. 42:2194–2206. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Peng JM, Bera R, Chiou CY, Yu MC, Chen TC,
Chen CW, Wang TR, Chiang WL, Chai SP, Wei Y, et al: Actin
cytoskeleton remodeling drives epithelial-mesenchymal transition
for hepatoma invasion and metastasis in mice. Hepatology.
67:2226–2243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Spitz F and Furlong EE: Transcription
factors: From enhancer binding to developmental control. Nat Rev
Genet. 13:613–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang D, Hou T, Li L, Chu Y, Zhou F, Xu Y,
Hou X, Song H, Zhu K, Hou Z, et al: Smad1 promotes colorectal
cancer cell migration through Ajuba transactivation. Oncotarget.
8:110415–110425. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang L, Liu Z, Tan J, Dong H and Zhang X:
Multispectral imaging reveals hyper active TGF-β signaling in
colorectal cancer. Cancer Biol Ther. 19:105–112. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gordziel C, Bratsch J, Moriggl R, Knösel T
and Friedrich K: Both STAT1 and STAT3 are favourable prognostic
determinants in colorectal carcinoma. Br J Cancer. 109:138–146.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu Y, Peng Y, Wu M, Zhang W, Zhang M, Xie
R, Zhang P, Bai Y, Zhao J, Li A, et al: Oncogene FOXK1 enhances
invasion of colorectal carcinoma by inducing epithelial-mesenchymal
transition. Oncotarget. 7:51150–51162. 2016.PubMed/NCBI
|