Introduction
Multicellular organisms develop complex genomes
through transcriptional regulation, which is crucial for spatial
and temporal cellular specialization to promote phenotypic
complexity. The interaction of regulatory DNA sequence motifs with
DNA-binding transcription factors induces transcriptional
regulation. These transcriptional factors serve a critical role in
the activation or repression of target genes. Zinc-finger proteins
are among the most commonly occurring DNA-binding motifs and
constitute 2–3% of the transcription factors in the human genome
(1). These proteins are associated
with several biological functions, including differentiation,
chromatin remodeling and development (2). The zinc finger, BED-type (ZBED) gene
family is widely expressed in vertebrate tissues. It comprises a
closely related group of genes that contribute to the regulation of
various functions by encoding regulatory proteins. For example, the
ZBED6 gene is involved in the regulation of diverse phenotypic
effects. ZBED6 binds to the conserved target motif of insulin-like
growth factor 2 and inhibits its expression, thereby promoting cell
proliferation, growth and development in placental mammals
(3). Furthermore, conflicting
phenotypic changes were induced by the inactivation of ZBED6 in
HCT116 and RKO human colorectal cancer cell lines, with
consequences of reduced and increased growth, respectively,
indicating that ZBED6 exhibits transcriptional modulating
properties, and that the effect of ZBED6 on tumor development is
dependent on the transcriptional state and genetic background of
its target genes (4). The ZBED3
protein interacts with axin, which is vital for Wnt/β-catenin
signal modulation in mammalian carcinogenesis and embryogenesis
(5). Notably, Fan et al
(6) reported higher expression
levels of ZBED3 in cancer tissues compared with normal lung tissues
and demonstrated that ZBED3 expression levels have
clinicopathological significance. Downregulation of ZBED3 by small
interfering RNA significantly inhibits expression of p120ctn-1 and
β-catenin in lung cancer cells; these results indicate that lung
cancer cell invasion may be induced by ZBED3, which functions by
regulating p120ctn-1 and β-catenin (6). Therefore, ZBED3 has potential
application in the management of cancer, particularly non-small
cell lung cancer. Furthermore, ZBED4 is restricted in glial Müller
cells and the cone photoreceptors of the human retina (7). It binds to DNA and RNA sequences and
effectively affects the transcription process of genes expressed in
the retina via G-rich promoters (8).
ZBED1 was identified as a human homolog of
Drosophila DNA replication-related element-binding factor
(DREF) via BLAST search. Also known as human DREF, Activator
(Ac)-like transposable element, TRAMP and KIAA0785, ZBED1 has been
found to play a critical role in cell proliferation via the
regulation of gene expression (9).
As ZBED1 has similar structural and functional properties to DREF,
the present review comprehensively describes recent progress on the
origin, structure and functions on ZBED1, specifically highlights
the similarities and differences between ZBED1 and DREF, and
discusses future research directions.
Origin of ZBED1
A large proportion of vertebrate genomes consist of
eukaryotic genome components and transposable elements (TEs), such
as DNA transposons and retrotransposons (10–12).
Several genes, particularly those that possess rapidly evolving
coding sequence properties, have been shown to contain functionally
important TEs that are involved in gene expression and regulatory
changes (11,13,14).
Notably, a quarter of analyzed human promoter regions have been
found to have TE-derived sequences that potentially function as
alternative promoters in several genes (13). TEs contribute entire functional genes
to the host genome through molecular domestication (11,15,16).
Domesticated TEs, which are immobile, often exist as single-copy
orthologs within the genomes of associated organisms (15). Since DNA transposons encode
multi-domain proteins with diverse functions, such as protein- and
DNA-binding affinity, they may be suitable for domestication to
perform host functions.
The hAT transposons and their associated
domesticated sequences form a large superfamily that is divided
into Buster and Ac families. These two families exhibit distinct
target-site selections and were named according to the first
identified transposon or transposon-like sequence in each family
(17). Through phylogenetic
methodology, Hayward et al (18) demonstrated that ZBED5, ZBED7, ZBED8
and ZBED9 are associated with the Buster family and are distinct
from other ZBEDs. The remaining ZBEDs form two monophyletic clades
within the Ac transposon family. Notably, ZBED2, ZBED3, ZBED4,
ZBED6 and ZBEDX form a monophyletic group with C7ORF29 and differ
from ZBED1 (Fig. 1). The ZBEDs
exhibit structural similarity via the zinc finger domain that
functions in DNA binding. Closely related ZBED molecules regulate a
variety of different host functions, indicating that ZBED protein
domains are particularly suitable for controlling host
functions.
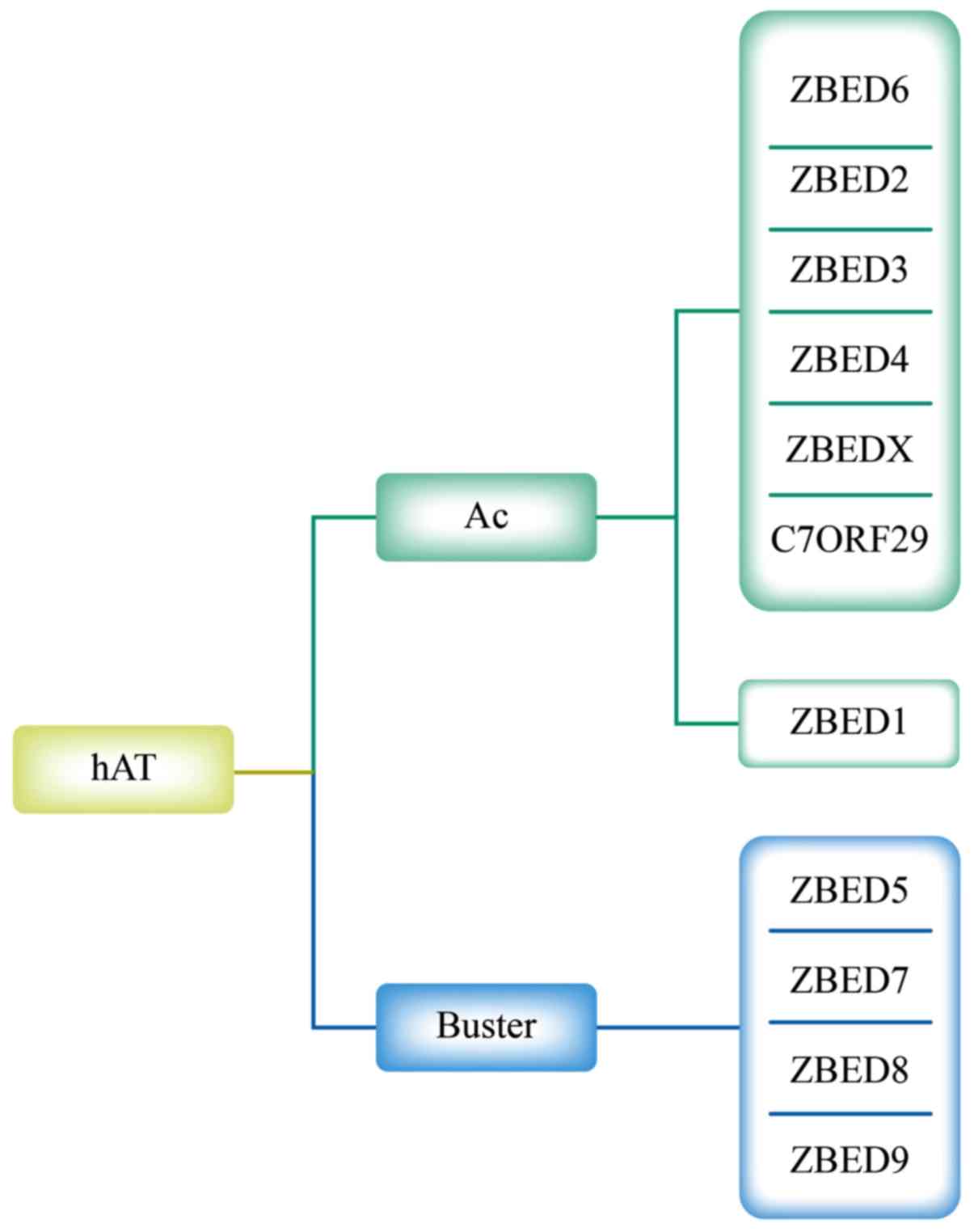 | Figure 1.Phylogenetic relationships of ZBED
genes with related sequences. The hAT transposons comprise Buster
and Ac families. ZBED6, ZBED2, ZBED3, ZBED4, ZBEDX, C7ORF29, and
ZBED1 belong to the Ac family. ZBED5, ZBED7, ZBED8, and ZBED9
belong to the Buster family. ZBED, zinc finger, BED-type; Ac,
Activator. |
Similar zinc-finger structure of DREF and
ZBED1
A palindromic 8 base-pair (bp) sequence 5′-TATCGATA
was identified as being common to the promoter regions of the
proliferating cell nuclear antigen (PCNA) and DNA polymerase α
genes in Drosophila (19).
This sequence is known as the DNA replication-related element (DRE)
and is vital for transcriptional activation. Subsequently, several
methods, including band mobility shift assays, were used to
identify specific DREF (20), and
the cDNA of DREF was then cloned (21). Notably, DREF was found to comprise a
polypeptide with 709 amino acid (aa) residues. The DREF gene of
Drosophila virilis (D. virilis) has been shown to
share 71% of its aa sequence identity with its D.
melanogaster homolog, with three highly conserved regions (CRs)
at the aa positions 14–182 (CR1; identity, 86.4%), 432–568 (CR2;
identity, 86.1%) and 636–730 (CR3; identity, 83.3%) of the D.
virilis DREF (22).
DREF is proposed to have originated from a
combination of the amino-terminal boundary element-associated
factor (BEAF) and DREF (BED) zinc finger domain protein and the
carboxy-terminal hATC domain (23).
Aravind (24) conducted sequence
analyses that led to the prediction of protein signature
CX2CXNHX3-5(H/C) for the BED
finger, a name derived from DREF protein and domesticated
Drosophila BEAF. The BED domain forms the zinc finger common
to fungal, animal and plant proteins. This domain has been
postulated to originate from transposons in cellular genes or to
have been acquired for the cellular functions of transposases at
independent occasions. A domain of BED zinc finger containing 50–60
aa residues has been found to contain a characteristic motif with
two highly conserved aromatic residues. It has a shared pattern of
histidines and cysteines that is considered to create a DNA-binding
zinc finger domain. Furthermore, the BED domains of BEAF and DREF
have been reported to exhibit DNA-binding properties (21,25).
ZBED1 comprises a polypeptide of 694 aa residues
that has 21% similarity and 22% identity when compared with DREF
(9). Three CRs between D.
virilis and D. melanogaster as aforementioned have aa
sequences highly similar to those of the ZBED1 polypeptide,
particularly the CR1 region, which is essential for DRE-binding and
dimerization. The ZBED1 binding sequence [hDRE;
5′-TGTCG(C/T)GA(C/T)A] was identified using the CASTing method and
found to overlap with the DREF (5′-TATCGATA) sequence (9). Furthermore, the 6-bp sequence
5′-TCG(C/T)GA at the center of ZBED1 was indicated to serve an
important role in ZBED1 binding (9).
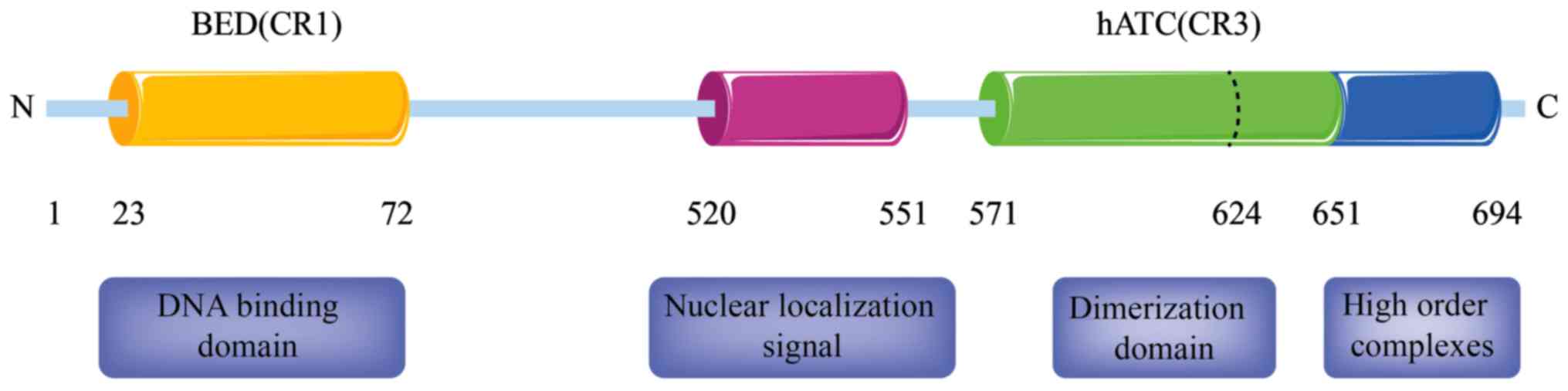
Two regions of ZBED1, CR1 and CR3, correspond with two domains that
are conserved within the family, namely the amino-terminal BED zinc
finger (aa 23–72) and the carboxyl-terminal hATC domain (26). The BED zinc finger is regarded as a
DNA-binding domain of chromatin boundary element-binding
transposase (24), whereas the hATC
domain of hAT transposase family members is considered as the
dimerization domain, as assessed through in vitro
cross-linking experiments and yeast two-hybrid assays using Ac and
housefly Hermes family members (27,28).
The DNA-binding protein ZBED1 has granular
structures and is predominantly distributed in the nucleus
(9). A study by Yamashita et
al (26) explored the mechanism
of action of ZBED1 using detailed mutagenesis analyses to
investigate the functions of the hATC domain. Trp-590, Trp-591 and
Leu-601, which are highly conserved hydrophobic aa residues in hATC
domains, were found to be crucial for the in vivo
self-association of ZBED1. Furthermore, through substitution and
truncated mutant evaluation, it was found that the hATC domain (aa
571–651) has two regions with distinct functions. The first region,
namely the amino-terminal region aa 571–624, is required for
various processes, including ZBED1 DNA-binding, nuclear
accumulation and self-association. The second region, namely the
carboxyl-terminal region aa 625–651, is vital for granular pattern
formation and self-association. These findings indicate that ZBED1
self-association via the hATC domain is required for nuclear
accumulation prior to nuclear importation. Furthermore, the aa
652–694 region has been identified to contain highly ordered
complexes of ZBED1 binding sequences, whereas the aa 520–551 region
is a classical nuclear localization signal (Fig. 2).
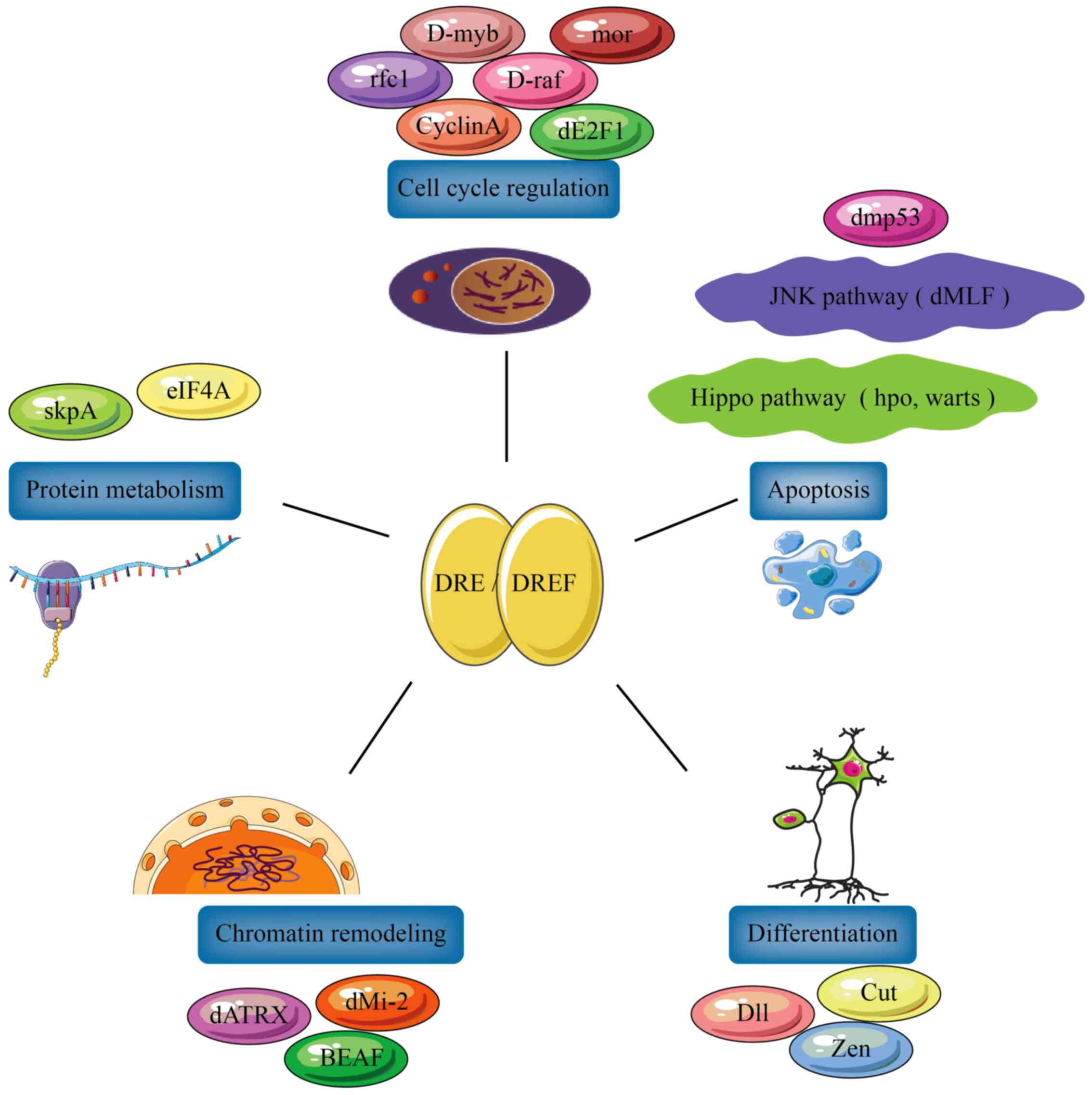
Multiple functions of DREF and ZBED1
Matsukage et al (29) used bioinformatics analysis to
determine the type and number of genes regulated by the DRE/DREF
system. They established that 456 DRE sequences and 156 genes with
DRE sequences are <1 kb upstream of their transcription
initiation sites. Most of these genes are associated with processes
required for cell proliferation (30), including chromatin remodeling, cell
cycle regulation and protein metabolism, while others are
associated with apoptosis and differentiation. This indicates that
the DRE/DREF system functions as a regulator of several cell
proliferation-associated genes (Fig.
3). Ohshima et al (9)
searched the GenBank® and EMBL databases to identify
genes controlled by ZBED1 protein. Through BLAST analysis, they
found >500 TGTCG(C/T)GA(T/C)A-like sequences in the human
genome. Notably, ZBED1 binding sequences exist in a series of genes
involved in DNA synthesis (DNA replication) and repair, protein
synthesis, regulation of chromatin structure and cell cycle
regulation, which seems to be consistent with DREF (31), as some of these genes have been shown
to be controlled by the DRE/DREF system (20,32–35).
These findings indicate that ZBED1 is a potential functional
homolog of Drosophila DREF and can regulate the expression
of genes involved in cell proliferation via a similar
mechanism.
A notable study indicates that DREF preferentially
binds to and activates housekeeping enhancers that are located
closely to ubiquitously expressed genes with specificity to its
core promoter, and suggests that the DRE motif is required and
sufficient for housekeeping enhancer function (36). The CGCG sequence is central to the
ZBED1 binding sequence TGTCGCGACA that occurs mostly in promoter
regions of housekeeping genes within the mammalian genome,
suggesting that ZBED1 could regulate numerous housekeeping genes
with CpG islands (37).
Chromatin remodeling
Chromatin-remodeling protein contains many conserved
motifs and is encoded by human alpha-thalassemia and mental
retardation X-linked syndrome (hATRX) gene. The ATRX protein
contains a helicase/ATPase domain at its carboxyl terminus, which
has catalytic subunits of chromatin remodeling complexes (38). Valadez-Graham et al (39) found that dATRX, a Drosophila
homolog of hATRX, interacts with transcription factor DREF in
vivo and in vitro. This results in transcriptional
regulation of the pannier (pnr) gene, which belongs to the GATA
family of transcription factors. Therefore, the transcriptional
factor DREF is proposed to associate with the chromatin remodeling
factor dATRX in the regulation of the pnr gene. These studies on
Drosophila may provide insights for further research
exploring the functional interaction of hATRX with ZBED1 in
humans.
Studies conducted by Hart et al (25,40)
suggested that DREF functions as a BEAF antagonist that is
necessary for boundary activity of the special chromatin structure
region of the Drosophila 87A7 hsp70 gene (25,40,41). The
boundary element sequence 5′-CGATA-3′, which is also present in DRE
(5′-TATCGATA-3′), binds to BEAF-32. This induces the blockade of
upstream regulatory elements such as enhancers (25), as demonstrated by chromatin
immunoprecipitation. Thus, it is suggested that BEAF may bind to
the same sequences as DREF and binding sequence competition between
BEAF and DREF may be crucial in the regulation of chromatin
boundary activities.
Drosophila Mi-2 (dMi-2), a
nucleosome-stimulated ATPase, employs energy from ATP hydrolysis to
deacetylate nucleosomal histones and mobilize histone octamers in
certain cases (42). A study
revealed that dMi-2 negatively regulates DREF by inhibiting DNA
binding activity when the C-terminal region of dMi-2 binds to DREF
(43). The chromatin remodeling
factor Mi-2 has also been reported to be a ZBED1-interacting
protein (43). The above findings
indicate that the activity of DREF is regulated via protein
complexes that determine the chromatin structure.
Protein metabolism
DREF has been indicated to directly regulate the
expression of the eukaryotic initiation factor 4A (eIF4A) gene
(44). The eIF4A gene belongs to the
DEAD-box family of ATP-dependent RNA helicases (45) and is proposed to function by
unwinding the secondary structure of 5′-untranslated regions of
mRNA in the cap(m7GpppN)-dependent initiation of protein
synthesis (46,47). Furthermore, eIF4A plays a significant
regulatory role in the initiation of translation (48). A study by Ida et al (44) showed that eIF4A gene promoter
activity and eIF4A mRNA levels decreased following the knockdown of
DREF in Drosophila S2 cells. Also, through a band mobility
shift assay, DRE sequences in the eIF4A gene promoter were found to
bind with DREF in vitro. These findings together indicate
that the eIF4A gene is regulated via the DREF pathway and that,
therefore, DREF regulates protein synthesis, particularly by
initiating translation.
DREF has been found to bind to the skpA gene locus
via DRE sequences. DRE1 and DRE2 sites present in the 5′-flanking
region of the skpA gene have been reported to serve vital roles in
skpA promoter activity in living flies and cultured cells (49). SKP1 protein is a key constituent of
the SKP1, cullin/CDC53, F-box protein (SCF) complex and functions
in protein degradation, linking the substrate recognition subunit
F-box protein to a cullin that then binds the ubiquitin-conjugating
enzyme. Notably, the SKP1 constituent among Drosophila SCF
ubiquitin ligases is referred to as skpA (50). Further study has shown that the
knockdown of DREF in tissues with highly distributed skpA abrogates
the skpA gene expression whereas the overexpression of DREF
activates endogenous skpA expression, indicating that skpA may be
effectively regulated via the DRE/DREF pathway at the
transcriptional level (49). The
available data, therefore, indicate that DREF enhances protein
degradation and protein synthesis by activating genes involved in
various processes, presumably through active protein metabolism in
proliferating cells.
Cell cycle regulation
A study revealed via luciferase transient expression
assays that DREs localized in the replication factor C (rfc)1 gene
promoter participate in transcriptional regulation. Subsequent
in vitro and in vivo assays in the study showed that
DRE sequences of the rfc1 gene are bound to DREF (51). RFC is a five-subunit protein complex
involved in DNA replication, of which RFC140 is the largest
subunit. The study conducted by Tsuchiya et al (51) also identified via phenotype
observation that the rfc1 gene encoding Drosophila RFC140
protein is vital for G1/S cell-cycle progression.
Furthermore, through immune-cytochemical experiments, the study
found that the cell cycle is closely associated with rfc1 gene
expression. These findings indicate that DREF effectively regulates
the rfc1 promoter involved in G1/S cell-cycle
regulation. Additionally, the DRE/DREF system has been found to
regulate several Drosophila genes associated with cell cycle
progression, including dE2F1, cyclin A and D-myb for progression
through the S phase (32,33,52), and
D-raf for progression through the G1 and M phases
(35).
It has been reported that DRE/DREF regulates the
transcription of the Drosophila osa and moira (mor) genes
(53). These two genes encode
subunits of the brahma (BRM) complex, which is an ATP-dependent
chromatin remodeling complex conserved from yeast to humans
(54,55). The BRM complex is thought to
negatively regulate entry into the S phase whereas, by contrast,
DREF promotes G1/S and S phase progression. Thus, DREF
functions simultaneously as a negative and positive regulator of
G1/S progression. This type of regulation may inhibit
excess induction of the S phase when cell cycle progression is
finely tuned.
Ohshima et al (9) used the western blot technique to
examine the fluctuation of ZBED1 protein levels during the cell
cycle. In this analysis, they used primary cultures from human
embryonic lung fibroblasts (HEL cells) that could easily be
transformed to a quiescent state when subjected to serum
deprivation and then returned to cell cycling after the addition of
10% fetal calf serum. Progression in the cell cycle was assessed
after the cells were released from serum starvation using propidium
iodine staining followed by flow cytometric analysis. The results
indicated that ZBED1 expression is induced at the G1/S
transition stage, and so potentially plays an important role in
G1/S progression. Therefore, it is hypothesized that
DREF/ZBED1 highly regulates G1/S progression as a
transcription factor, thereby promoting cell proliferation.
Apoptosis
Although DREF serves a role in the regulation of
cell proliferation, it also activates various apoptosis-inducing
genes. Cell proliferation is restricted via the Hippo pathway
through cell cycle arrest and the induction of apoptosis (56–58).
Notably, it has been reported that the DRE/DREF pathway is required
for the transcriptional activation of the hpo gene to positively
regulate the Hippo pathway (59).
Warts, a tumor suppressor gene that encodes a core kinase in the
Hippo pathway, is also controlled through the DRE/DREF pathway
(60). Using in vivo
chromatin immunoprecipitation assays, DREF was demonstrated to
selectively bind to the warts gene promoter region containing DREs,
and endogenous warts mRNA expression was shown to be reduced in S2
cells following the knockdown of DREF (60). These findings confirm the association
between DRE/DREF and the Hippo pathway. Furthermore, in addition to
Hippo pathway-associated genes, DREF also associates with
Drosophila myeloid leukemia factor in thorax development to
positively regulate the basket gene and thereby activate the JNK
signaling pathway (61,62), which induces apoptosis and protects
the genome.
Studies indicate that the regulation of
Drosophila p53 (dmp53) gene expression is highly
attributable to the binding of the DRE-containing region of dmp53
to the DREF and that DREF affects apoptosis through the
transcriptional activation of dmp53 (63). The dmp53 gene has limited sequence
similarity to the human p53 gene (64); however, the function and structure of
p53 are conserved in flies and mammals (65,66). P53
is a critical regulatory protein required for diverse cellular
metabolic processes, including cell cycle arrest, DNA repair and
apoptosis (67). Nearly all common
human cancers possess mutations or loss of function of the p53 gene
(68,69). The p53 gene, regarded as the
‘guardian of the genome’, acts as a tumor suppressor gene.
Therefore, the finding that DREF positively regulates the
expression of the dmp53 gene indicates the role of the former in
the fine-tuning of cell proliferation.
Through the database searching of other tumor
suppressor genes, including Rbf, APC, Brca2, NF1 and Vhl, DRE and
DRE-like sequences have been identified to be regulated by DREF in
upstream regions of Drosophila (63). The activation of oncogenes such as
Ras or Myc through increased proliferation simultaneously induces
apoptosis (70). Apoptosis then
represses inappropriate cell proliferation, a function that acts as
a mechanism to prevent the proliferation of damaged cells (71). Therefore, DREF is potentially
critical in maintaining tissue kinetics and the balance between
cell proliferation and apoptosis.
Differentiation
For cells to shift from a proliferating state to a
resting state, it is necessary to coordinatively shut down several
cell proliferation-associated genes. This process is controlled
through differentiation signals. DREF as a major transcription
factor of proliferation-related genes is thus a potential mediator
of this repression (72). Some
differentiation signals have been found to target DRE/DREF. The
zerknullt (zen) gene is expressed in the dorsal region of the early
embryo at the cellular blastoderm stage. It encodes a
homeodomain-containing protein Zen that participates in
differentiation of the optic lobe and the amnioserosa (73). Notably, when DREF activity is
reduced, Zen expression in cultured cells represses DRE-containing
genes (74). Therefore, the DRE/DREF
system appears to be a point of intersection between growth and
differentiation signaling pathways. Another homeodomain protein,
Distal-less (Dll), has been found to negatively regulate
Drosophila DREF activity (75). The DNA-binding domain of DREF was
revealed to selectively bind to Dll and thereby inhibit its
transcriptional activity via electrophoretic mobility shift assays.
Co-immunoprecipitation assays for Dll and DREF were also conducted;
however, they did not yield positive results, indicating that
interactions between Dll and DREF may be transient.
DREF has been shown to bind to its DRE sequence
thereby promoting the expression of dPCNA in cell proliferation,
whereas Cut, which is a Drosophila homolog of the mammalian
CCAAT-displacement protein (CDP)/Cux, functions as a
transcriptional repressor of the dPCNA gene by binding to the
promoter region during cell differentiation (76). The dPCNA promoter was shown to be
activated by DREF and repressed by Cut via the measurement of dPCNA
promoter activity in transient luciferase expression assays.
Moreover, Cut and DREF were demonstrated to be localized in genomic
regions containing the dPCNA promoter, which initiates Cut-induced
differentiation, using chromatin immunoprecipitation assays. These
findings indicate that the DRE sequence binds to DREF during cell
proliferation, which causes the state of expression of the dPCNA
gene to become ‘on’. The dPCNA promoter recruits Cut to its region
when the cell is in a differentiated state. Cut then associates
with other factors to inhibit expression of dPCNA gene. Therefore,
we suggest that during cell proliferation, the
differentiation-coupled reduction could be linked to DREF, which is
a key regulatory factor for proliferation-associated genes.
ZBED1 as a transcription factor for cell
proliferation
To examine whether ZBED1 regulates human DNA
replication-related genes, Ohshima et al (9) assessed the histone H1 gene, the
expression of which is stringently coupled with DNA replication.
This gene has a single 10-bp sequence in its promoter region that
completely matches with the ZBED1 binding sequence (77,78).
Co-transfection experiments indicate that the activity of the human
histone H1 gene promoter is stimulated when it specifically binds
to ZBED1, and RNA interference experiments targeting ZBED1 indicate
that transcription of the histone H1 gene is likely under the
control of ZBED1 in the G1/S phase during the cell cycle
(9).
Notably, a study established that 22 of the 79 human
ribosomal protein (RP) genes have sequences similar to that of hDRE
in their transcriptional start sites <200 bp upstream. The study
also demonstrated that ZBED1 potentially binds to the hDRE-like
sequences in the RP genes in vivo and in vitro, and
the hDRE-like sequences function as positive elements for RP gene
transcription (79). In a similar
manner to ZBED1 expression, RP gene expression is enhanced in the
late G1 to S phases, whereas a reduction in the
expression of the RP gene occurs when ZBED1 is depleted, thus
impairing cell proliferation and the G1/S transition in
normal human fibroblasts. These findings indicate that ZBED1
significantly regulates cell proliferation at the transcriptional
level via the histone H1 and several RP genes. Also, ZBED1 has a
key role in the cell cycle-dependent regulation of these genes.
Furthermore, a study by Yamashita et al (80) explored the underlying transcriptional
regulation mechanism and demonstrated that ZBED1 has a slight
ubiquitin-like modifier (SUMO) ligase activity and may stimulate
transcriptional activation by specifically SUMOylating Mi2α. This
results in the dissociation of Mi2α from the gene loci, thus
maintaining active states of housekeeping target genes for
ZBED1.
Conclusions and future perspectives
The present review highlights that Drosophila
DREF has different structural features than its human ortholog
ZBED1. However, there are notable similarities between the two
species based on protein functions associated with, for example,
the regulation of DNA replication, chromatin structure, protein
synthesis and the cell cycle. Furthermore, ZBED1 potentially
participates in the regulation of the expression of housekeeping
genes within the mammalian genome. Also, the potential regulatory
sites of different genes for DREF binding are likely to be
conserved between Drosophila and humans. Therefore, numerous
other genes may be regulated similarly in humans via the ZBED1
pathway. However, further research is essential to identify
substrates of ZBED1 other than Mi2α in order to clarify the
underlying molecular mechanism and comprehensively analyze the
effect of ZBED1 in humans. Additionally, ZBED3 and ZBED6 are
associated with tumor development. A recent study has shown that
ZBED1 is upregulated in gastric cancer cells, thereby promoting
their proliferation and decreasing their chemosensitivity, although
the molecular mechanism requires full elucidation (81). Therefore, it is concluded that ZBED1
may be a novel cancer biomarker and therapeutic target in the
future. However, further investigation is required to verify
this.
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81402458), the Basic
Research Project of Shanxi Province (grant no. 2014021037-4), the
Graduate Program of Scientific Research Foundation of Yunnan
Provincial Department of Education (grant no. 2016YJS057) and the
Science and Technology Research Project of Colleges and
Universities of Hebei Province (grant no. QN2020234).
Availability of data and materials
Not applicable.
Authors' contributions
YJ, RL, XS and GZ conceived and designed the review.
YJ, ZZ, JR, XS and GZ were involved in the collection and collation
of references. YJ and ZZ drew the figures. YJ, RL and GZ wrote the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lander ES, Linton LM, Birren B, Nusbaum C,
Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al:
Initial sequencing and analysis of the human genome. Nature.
409:860–921. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Razin SV, Borunova VV, Maksimenko OG and
Kantidze OL: Cys2His2 zinc finger protein family: Classification,
functions, and major members. Biochemistry (Mosc). 77:217–226.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Markljung E, Jiang L, Jaffe JD, Mikkelsen
TS, Wallerman O, Larhammar M, Zhang X, Wang L, Saenz-Vash V, Gnirke
A, et al: ZBED6, a novel transcription factor derived from a
domesticated DNA transposon regulates IGF2 expression and muscle
growth. PLoS Biol. 7:e10002562009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Akhtar Ali M, Younis S, Wallerman O, Gupta
R, Andersson L and Sjöblom T: Transcriptional modulator ZBED6
affects cell cycle and growth of human colorectal cancer cells.
Proc Natl Acad Sci USA. 112:7743–7748. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen T, Li M, Ding Y, Zhang LS, Xi Y, Pan
WJ, Tao DL, Wang JY and Li L: Identification of zinc-finger BED
domain-containing 3 (Zbed3) as a novel Axin-interacting protein
that activates Wnt/beta-catenin signaling. J Biol Chem.
284:6683–6689. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fan C, Jiang G, Zhang X, Miao Y, Lin X,
Luan L, Xu Z, Zhang Y, Zhao H, Liu D, et al: Zbed3 contributes to
malignant phenotype of lung cancer via regulating β-catenin and
P120-catenin 1. Mol Carcinog. 1 (54 Suppl):E138–E147. 2015.
View Article : Google Scholar
|
|
7
|
Saghizadeh M, Akhmedov NB, Yamashita CK,
Gribanova Y, Theendakara V, Mendoza E, Nelson SF, Ljubimov AV and
Farber DB: ZBED4, a BED-type zinc-finger protein in the cones of
the human retina. Invest Ophthalmol Vis Sci. 50:3580–3588. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mokhonov VV, Theendakara VP, Gribanova YE,
Ahmedli NB and Farber DB: Sequence-specific binding of recombinant
Zbed4 to DNA: Insights into Zbed4 participation in gene
transcription and its association with other proteins. PLoS One.
7:e353172012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ohshima N, Takahashi M and Hirose F:
Identification of a human homologue of the DREF transcription
factor with a potential role in regulation of the histone H1 gene.
J Biol Chem. 278:22928–22938. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Deininger PL and Batzer MA: Mammalian
retroelements. Genome Res. 12:1455–1465. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sinzelle L, Izsvak Z and Ivics Z:
Molecular domestication of transposable elements: From detrimental
parasites to useful host genes. Cell Mol Life Sci. 66:1073–1093.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
van de Lagemaat LN, Landry JR, Mager DL
and Medstrand P: Transposable elements in mammals promote
regulatory variation and diversification of genes with specialized
functions. Trends Genet. 19:530–536. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jordan IK, Rogozin IB, Glazko GV and
Koonin EV: Origin of a substantial fraction of human regulatory
sequences from transposable elements. Trends Genet. 19:68–72. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kashkush K, Feldman M and Levy AA:
Transcriptional activation of retrotransposons alters the
expression of adjacent genes in wheat. Nat Genet. 33:102–106. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feschotte C and Pritham EJ: DNA
transposons and the evolution of eukaryotic genomes. Annu Rev
Genet. 41:331–368. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Volff JN: Turning junk into gold:
Domestication of transposable elements and the creation of new
genes in eukaryotes. Bioessays. 28:913–922. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arensburger P, Hice RH, Zhou L, Smith RC,
Tom AC, Wright JA, Knapp J, O'Brochta DA, Craig NL and Atkinson PW:
Phylogenetic and functional characterization of the hAT transposon
superfamily. Genetics. 188:45–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hayward A, Ghazal A, Andersson G,
Andersson L and Jern P: ZBED evolution: Repeated utilization of DNA
transposons as regulators of diverse host functions. PLoS One.
8:e599402013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hirose F, Yamaguchi M, Nishida Y, Masutani
M, Miyazawa H, Hanaoka F and Matsukage A: Structure and expression
during development of Drosophila melanogaster gene for DNA
polymerase alpha. Nucleic Acids Res. 19:4991–4998. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hirose F, Yamaguchi M, Handa H, Inomata Y
and Matsukage A: Novel 8-base pair sequence (Drosophila DNA
replication-related element) and specific binding factor involved
in the expression of Drosophila genes for DNA polymerase alpha and
proliferating cell nuclear antigen. J Biol Chem. 268:2092–2099.
1993.PubMed/NCBI
|
|
21
|
Hirose F, Yamaguchi M, Kuroda K, Omori A,
Hachiya T, Ikeda M, Nishimoto Y and Matsukage A: Isolation and
characterization of cDNA for DREF, a promoter-activating factor for
Drosophila DNA replication-related genes. J Biol Chem.
271:3930–3937. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Takahashi Y, Hirose F, Matsukage A and
Yamaguchi M: Identification of three conserved regions in the DREF
transcription factors from Drosophila melanogaster and Drosophila
virilis. Nucleic Acids Res. 27:510–516. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smit AF: Interspersed repeats and other
mementos of transposable elements in mammalian genomes. Curr Opin
Genet Dev. 9:657–663. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Aravind L: The BED finger, a novel
DNA-binding domain in chromatin-boundary-element-binding proteins
and transposases. Trends Biochem Sci. 25:421–423. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hart CM, Cuvier O and Laemmli UK: Evidence
for an antagonistic relationship between the boundary
element-associated factor BEAF and the transcription factor DREF.
Chromosoma. 108:375–383. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamashita D, Komori H, Higuchi Y,
Yamaguchi T, Osumi T and Hirose F: Human DNA replication-related
element binding factor (hDREF) self-association via hATC domain is
necessary for its nuclear accumulation and DNA binding. J Biol
Chem. 282:7563–7575. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Essers L, Adolphs RH and Kunze R: A highly
conserved domain of the maize activator transposase is involved in
dimerization. Plant Cell. 12:211–224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Michel K, O'Brochta DA and Atkinson PW:
The C-terminus of the Hermes transposase contains a protein
multimerization domain. Insect Biochem Mol Biol. 33:959–970. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matsukage A, Hirose F, Yoo MA and
Yamaguchi M: The DRE/DREF transcriptional regulatory system: A
master key for cell proliferation. Biochim Biophys Acta.
1779:81–89. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tue NT, Yoshioka Y, Mizoguchi M, Yoshida
H, Zurita M and Yamaguchi M: DREF plays multiple roles during
Drosophila development. Biochim Biophys Acta Gene Regul Mech.
1860:705–712. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matsukage A, Hirose F, Hayashi Y, Hamada K
and Yamaguchi M: The DRE sequence TATCGATA, a putative
promoter-activating element for Drosophila melanogaster
cell-proliferation-related genes. Gene. 166:233–236. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sawado T, Hirose F, Takahashi Y, Sasaki T,
Shinomiya T, Sakaguchi K, Matsukage A and Yamaguchi M: The DNA
replication-related element (DRE)/DRE-binding factor system is a
transcriptional regulator of the Drosophila E2F gene. J Biol Chem.
273:26042–26051. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ohno K, Hirose F, Sakaguchi K, Nishida Y
and Matsukage A: Transcriptional regulation of the Drosophila CycA
gene by the DNA replication-related element (DRE) and DRE binding
factor (DREF). Nucleic Acids Res. 24:3942–3946. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takahashi Y, Yamaguchi M, Hirose F,
Cotterill S, Kobayashi J, Miyajima S and Matsukage A: DNA
replication-related elements cooperate to enhance promoter activity
of the drosophila DNA polymerase alpha 73-kDa subunit gene. J Biol
Chem. 271:14541–14547. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ryu JR, Choi TY, Kwon EJ, Lee WH, Nishida
Y, Hayashi Y, Matsukage A, Yamaguchi M and Yoo MA: Transcriptional
regulation of the Drosophila-raf proto-oncogene by the DNA
replication-related element (DRE)/DRE-binding factor (DREF) system.
Nucleic Acids Res. 25:794–799. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zabidi MA, Arnold CD, Schernhuber K,
Pagani M, Rath M, Frank O and Stark A: Enhancer-core-promoter
specificity separates developmental and housekeeping gene
regulation. Nature. 518:556–559. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Bird AP: CpG-rich islands and the function
of DNA methylation. Nature. 321:209–213. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Argentaro A, Yang JC, Chapman L, Kowalczyk
MS, Gibbons RJ, Higgs DR, Neuhaus D and Rhodes D: Structural
consequences of disease-causing mutations in the ATRX-DNMT3-DNMT3L
(ADD) domain of the chromatin-associated protein ATRX. Proc Natl
Acad Sci USA. 104:11939–11944. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Valadez-Graham V, Yoshioka Y, Velazquez O,
Kawamori A, Vazquez M, Neumann A, Yamaguchi M and Zurita M:
XNP/dATRX interacts with DREF in the chromatin to regulate gene
expression. Nucleic Acids Res. 40:1460–1474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hart CM, Zhao K and Laemmli UK: The scs'
boundary element: Characterization of boundary element-associated
factors. Mol Cell Biol. 17:999–1009. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Royzman I, Whittaker AJ and Orr-Weaver TL:
Mutations in Drosophila DP and E2F distinguish G1-S progression
from an associated transcriptional program. Genes Dev.
11:1999–2011. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Brehm A, Langst G, Kehle J, Clapier CR,
Imhof A, Eberharter A, Muller J and Becker PB: dMi-2 and ISWI
chromatin remodelling factors have distinct nucleosome binding and
mobilization properties. EMBO J. 19:4332–4341. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hirose F, Ohshima N, Kwon EJ, Yoshida H
and Yamaguchi M: Drosophila Mi-2 negatively regulates dDREF by
inhibiting its DNA-binding activity. Mol Cell Biol. 22:5182–5193.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ida H, Yoshida H, Nakamura K and Yamaguchi
M: Identification of the Drosophila eIF4A gene as a target of the
DREF transcription factor. Exp Cell Res. 313:4208–4220. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Linder P, Lasko PF, Ashburner M, Leroy P,
Nielsen PJ, Nishi K, Schnier J and Slonimski PP: Birth of the
D-E-A-D box. Nature. 337:121–122. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Blum S, Schmid SR, Pause A, Buser P,
Linder P, Sonenberg N and Trachsel H: ATP hydrolysis by initiation
factor 4A is required for translation initiation in Saccharomyces
cerevisiae. Proc Natl Acad Sci USA. 89:7664–7668. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pestova TV, Kolupaeva VG, Lomakin IB,
Pilipenko EV, Shatsky IN, Agol VI and Hellen CU: Molecular
mechanisms of translation initiation in eukaryotes. Proc Natl Acad
Sci USA. 98:7029–7036. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Pestova TV, Shatsky IN and Hellen CU:
Functional dissection of eukaryotic initiation factor 4F: The 4A
subunit and the central domain of the 4G subunit are sufficient to
mediate internal entry of 43S preinitiation complexes. Mol Cell
Biol. 16:6870–6878. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Phuong Thao DT, Ida H, Yoshida H and
Yamaguchi M: Identification of the Drosophila skpA gene as a novel
target of the transcription factor DREF. Exp Cell Res.
312:3641–3650. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Seol JH, Shevchenko A, Shevchenko A and
Deshaies RJ: Skp1 forms multiple protein complexes, including RAVE,
a regulator of V-ATPase assembly. Nat Cell Biol. 3:384–391. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tsuchiya A, Inoue YH, Ida H, Kawase Y,
Okudaira K, Ohno K, Yoshida H and Yamaguchi M: Transcriptional
regulation of the Drosophila rfc1 gene by the DRE-DREF pathway.
FEBS J. 274:1818–1832. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sharkov NV, Ramsay G and Katzen AL: The
DNA replication-related element-binding factor (DREF) is a
transcriptional regulator of the Drosophila myb gene. Gene.
297:209–219. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Nakamura K, Ida H and Yamaguchi M:
Transcriptional regulation of the Drosophila moira and osa genes by
the DREF pathway. Nucleic Acids Res. 36:3905–3915. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mohrmann L and Verrijzer CP: Composition
and functional specificity of SWI2/SNF2 class chromatin remodeling
complexes. Biochim Biophys Acta. 1681:59–73. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bouazoune K and Brehm A: ATP-dependent
chromatin remodeling complexes in Drosophila. Chromosome Res.
14:433–449. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
He M, Zhou Z, Shah AA, Hong Y, Chen Q and
Wan Y: New insights into posttranslational modifications of Hippo
pathway in carcinogenesis and therapeutics. Cell Div. 11:42016.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Meng Z, Moroishi T and Guan KL: Mechanisms
of Hippo pathway regulation. Genes Dev. 30:1–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ye S and Eisinger-Mathason TS: Targeting
the Hippo pathway: Clinical implications and therapeutics.
Pharmacol Res. 103:270–278. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Vo N, Horii T, Yanai H, Yoshida H and
Yamaguchi M: The Hippo pathway as a target of the Drosophila
DRE/DREF transcriptional regulatory pathway. Sci Rep. 4:71962014.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Fujiwara S, Ida H, Yoshioka Y, Yoshida H
and Yamaguchi M: The warts gene as a novel target of the Drosophila
DRE/DREF transcription pathway. Am J Cancer Res. 2:36–44.
2012.PubMed/NCBI
|
|
61
|
Yanai H, Yoshioka Y, Yoshida H, Nakao Y,
Plessis A and Yamaguchi M: Drosophila myeloid leukemia factor acts
with DREF to activate the JNK signaling pathway. Oncogenesis.
3:e982014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yoshioka Y, Nguyen TT, Fujiwara S, Matsuda
R, Valadez-Graham V, Zurita M and Yamaguchi M: Drosophila DREF
acting via the JNK pathway is required for thorax development.
Genesis. 50:599–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Trong-Tue N, Thao DT and Yamaguchi M: Role
of DREF in transcriptional regulation of the Drosophila p53 gene.
Oncogene. 29:2060–2069. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Brodsky MH, Nordstrom W, Tsang G, Kwan E,
Rubin GM and Abrams JM: Drosophila p53 binds a damage response
element at the reaper locus. Cell. 101:103–113. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jin S, Martinek S, Joo WS, Wortman JR,
Mirkovic N, Sali A, Yandell MD, Pavletich NP, Young MW and Levine
AJ: Identification and characterization of a p53 homologue in
Drosophila melanogaster. Proc Natl Acad Sci USA. 97:7301–7306.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Ollmann M, Young LM, Di Como CJ, Karim F,
Belvin M, Robertson S, Whittaker K, Demsky M, Fisher WW, Buchman A,
et al: Drosophila p53 is a structural and functional homolog of the
tumor suppressor p53. Cell. 101:91–101. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Nicolai S, Rossi A, Di Daniele N, Melino
G, Annicchiarico-Petruzzelli M and Raschella G: DNA repair and
aging: The impact of the p53 family. Aging (Albany NY).
7:1050–1065. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Kim S and An SS: Role of p53 isoforms and
aggregations in cancer. Medicine (Baltimore). 95:e39932016.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Chen J: The Cell-cycle arrest and
apoptotic functions of p53 in tumor initiation and progression.
Cold Spring Harb Perspect Med. 6:a0261042016. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Green DR and Evan GI: A matter of life and
death. Cancer Cell. 1:19–30. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wu S, Huang J, Dong J and Pan D: hippo
encodes a Ste-20 family protein kinase that restricts cell
proliferation and promotes apoptosis in conjunction with salvador
and warts. Cell. 114:445–456. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Matsukage A, Hirose F and Yamaguchi M:
Transcriptional regulation of DNA replication-related genes in cell
growth, differentiation and oncogenesis. Jpn J Cancer Res. 85:1–8.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Doyle HJ, Kraut R and Levine M: Spatial
regulation of zerknullt: A dorsal-ventral patterning gene in
Drosophila. Genes Dev. 3:1518–1533. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Hirose F, Yamaguchi M and Matsukage A:
Repression of regulatory factor for Drosophila DNA
replication-related gene promoters by zerknullt homeodomain
protein. J Biol Chem. 269:2937–2942. 1994.PubMed/NCBI
|
|
75
|
Hayashi Y, Kato M, Seto H and Yamaguchi M:
Drosophila distal-less negatively regulates dDREF by inhibiting its
DNA binding activity. Biochim Biophys Acta. 1759:359–366. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Seto H, Hayashi Y, Kwon E, Taguchi O and
Yamaguchi M: Antagonistic regulation of the Drosophila PCNA gene
promoter by DREF and Cut. Genes Cells. 11:499–512. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
van Wijnen AJ, Wright KL, Massung RF,
Gerretsen M, Stein JL and Stein GS: Two target sites for protein
binding in the promoter region of a cell cycle regulated human H1
histone gene. Nucleic Acids Res. 16:571–592. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Albig W, Meergans T and Doenecke D:
Characterization of the H1.5 gene completes the set of human H1
subtype genes. Gene. 184:141–148. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yamashita D, Sano Y, Adachi Y, Okamoto Y,
Osada H, Takahashi T, Yamaguchi T, Osumi T and Hirose F: hDREF
regulates cell proliferation and expression of ribosomal protein
genes. Mol Cell Biol. 27:2003–2013. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Yamashita D, Moriuchi T, Osumi T and
Hirose F: Transcription Factor hDREF Is a Novel SUMO E3 Ligase of
Mi2α. J Biol Chem. 291:11619–11634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Jiang S, Wang Y, Xiong Y, Feng Y, Tang J
and Song R: High expression of ZBED1 affects proliferation and
apoptosis in gastric cancer. Int J Clin Exp Pathol. 11:4019–4025.
2018.PubMed/NCBI
|