Introduction
Liver cancer is one of the most aggressive
malignancies and is responsible for ~830,000 deaths worldwide in
2020 (1). The diagnosis of this
disease is typically made when the liver cancer is already at an
advanced stage, rendering it unresectable since it has already
spread. Therefore, surgical options are limited. In such cases,
chemotherapy is the only viable option, but drug resistance remains
the main hindrance to its efficacy (2). Since the property of cancer stemness
is closely associated with tumor initiation, self-renewal and
differentiation into bulk tumor cells, it has been proposed to be a
target for cancer treatment (3).
Accumulating evidence supports the notion that the stemness trait
of tumors is fundamentally responsible for cancer metastasis,
recurrence and chemoresistance (4,5).
Various survival pathways, such as the Wnt/β-catenin and STAT3
signaling pathways, have been documented to be activated in liver
cancer cells expressing CD133 (a stem cell marker), which impart
resistance to chemotherapy (6). Kim
et al (7) previously
demonstrated that cancer stem cell subpopulations serve a role in
chemotherapy resistance in liver cancer by increasing plasticity,
an ability that allows cells to maintain stability through changes
in their environment, which is associated with cellular ‘stemness’.
Therefore, understanding the mechanisms underlying cancer stemness
and chemotherapy resistance is important for providing insights
into the development of effective and prospective therapeutic
strategies against liver cancer.
Over the past decades, microRNAs (miR or miRNAs)
have been widely studied as potential mediators of numerous human
diseases, including uremia and cancer (8). As a type of non-coding RNA that are
18–25 nucleotides length, miRNAs can suppress the transcription and
subsequently the translation of target mRNAs by binding to its 3′
untranslated region, which in turn lead to changes in the
physiological and pathophysiological processes downstream, such as
the cell cycle, differentiation and autophagy (9). miRNAs, such as miR-221, miR-106b and
miR-21, have been demonstrated to be oncogenic miRNAs that can
regulate the malignant behaviors of tumor cells to participate in
the development and growth of liver tumors (10–12).
However, numerous downregulated miRNAs have also been identified to
be tumor suppressors in liver cancer, including miR-122 (13). miR-122 is the predominant miRNA
expressed in the liver and accounts for >50% of the total
hepatic miRNA content in adult humans (14). It has been implicated in the
regulation of various biological processes in the liver, including
hepatic inflammation and lipid metabolism (15,16).
The downregulation of miR-122 expression has been reported to serve
an oncogenic role in the liver (17). Xu et al (18) previously found that circulating
miR-122 levels are significantly associated with the overall
survival rate of patients with liver cancer, suggesting that
miR-122 can be applied as a reliable prognostic marker in liver
cancer. It has also been reported that circulating levels of
miR-122 can be an indicator of the response to transarterial
chemoembolization treatment in a patient with liver cancer
(19). However, the mechanisms by
which the downregulation of miR-122 can induce liver cancer remain
largely unclear.
Small ubiquitin-like modifier (SUMO) protein
isoforms can be reversibly linked to lysine residues that reside
within specific motifs in thousands of target substrates, leading
to alterations in stability, solubility, localization and
interaction profile (20).
SUMOylation has been previously reported to be a key form of
post-translational modification involved in liver cancer
progression (21,22). Therefore, in the present study,
bioinformatics online tools were applied to identify the potential
target genes of miR-122. The SUMOylation-related genes were
selected to further analyze the respective roles and underlying
mechanisms as well as miR-122 in liver cancer cell stemness and
chemoresistance.
Materials and methods
Bioinformatics analysis
The encyclopedia of RNA interactomes (ENCORI;
http://rnasysu.com/encori/), previously
known as StarBase (version 2.0) (23), was utilized to predict the potential
target genes of miR-122. Briefly, in the query page of miRNA-mRNA
interactions in miRNA-Target module, miR-122 was selected in order
to browse all miR-122-target interactions. Among the potential
targets, SUMOylation-related genes (SENP1, SENP2, SUMO1 and SUMO3)
were selected to analyze their correlation with miR-122 in liver
cancer based on the pan-cancer platform in ENCORI. Next, SENP1 was
selected for further analysis, as it has the strongest negative
correlation with miR-122 among the four selected genes.
Cell culture and transfections
The human liver cancer cell line HepG2 was purchased
from American Type Culture Collection and was maintained in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) containing 10% FBS (Gibco)
and 1% penicillin/streptomycin in a humidified environment of 5%
CO2 at 37°C. After cell confluence reached 80–90%, the
cultured cells were digested with 0.25% trypsin (w/v) and
sub-cultured at a ratio of 1:4. All cell lines were tested with
MycoAlert® Mycoplasma Detection Kit (Lonza Group, Ltd.)
every 3 months. Cell line verification was performed using single
tandem repeats profiling before the initiation of the present
study. HepG2 cells were treated with 10 µM proteasome inhibitors
MG132 (MilliporeSigma) for 6 h before the in vitro
SUMOylation assay.
Cells were divided into the following four groups
after transfection: i) Negative control (NC) group, which was
transfected with NC agents, namely NC mimics and/or empty vectors
(pcDNA3.1 vector; Invitrogen); ii) miR-122 mimic group; iii) SENP1
overexpression group (the pcDNA3.1 vector subcloned SENP1 cDNA
fragment: Forward, 5′-AAGAAGATCTTATGGATGATATTGCTGATAGGATGAGG-3′ and
reverse, 5′-GCCCGTCGAACTCATCACAAGAGTTTTCGGTGGAG-3′; accession no.
AF149770); and iv) miR-122 mimic + SENP1 overexpression group,
which was co-transfected with the miR-122 mimic and the SENP1
overexpression vector. miR-122 mimic (5′-CAAACACCAUUGUCACACUCCA-3′)
and NC mimic (5′-CAGUACUUUUGUGUAGUACAA-3′)
(pGCMV/EGFP/miR/Blasticidin plasmid backbone, cat. no. C09002) were
purchased from Shanghai GenePharma Co., Ltd. Non-transfected cells
were defined as the control group. In addition, HepG2 cells were
also subjected to the transfection of FLAG-SENP1 (pFlag-CMV plasmid
backbone; Addgene) and Myc-β-catenin (pSB1C3 plasmid backbone;
Addgene) in order to investigate the interaction between SENP1 and
β-catenin. In brief, cells were seeded into six-well plates and
transiently transfected with the aforementioned plasmids (empty
vector and SENP1 overexpression vector, 2 µg; FLAG-SENP1 and
Myc-β-catenin, 4 µg; miR-122 mimic and NC mimics, 50 nM; Agomir-122
and Agomir NC, 50 nM) using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 48 h according to the
manufacturer's protocol. After 48 h, transfection efficiency was
determined by reverse transcription quantitative PCR (RT-qPCR).
Subsequent experiments were performed 48 h after transfection.
RT-qPCR
The total RNA extraction from the cells with using
TRIzol® (Thermo Fisher Scientific, Inc.) and subsequent
reverse transcription with the PrimeScript RT reagent (cat. no.
RR037B; Takara Bio, Inc.) was performed according to the
manufacturer's instructions. Afterwards, qPCR was performed using
SYBR green reagent (Takara Bio, Inc.) on the 7500 real-time PCR
system. The thermocycling conditions were as follows: Initial
denaturation at 95°C for 45 sec, followed by 40 cycles of 95°C for
15 sec and 60°C for 15 sec. The present study utilized U6 and GAPDH
as the internal reference genes to quantify miRNA and mRNA
expression, respectively, using the 2−ΔΔCq method
(24). Primer sequences used in the
present study are listed in Table
I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Sequence
(5′-3′) |
|---|
| MicroRNA-122 | Forward,
ACAGTGGAGTGTGACAATG |
|
| Reverse,
TCCAGTTTTTTTTTTTTTTTCAAACAC |
| Sentrin-specific
protease 1 | Forward,
TTGGCCAGAGTGCAAATGG |
|
| Reverse,
TCGGCTGTTTCTTGATTTTTGTAA |
| Oct3/4 | Forward,
CTTGCTGCAGAAGTGGGTGGAGGAA |
|
| Reverse,
CTGCAGTGTGGGTTTCGGGCA |
| Nanog | Forward,
AATACCTCAGCCTCCAGCAGATG |
|
| Reverse,
TGCGTCACACCATTGCTATTCTTC |
| B lymphoma Mo-MLV
insertion region 1 homolog | Forward,
TGGAGAAGGAATGGTCCACTTC |
|
| Reverse,
GTGAGGAAACTGTGGATGAGGA |
| Notch | Forward,
CCTGAGGGCTTCAAAGTGTC |
|
| Reverse
CGGAACTTCTTGGTCTCCAG |
| U6 | Forward,
CTCGCTTCGGCAGCACA |
|
| Reverse,
AACGCTTCACGAATTTGCGT |
| GAPDH | Forward,
GAGTCAACGGATTTGGTCGT |
|
| Reverse,
TTGATTTTGGAGGGATCTC |
Dual-luciferase reporter assay
The dual-luciferase reporter assay was performed to
verify if miR-122 can directly bind to the SENP1 mRNA 3′
untranslated region (3′ UTR). Partial sequences of the SENP1 3′UTR
possessing the wild-type (WT) or mutant (MUT) miR-122 targeting
site were cloned into the luciferase reporter pmirGLO vector
(Promega Corporation) to construct the SENP1 WT and SENP1 MUT
plasmids. After constructing the indicated plasmids, HepG2 cells
were co-transfected with the SENP1 WT or SENP1 MUT plasmid and the
miR-122 mimic or NC mimic using Lipofectamine 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C. The final
concentration of the miR-122 mimic or NC mimic was 50 nM, while
that of SENP1 WT or SENP1 MUT plasmid was 2 µg. After 48 h, the
luciferase activity was assessed using the dual-luciferase reporter
assay kit (Promega Corporation; cat. no. E1960). Firefly luciferase
activity was normalized to Renilla luciferase activity.
Western blotting
Transfected cells were lysed with the RIPA buffer
(Beyotime Institute of Biotechnology) containing PMSF and a
protease inhibitor on ice for 20 min to acquire the total proteins.
After determining the concentration of the total proteins using a
bicinchoninic acid kit (Thermo Fisher Scientific, Inc.), equivalent
amounts of protein (30 µg protein/lane) were separated on 10% gels
using SDS-PAGE and transferred onto PVDF membranes. Thereafter, the
membranes were blocked with skimmed milk (5%) at room temperature
for 1 h, followed by incubation with the primary antibodies at 4°C
overnight. The membranes were then rinsed twice with TBS with 0.1%
Tween 20 prior to incubating with the secondary antibody at room
temperature for 1 h. Finally, using an enhanced chemiluminescent
substrate kit (Thermo Fisher Scientific, Inc.; cat. no. 34577), the
protein bands were visualized and the intensities were measured
using the Image J 6.0 software (National Institutes of Health).
Protein expression levels for each sample were normalized to GAPDH.
Detailed information regarding the antibodies used in the present
study is listed in Table II.
 | Table II.Detailed information of the
antibodies used in western blotting. |
Table II.
Detailed information of the
antibodies used in western blotting.
| Antibody | Manufacturer | Cat. no. | Dilution |
|---|
| Sentrin-specific
protease 1 | ProteinTech Group,
Inc. | 25349-1-AP | 1:2,000 |
| Multidrug
resistance protein | Beyotime Institute
of Biotechnology | AF7503 | 1:1,000 |
| P-glycoprotein | Beyotime Institute
of Biotechnology | AF2245 | 1:1,000 |
| Wnt1 | Beyotime Institute
of Biotechnology | AF8349 | 1:2,000 |
| β-catenin | Beyotime Institute
of Biotechnology | AC106 | 1:1,000 |
| Flag | Sigma-Aldrich | F2555 | 1:250 |
| Myc | Thermo Fisher
Scientific, Inc. | PA1-981 | 1:2,000 |
| GAPDH | Beyotime Institute
of Biotechnology | AF1186 | 1:1,000 |
| Goat anti-rabbit
IgG H&L HRP conjugate | Thermo Fisher
Scientific, Inc. | 31460 | 1:15,000 |
Sphere forming assay
A total of 1,000 cells per well were plated into
six-well ultra-low attachment plates and grown in DMEM/F12 medium
(Thermo Fisher Scientific, Inc.) supplemented with 5 µg/ml insulin
(MilliporeSigma), 20 ng/ml basic fibroblast growth factor
(Invitrogen; Thermo Fisher Scientific, Inc.) and 20 ng/ml epidermal
growth factor (Invitrogen; Thermo Fisher Scientific, Inc.). The
plates were then incubated at 37°C for 15 days. The total numbers
of spheroids with a diameter >50 µm in each well were then
counted using a light microscope at ×100 magnification.
Flow cytometry
Anti-CD24 conjugated to phycoerythrin (PE) (5
µl/test; Thermo Fisher Scientific, Inc.; cat. no 12-0247-42) was
used for the present study. After incubating in PBS with 2% FBS
followed by CD24-PE antibodies at room temperature in the dark for
30 min, the labeled HepG2 cells (1×106 cells per aliquot
of incubation) were analyzed using the BD FACSCanto II analyzer
flow cytometer (BD Biosciences), and the results were analyzed
using FlowJo software (version 10.8.1; FlowJo LLC).
Cell Counting kit 8 (CCK-8) assay
The transfected cells were seeded into 96-well
plates at a density of 8×103 cells/well after being
digested with 0.25% trypsin-EDTA solution (MilliporeSigma) and
resuspended. After complete adherence (24 h of incubation), the
cells were treated with DMEM containing various concentrations of
doxorubicin (DOX) (0, 5, 15 and 30 µM) or sorafenib (0, 10, 20 and
40 µM) for 24 h. The cells that had 0.1% DMSO added were used as
the experimental control, whereas the wells containing only DMEM
were used as the blank group. After 24 h incubation at 37°C, the
medium was replaced with the CCK-8 reagent (10%, v/v, dissolved in
DMEM; Dojindo Molecular Technologies, Inc.). After incubating for 2
h at 37°C, the absorbance was detected using a microplate reader at
450 nm.
Colony formation assay
The transfected cells were seeded into six-well
plates at a density of 2.5×102 cells/well. After
cultivating for 2 weeks at 37°C, the cell colonies (>50 cells)
were fixed with 4% paraformaldehyde for 10 min at room temperature
and stained with 1% crystal violet for 10 min at room temperature.
The dishes were then gently washed, photographed and counted using
a BX51 fluorescence microscope at ×40 magnification (Olympus,
Corporation).
Transwell assay
The migration of transfected cells was evaluated
using Transwell chambers (8-µm pore size; Corning, Inc.). Briefly,
5×104 transfected cells were seeded into the upper
chambers containing 200 µl DMEM without FBS. Simultaneously, 700 µl
DMEM containing FBS was added to the lower chamber. After
incubation for 24 h at 37°C, the cells remaining in the upper
chamber were wiped using a cotton swab, before the cells that
traversed the membrane to the lower chamber were fixed in 4%
paraformaldehyde for 10 min at room temperature and stained with
0.1% crystal violet for 15 min at room temperature. The stained
cells were imaged and counted using a BX51 fluorescence
microscope.
Co-immunoprecipitation (Co-IP)
Transfected cells were lysed with 200 µl of RIPA
buffer (Beyotime Institute of Biotechnology) containing PMSF and a
protease inhibitor. The lysate (800 µl) was subsequently incubated
with 2 µg of antibody against SENP1 (ProteinTech Group, Inc.; cat.
no 25349-1-AP), β-catenin (Beyotime Institute of Biotechnology;
cat. no. AC106), Flag (MilliporeSigma; cat. no. F2555) or IgG
(Abcam; cat. no. ab6715) with gentle rotation overnight at 4°C,
before being subsequently incubated with Pierce™ Protein
A/G Magnetic Agarose Beads (20 µl; Thermo Fisher Scientific, Inc.;
cat. no. 78609) for 2 h. The beads coupling with the
immune-complexes were centrifuged for 3 min at 4°C and 200 × g to
sink the agarose bead to the bottom of the tube. The supernatant
was removed carefully, and the agarose beads were washed with lysis
buffer before the proteins were eluted in SDS-PAGE buffer with
centrifugation at 1,000 × g for 1 min at room temperature.
Thereafter, eluted proteins were separated on 10% gels using
SDS-PAGE. The interacting proteins were detected by western blot
analysis.
In vitro SUMOylation assay
In vitro SUMOylation assays were conducted
using the SUMOylation kit (Enzo Life Sciences, Inc.; cat. no.
BML-UW8955) as per the manufacturer's protocols. In brief, the
reaction was performed using β-catenin with a reaction mixture
containing SUMO protein, SUMOylation enzyme, SUMOylation buffer and
Mg-ATP for 1 h at 30°C as per kit protocol. After the incubation,
protein SUMOylation was identified by immunoblotting using the
anti-SUMO1 antibody (1:1,000 dilution) provided with the kit. The
Goat anti-rabbit IgG H&L HRP conjugate secondary antibody
(1:15,000 dilution; Thermo Fisher Scientific, Inc.; cat. no. 31460)
was exploited in this assay at room temperature for 1 h.
Cycloheximide (CHX) chase assay
After 48 h post-transfection, CHX (20 µg/ml;
Beyotime Institute of Biotechnology) was added to the cell medium
for incubation at 37°C. At the designated time points (0, 2, 4 and
8 h), cells were collected and lysed with RIPA buffer (Beyotime
Institute of Biotechnology), and the protein levels of β-catenin
were detected by western blot analysis.
In vivo study
All procedures regarding animals in the present
study were approved by the Institutional Animal Care and Use
Committee at the Nan'an District People's Hospital of Chongqing
(approval no. 2020-0925; Chongqing, China) and conducted in
accordance with the AVMA guidelines. A total of 84 BALB/c nude mice
(female, 6–8 weeks old, 18–22 g) from the Animal Laboratory of
Chongqing Medical University were maintained under
specific-pathogen-free conditions with a 12/12 h light/dark cycle,
40–60% humidity, 24–26°C temperature conditions and free access to
food and water, and allowed to acclimatize for 1 week before they
were subjected to subsequent experiments.
Initially, HepG2 cells were stably transfected with
either 50 nM Agomir NC (5′-UUUGUACUACACAAAAGUACUG-3′) or 50 nM
Agomir-122 (5′-UGGAGUGUGACAAUGGUGUUUG-3′), synthesized by Guangzhou
RiboBio Co., Ltd., using Lipofectamine 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) before being subcutaneously injected
(suspended in PBS) into the mice. To determine the in vivo
tumor-initiating capacity of miR-122, four dilutions of HepG2 cells
with either Agomir NC or Agomir-122 (500, 5,000, 50,000, or 500,000
cells) in PBS were subcutaneously injected into the mice (eight
groups, n=8/group) and allowed to grow for 4 weeks. Extreme
limiting dilution analysis (ELDA) software version 5.6.1.5980
(https://bioinf.wehi.edu.au/software/elda/) (25) was utilized to calculate the
tumor-initiating cell frequency.
The remaining mice were randomly divided into the
following four groups (n=5/group): i) Agomir NC + saline; ii)
Agomir NC + DOX; iii) Agomir-122 + saline; and iv) Agomir-122 + DOX
groups. Mice from both the Agomir NC + saline and Agomir NC + DOX
groups were injected with 3×104 Agomir NC transfected
cells before being exposed to saline or 1 mg/kg DOX twice a week in
accordance with their group. Mice from both the Agomir-122 + saline
and Agomir-122 + DOX groups were injected with 3×104
miR-122-overexpressing HepG2 cells and received saline or 1 mg/kg
DOX twice a week in accordance with their group. Tumor formation
monitoring began on day 7, and tumor growth was checked every 3
days with caliper measurements. Tumor volume was calculated
according to the following formula: Volume=(L × W2)/2,
where W represents the width and L represents the length. On day
25, after anesthesia with 1% sodium pentobarbital intraperitoneal
injection (30 mg/kg), all mice were sacrificed by cervical
dislocation and tumors were harvested for protein extraction. A
comprehensive judgment on death was made by observing signs of
respiration, heartbeat and pupil and nerve reflexes.
Statistical analysis
All statistical analysis was conducted using the
GraphPad Prism 8.0.1 software (GraphPad Software, Inc.; Dotmatics).
All experiments were repeated at least three times, and all data
were presented as mean ± standard deviation. Students' unpaired
t-test or one-way analysis of variance followed by Bonferroni
post-hoc test was performed to analyze the difference between the
groups in the present study. RNA expression correlations were
analyzed by Pearson's correlation coefficients based on ENCORI
online database. ELDA online software was used to calculate the
cancer cell stem frequency and statistical significance was
assessed using the χ2 test. P<0.05 was considered to
indicate a statistically significant difference.
Results
SENP1 is predicted and confirmed as a
direct target of miR-122
An increasing number of studies support the key
regulatory role of miR-122 in the progression of liver cancer
(17–19). However, the mechanism underlying the
regulation of miR-122 in liver cancer is still not fully revealed.
Considering that SUMOylation has been shown to play a crucial role
in various processes of liver cancer (21,26,27),
we speculate whether the regulatory role of miR-122 in liver cancer
is partly related to SUMOylation. Based on the ENCORI database,
1,278 potential target genes of miR-122 were predicted. To explore
the relationship between miR-122 and SUMOylation in liver cancer,
SUMOylation-related genes (SENP1-7 and SUMO1-5) were selected for
subsequent analysis, and four SUMOylation-related genes, namely
SENP1, SENP2, SUMO1 and SUMO3, were predicted as the potential
targets of miR-122. Among these four targets, SENP1 was found to be
the gene with expression levels most correlated to miR-122
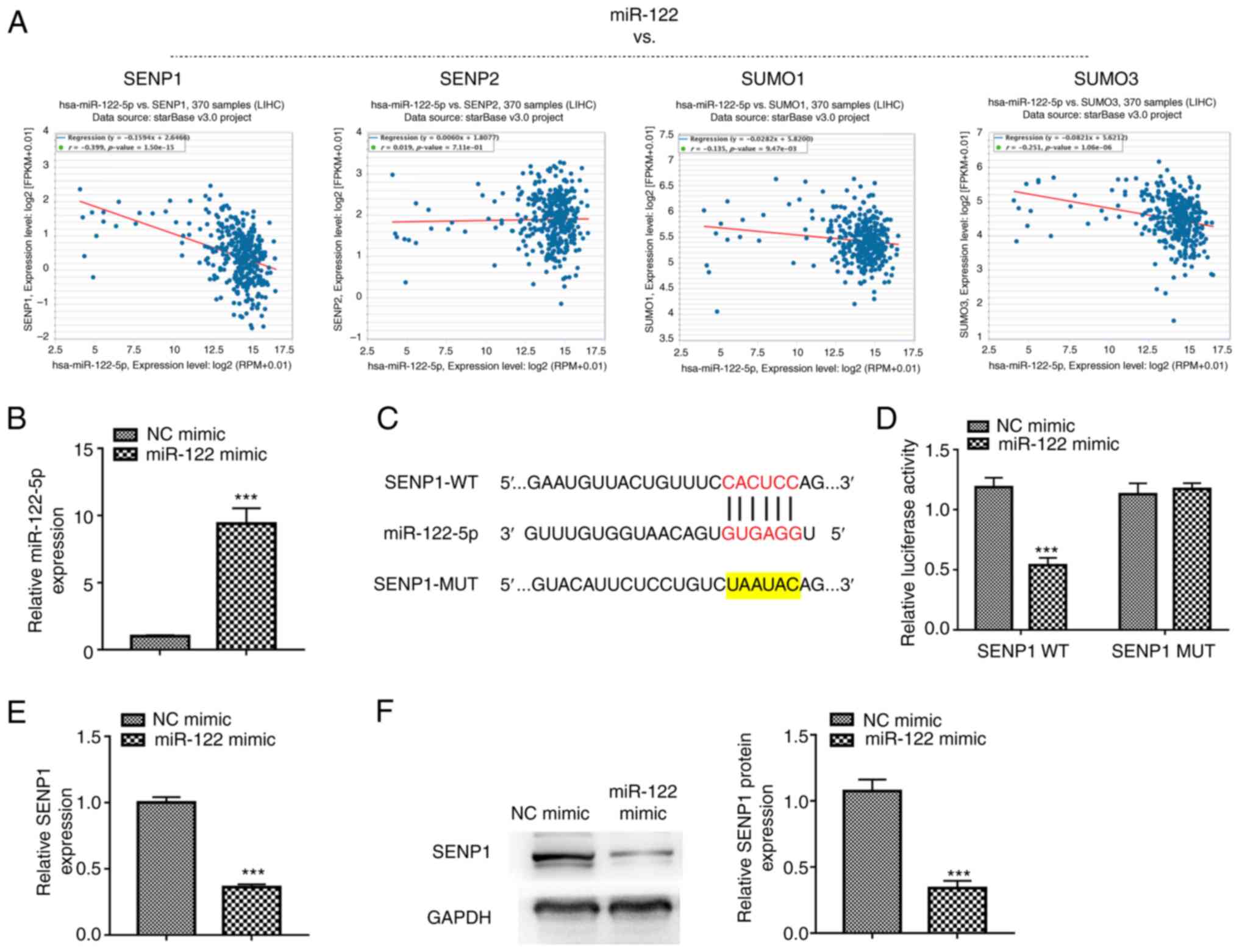
expression levels in liver cancer (r=−0.339, P<0.001; Fig. 1A), therefore, SENP1 was selected for
further study. RT-qPCR demonstrated that the miR-122 mimic
transfection successfully caused a significant overexpression of
miR-122 in HepG2 cells (Fig. 1B).
The direct binding sites between miR-122 and SENP1 mRNA 3′ UTR were
predicted by ENCORI (Fig. 1C). To
further identify whether miR-122 directly binds to the 3′UTR of
SENP1, a dual-luciferase reporter assay was performed and showed
that overexpression of miR-122 inhibited the luciferase activity of
the reporter gene in the WT construct but not in the SENPT-MUT
construct (Fig. 1D). The expression
levels of SENP1 were detected further in cells after transfecting
them with either the miR-122 or NC mimics to understand the effect
of miR-122 on SENP1 expression. The overexpression of miR-122
significantly led to the increase in the expression of SENP1 at
both mRNA and protein levels (Fig. 1E
and F), suggesting that SENP1 is a direct target of miR-122 in
liver cancer cells.
Stem properties impaired by miR-122
are restored by overexpressing SENP1 in HepG2 cells
Before investigating the roles of miR-122/SENP1 in
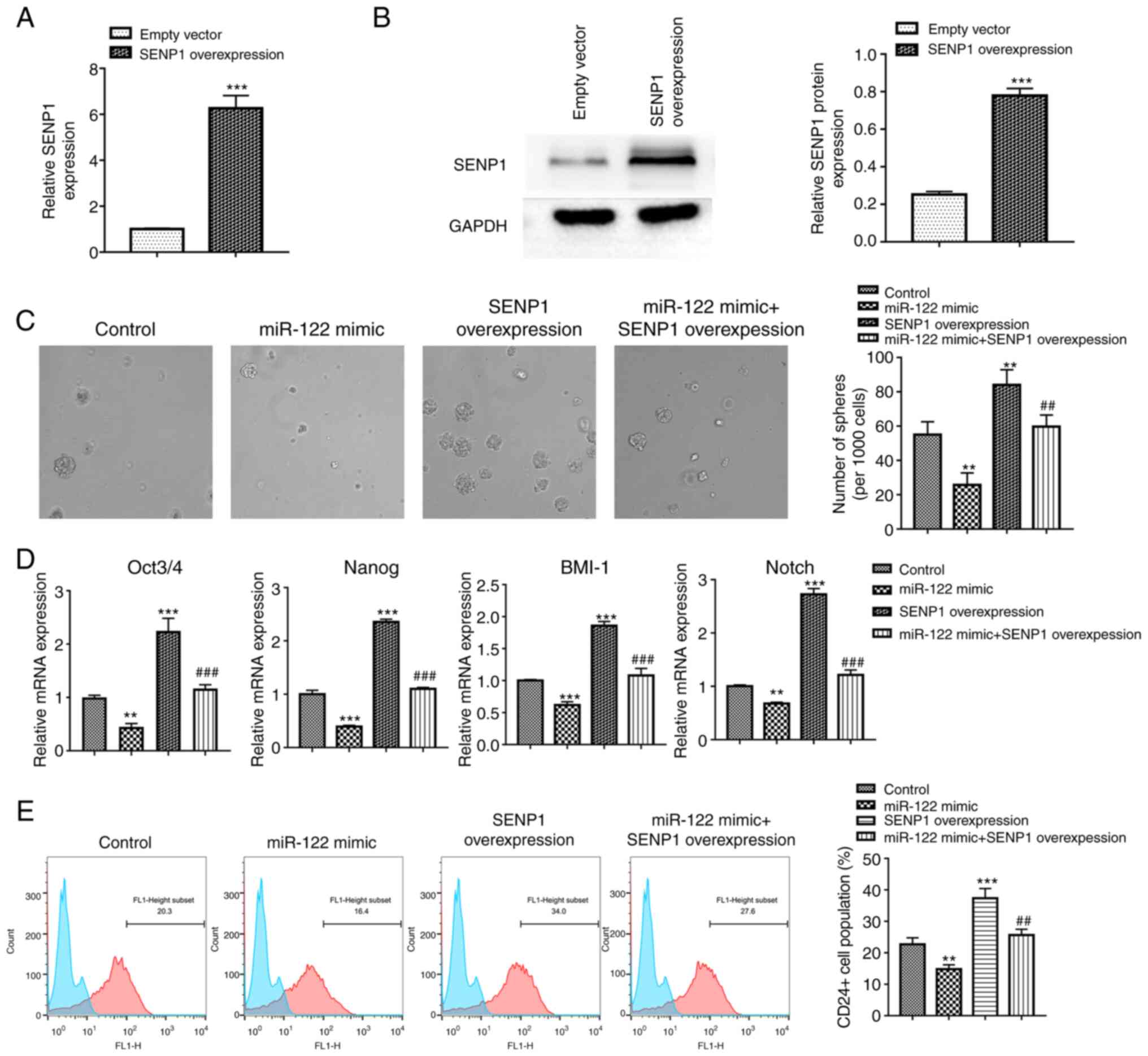
liver cancer stemness, the transfection efficiency of the SENP1
overexpression vector was evaluated by RT-qPCR and western
blotting. The SENP1 mRNA expression in the SENP1 overexpression
group was 6.3-fold greater compared with that of the empty vector
group, and the expression of SENP1 protein also showed a similar
trend (Fig. 2A and B). These
results indicated that the SENP1 was successfully overexpressed in
HepG2. For the sphere formation assay, the number of hepatospheres
formed from HepG2 cells were found to be significantly decreased
after transfection with an miR-122 mimic, but was reversed by
co-transfecting with the SENP1 overexpression vector (Fig. 2C). Detection of the expression of
stemness-related genes Oct3/4, Nanog, B lymphoma Mo-MLV insertion
region 1 homolog and Notch1 by RT-qPCR revealed a similar tendency
as the sphere formation assay, showing that the miR-122 mimic led
to a significant decrease in the expression levels of Oct3/4,
Nanog, B lymphoma Mo-MLV insertion region 1 homolog and Notch,
while the changes were reversed by co-transfection with the SENP1
overexpression vector (Fig. 2D).
Collectively, this suggests that miR-122 suppressed the stemness
properties of HepG2 cells, which could be reversed by the
overexpression of SENP1. To further validate these results, flow
cytometry assays were performed to analyzed CD24, a known marker of
liver cancer stem cells. The results demonstrated that the
overexpression of miR-122 caused a significant decrease in the
CD24+ cell population (Fig.
2E). By contrast, co-transfection with the SENP1 overexpression
vector significantly reversed this effect (Fig. 2E). These findings suggest that
miR-122 can regulate stemness properties in HepG2 cells through
SENP1.
SENP1 reverses drug sensitivity in
miR-122 overexpressing HepG2 cells
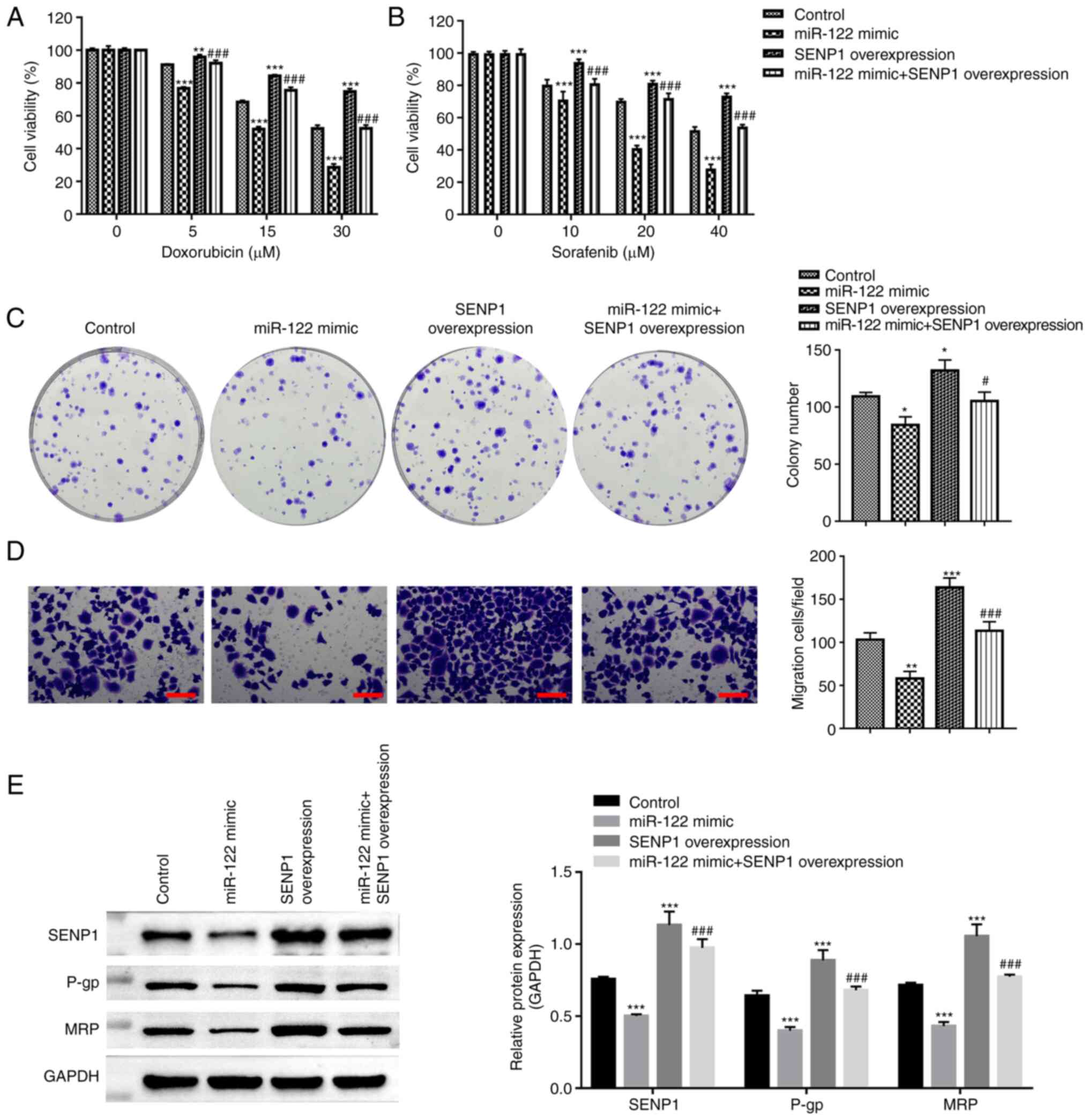
To address whether miR-122/SENP1 can regulate the
chemoresistance of liver cancer, CCK-8, colony formation and
Transwell assays were performed after treating the HepG2 cells with
DOX or sorafenib (a multikinase inhibitor used as a first-line
systemic treatment for liver cancer). The chemosensitivity to
sorafenib and DOX in the HepG2 cells was found to be increased
after transfection with miR-122 mimics, whilst SENP1
co-transfection abolished this response (Fig. 3A and B). The overexpression of
miR-122 resulted in the formation of less colonies, reduced
migratory ability, reduced the expression of drug-resistant
P-glycoprotein (P-gp) and multidrug resistance protein in HepG2
cells (Fig. 3B-D). Conversely,
SENP1 co-transfection reversed these inhibitory effects originally
induced by the miR-122 mimics (Fig.
3C-E). The results of the present study suggest that the
miR-122/SENP1 axis is associated with the chemoresistance of HepG2
cells.
miR-122 regulates the Wnt/β-catenin
signaling pathway through the de-SUMOylation effect of SENP1 on
β-catenin
It has been frequently reported that the
Wnt/β-catenin signaling pathway serves important roles in processes
associated with chemoresistance and stemness (28). The role of the miR-122/SENP1 axis in
the stemness and chemoresistance of HepG2 cells prompted the
subsequent investigation into their potential effects on this
pathway. Compared with those in the control group, the expression
levels of both Wnt1 and β-catenin were significantly lower in
miR-122 overexpressing cells, but higher in SENP1-overexpressing
cells (Fig. 4A). Co-transfection
with the SENP1 overexpression vector reversed the suppression
induced by the miR-122 mimics on Wnt1 and β-catenin expression
(Fig. 4A). Since SENP1 is a
de-SUMOylation enzyme and the SUMOylation of β-catenin has been
implicated in liver cancer growth (26), the effect of SENP1 on the
SUMOylation of β-catenin and stability in liver cancer cells was
next examined. The interaction between endogenous SENP1 and
β-catenin was confirmed in HepG2 cells (Fig. 4B). Following the transfection with
SENP1 or/and Myc-tagged β-catenin, the expressed FLAG-SENP1 and
Myc-β-catenin were detected by the anti-FLAG and anti-Myc
antibodies, further demonstrating the interaction between β-catenin
and SENP1 in in vitro settings (Fig. 4C). After overexpressing SENP1 in
cells by transfection, the levels of SUMOylation of β-catenin were
markedly reduced (Fig. 4D). In
addition, the half-life of β-catenin isolated from HepG2 was
markedly prolonged in the SENP1 overexpressing cells (Fig. 4E). These results collectively
suggest that SENP1 promotes β-catenin stability via its
de-SUMOylation function, thereby regulating the Wnt/β-catenin
signaling pathway.
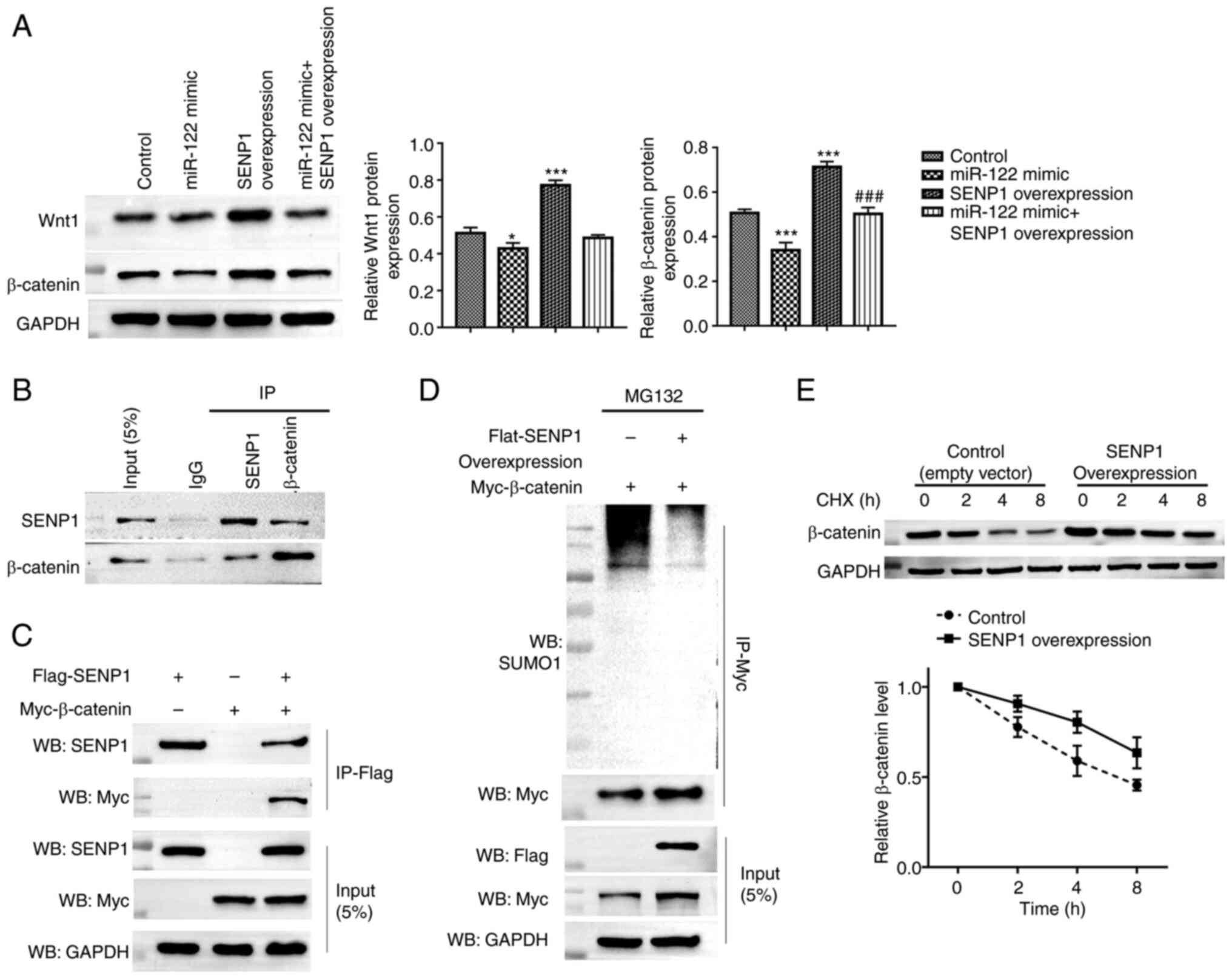 | Figure 4.miR-122/SENP1 axis regulates the
Wnt/β-catenin signaling pathway through the de-SUMOylation effect
of SENP1 on β-catenin. (A) Western blotting was performed to
measure the Wnt1 and β-catenin protein expressions in HepG2 cells
transfected with the miR-122 mimic and/or SENP1 overexpression
vector. Untransfected cells were considered as the control. (B)
HepG2 cell lysates were immunoprecipitated with control IgG,
anti-SENP1 and anti-β-catenin antibodies. The immunoprecipitates
were subsequently immunoblotted with anti-SENP1 and anti-β-catenin
antibodies. (C) FLAG-tagged SENP1 and/or Myc-tagged β-catenin were
transfected into HepG2 cells before being lysed and
immunoprecipitated with anti-FLAG antibodies. The
immunoprecipitates were subsequently immunoblotted with anti-SENP1
and anti-Myc antibodies. The whole-cell lysate of HepG2 cells
served as input. The lysate from HepG2 cells transfected with
FLAG-tagged SENP1 or Myc-tagged β-catenin alone served as the
negative control. (D) HepG2 cells were transfected with the
indicated constructs and treated with MG132 for 6 h before in
vitro SUMOylation assay. (E) HepG2 cells were transfected with
either the empty or SENP1 overexpression vectors. After 48 h, cells
were treated with CHX for 0, 2, 4 and 8 h, before being harvested,
lysed and the proteins detected by western blot analysis for
β-catenin. *P<0.05 and ***P<0.001 vs. the control;
###P<0.001 vs. the miR-122. miR, microRNA; SENP1,
sentrin-specific protease 1; IgG, immunoglobulin G; CHX,
cycloheximide. |
Overexpressing miR-122 reduces liver
cancer stemness and chemoresistance by downregulating
SENP1/β-catenin expression in vivo
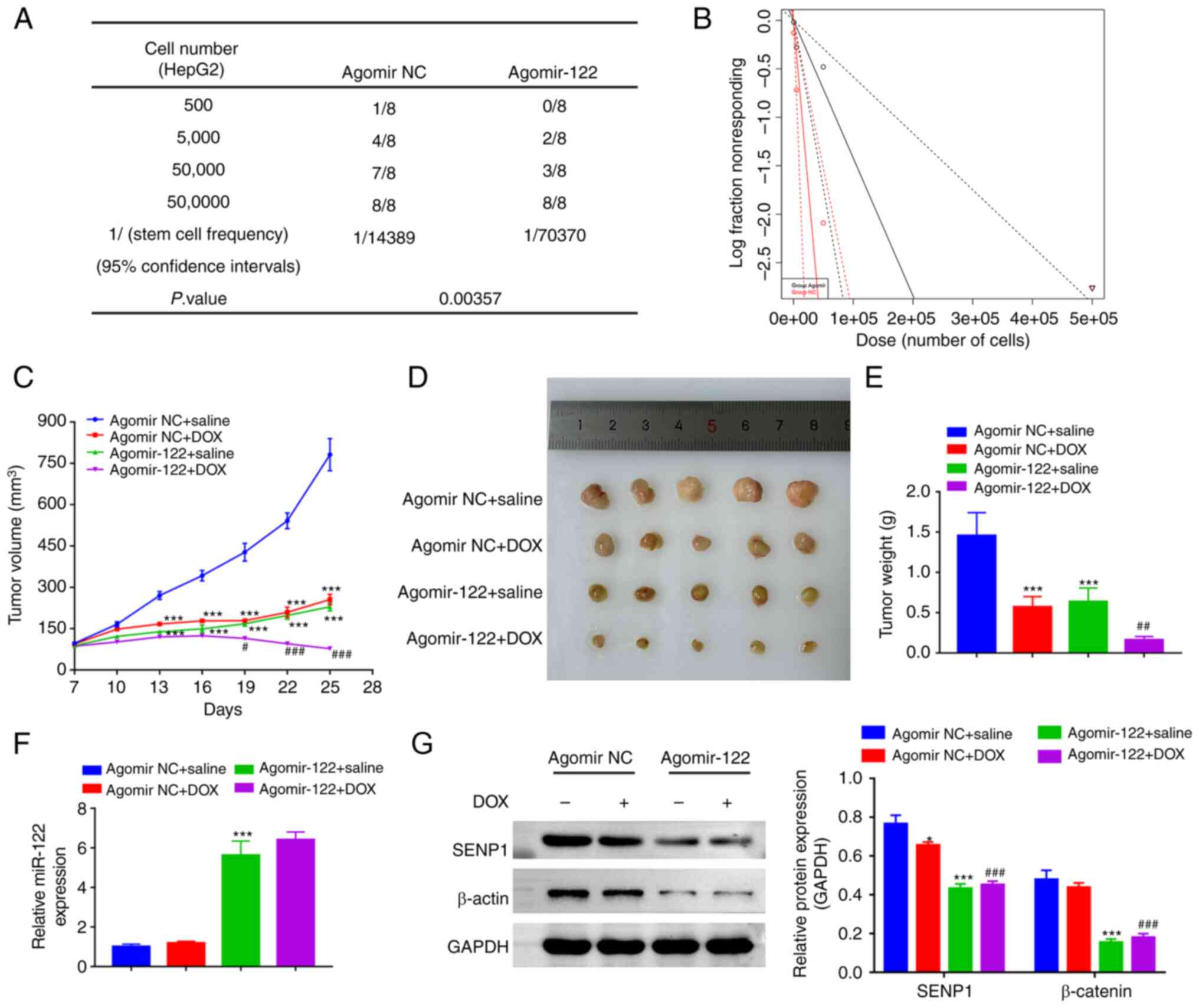
To investigate whether miR-122 can suppress the
tumor initiation frequency in vivo, a limiting dilution
experiment was performed using three different dilutions of HepG2
cells transfected with Agomir NC or Agomir-122. In total, 4 weeks
after inoculation, the tumors were collected for ELDA. The results
showed that the miR-122-overexpressing HepG2 cells exhibited
significantly lower stem cell frequency (1/70,370), compared with
that in the agomir NC group (1/14,389) (Fig. 5A and B). Xenografts of the
miR-122-overexpressing HepG2 cells demonstrated a lower growth rate
and superior responses to DOX compared with those in the cells
transfected with Agomir NC (Fig.
5C-E). The expression levels of miR-122 in the Agomir-122 group
was significantly higher compared with those in the Agomir NC group
(Fig. 5F). In addition,
miR-122-overexpressing tumors exhibited a significantly decreased
expression levels of SENP1 and β-catenin compared with those in the
control tumors (Fig. 5G), which in
agreement with the in vitro analysis.
Discussion
As one of the most aggressive malignancies, liver
cancer is the second leading cause of cancer mortality and the
fifth most commonly diagnosed cancer, with >410,000 new cases in
China in 2020 (29). Despite the
progress achieved in liver cancer therapy over the last few
decades, the prognosis of patients with liver cancer remains poor
(30). Therefore, potentially novel
therapeutic targets for improving the clinical outcomes is of
considerable importance for patients with liver cancer.
miRNAs may either serve as tumor suppressors or
oncogenes in liver cancer by regulating the expression of key
regulatory genes associated with cancer occurrence and progression
(31). Among them, miR-122 has been
identified to be a tumor suppressor miRNA in multiple malignancies,
including liver cancer (17).
However, the specific mechanism underlying its suppressive role in
liver cancer remain to be fully elucidated.
Post-translational protein modifications, such as
phosphorylation and ubiquitination, can modulate the stability,
activity, interactions and subcellular localization of their target
proteins, which in turn can alter subsequent biological processes.
SUMOylation is another important type of reversible
post-translational protein modification that is mediated by a
family of ubiquitin-like small proteins (SUMO1-5), which serves to
modulate protein stability and function (32). By contrast, SUMOylation can be
directly reversed by a group of SENPs, which de-SUMOylate the
proteins (33). Accumulating
evidence has demonstrated the causal relationship between
SUMOylation and liver cancer (27).
The protein inhibitor of activated STAT4, a pivotal component of
the TGFβ pathway, was found to be highly expressed and correlated
with poor prognosis in patients with liver cancer. In addition, it
was found to contribute to tumorigenicity and metastasis by
promoting the SUMOylation of its target proteins (34). The expression level of
SUMO-activating enzyme subunit 1 was also found to be positively
associated with liver cancer progression and metastasis (35). Therefore, it was hypothesized in the
present study that miR-122 may participate in the malignant
processes of liver cancer by regulating SUMOylation. A
bioinformatic online tool was first used to screen out the genes
involved in SUMOylation targeted by miR-122 in liver cancer.
The present study initially found that SENP1 is a
direct target of miR-122, where the miR-122/SENP1 axis was involved
in regulating the stemness properties, chemoresistance,
proliferation and migration of HepG2 cells. This was then observed
to be at least partially due to the de-SUMOylation function of
SENP1 on β-catenin, which is part of the Wnt/β-catenin signaling
pathway. To elucidate the role of the miR-122/SENP1 axis in the
regulation of the malignant phenotype, the present study modulated
their expression by transfecting with miR-122 mimics alone or
together with SENP1 overexpression vectors in HepG2 cells. Cancer
cells with ‘stemness’ characteristics are the major drivers of
tumor growth, invasion and treatment failure (36). Data in the present study showed that
miR-122 negatively regulated the stemness properties, which was
demonstrated by the decreased number of tumor spheres formed,
decreased expression of stem markers and reduced stem cell
population (HepG2 cells expressing the CD24+ marker),
after miR-122 overexpression in HepG2 cells. SENP1 co-transfection
reversed the aforementioned effects. This suggests that SENP1 can
facilitate the stemness property in liver cancer. Similarly, a
previous study demonstrated that SENP1 enhanced liver cancer
stemness through the de-SUMOylation of hypoxia-inducible factor 1-α
under hypoxia (37). Importantly,
it was observed that the downregulated self-renewal capacity
phenotype of HepG2 cells transfected with miR-122 mimics was
reversed after overexpressing SENP1, suggesting that the role of
miR-122 in the stemness of liver cancer can be mediated by
SENP1.
Consistent with previous reports (37,38),
data in the present study showed a significant suppressive effect
of miR-122 but an enhancement effect of SENP1 on cell
proliferation, migration and multidrug resistance in liver cancer.
It has been previously shown that the overexpression of miR-122 can
increase both sorafenib (39) and
DOX (40) sensitivity in HepG2
cells. Consistent with previous studies, the overexpression of
miR-122 increased the sensitivity of HepG2 cells to DOX and
sorafenib. However, this effect was reversed by co-transfection
with the SENP1 overexpression vector. These data collectively
suggest that the miR-122/SENP1 axis can contribute to liver cancer
stemness, drug sensitivity, cell proliferation and migration.
The Wnt/β-catenin signaling pathway has been
implicated in the occurrence, development and progression of
multiple cancers, including liver cancer (41). The important role of the
Wnt/β-catenin signaling pathway prompts its consideration as a
possible mechanism regulated by the miR-122/SENP1 axis. Results in
the present study showed that SENP1 overexpression significantly
decreased the SUMOylation of β-catenin, a key molecule in this
pathway to increase its stability, rendering the upregulation of
β-catenin. By contrast, the overexpression of miR-122 led to a
decrease in SENP1 expression, which subsequently increased
β-catenin degradation. Subsequently, results from in vivo
experiments confirmed the findings of the present study that
miR-122 could suppress stemness, chemoresistance and β-catenin
expression in liver cancer by regulating SENP1.
Collectively, the present study explored the role of
the miR-122/SENP1 axis in liver cancer, demonstrating that it can
serve a role in the stemness, chemoresistance, cell proliferation
and migration of this type. Specifically, it can activate the
Wnt/β-catenin signaling pathway through the de-SUMOylation of
β-catenin by SENP1. However, all functional studies that determined
the effect of the miR-122/SENP1 axis in the present study are based
on HepG2 cells in culture. The true biological functions of the
miR-122/SENP1 axis and its role in hepatocarcinogenesis should be
elucidated in further investigations using knock-out mice in the
future.
In summary, the findings of the present study
demonstrated that the miR-122/SENP1 axis contributes to the
stemness, proliferation, migration and chemoresistance of liver
cancer cells by regulating the Wnt/β-catenin signaling pathway
through the de-SUMOylation of β-catenin by SENP1. This may serve as
a scientific foundation for the potential therapeutic exploitation
of the miR-122/SENP1 axis for treating patients with liver
cancer.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Author contributions
JD and BW confirm the authenticity of all the raw
data. JD, BW and YH designed the study and wrote original draft. JD
performed the bioinformatics analysis and animal experiments, and
analyzed the data. YH, XC and QSY performed the cell experiments.
BW reviewed and edited the manuscript. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
All procedures regarding animals in the present
study were performed in accordance with relevant guidelines and
regulations with the approval by the Institutional Animal Care and
Use Committee at the Nan'an District People's Hospital of Chongqing
(approval no. 2020-0925). The euthanasia method used was approved
by the Institutional Animal Care and Use Committee at the Nan'an
District People's Hospital of Chongqing.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lohitesh K, Chowdhury R and Mukherjee S:
Resistance a major hindrance to chemotherapy in hepatocellular
carcinoma: An insight. Cancer Cell Int. 18:442018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Phi LTH, Sari IN, Yang YG, Lee SH, Jun N,
Kim KS, Lee YK and Kwon HY: Cancer stem cells (CSCs) in drug
resistance and their therapeutic implications in cancer treatment.
Stem Cells Int. 2018:54169232018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chang JC: Cancer stem cells: Role in tumor
growth, recurrence, metastasis, and treatment resistance. Medicine
(Baltimore). 95 (1 Suppl 1):S20–S25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Adhikari AS, Agarwal N and Iwakuma T:
Metastatic potential of tumor-initiating cells in solid tumors.
Front Biosci (Landmark Ed). 16:1927–1938. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee TK, Cheung VC and Ng IO: Liver
tumor-initiating cells as a therapeutic target for hepatocellular
carcinoma. Cancer Lett. 338:101–109. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim HM, Haraguchi N, Ishii H, Ohkuma M,
Okano M, Mimori K, Eguchi H, Yamamoto H, Nagano H, Sekimoto M, et
al: Increased CD13 expression reduces reactive oxygen species,
promoting survival of liver cancer stem cells via an
epithelial-mesenchymal transition-like phenomenon. Ann Surg Oncol.
19 (Suppl 3):S539–S548. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Subramanian S and Steer CJ: Special issue:
MicroRNA regulation in health and disease. Genes (Basel).
10:4572019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vasudevan S: Posttranscriptional
upregulation by microRNAs. Wiley Interdiscip Rev RNA. 3:311–330.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Giovannini C, Minguzzi M, Baglioni M,
Fornari F, Giannone F, Ravaioli M, Cescon M, Chieco P, Bolondi L
and Gramantieri L: Suppression of p53 by Notch3 is mediated by
cyclin G1 and sustained by MDM2 and miR-221 axis in hepatocellular
carcinoma. Oncotarget. 5:10607–10620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun C, Yao X, Jiang Q and Sun X: miR-106b
targets DAB2 to promote hepatocellular carcinoma cell proliferation
and metastasis. Oncol Lett. 16:3063–3069. 2018.PubMed/NCBI
|
|
12
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Simerzin A, Zorde-Khvalevsky E, Rivkin M,
Adar R, Zucman-Rossi J, Couchy G, Roskams T, Govaere O, Oren M,
Giladi H and Galun E: The liver-specific microRNA-122*, the
complementary strand of microRNA-122, acts as a tumor suppressor by
modulating the p53/mouse double minute 2 homolog circuitry.
Hepatology. 64:1623–1636. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Girard M, Jacquemin E, Munnich A, Lyonnet
S and Henrion-Caude A: miR-122, a paradigm for the role of
microRNAs in the liver. J Hepatol. 48:648–656. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cheung O, Puri P, Eicken C, Contos MJ,
Mirshahi F, Maher JW, Kellum JM, Min H, Luketic VA and Sanyal AJ:
Nonalcoholic steatohepatitis is associated with altered hepatic
MicroRNA expression. Hepatology. 48:1810–1820. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hsu SH, Wang B, Kota J, Yu J, Costinean S,
Kutay H, Yu L, Bai S, La Perle K, Chivukula RR, et al: Essential
metabolic, anti-inflammatory, and anti-tumorigenic functions of
miR-122 in liver. J Clin Invest. 122:2871–2883. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tsai WC, Hsu SD, Hsu CS, Lai TC, Chen SJ,
Shen R, Huang Y, Chen HC, Lee CH, Tsai TF, et al: MicroRNA-122
plays a critical role in liver homeostasis and
hepatocarcinogenesis. J Clin Invest. 122:2884–2897. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xu Y, Bu X, Dai C and Shang C: High serum
microRNA-122 level is independently associated with higher overall
survival rate in hepatocellular carcinoma patients. Tumour Biol.
36:4773–4776. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kim SS, Nam JS, Cho HJ, Won JH, Kim JW, Ji
JH, Yang MJ, Park JH, Noh CK, Shin SJ, et al: Plasma micoRNA-122 as
a predictive marker for treatment response following transarterial
chemoembolization in patients with hepatocellular carcinoma. J
Gastroenterol Hepatol. 32:199–207. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Celen AB and Sahin U: Sumoylation on its
25th anniversary: Mechanisms, pathology, and emerging concepts.
FEBS J. 287:3110–3140. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zubiete-Franco I, García-Rodríguez JL,
Lopitz-Otsoa F, Serrano-Macia M, Simon J, Fernández-Tussy P,
Barbier-Torres L, Fernández-Ramos D, Gutiérrez-de-Juan V, López de
Davalillo S, et al: SUMOylation regulates LKB1 localization and its
oncogenic activity in liver cancer. EBioMedicine. 40:406–421. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qin Y, Bao H, Pan Y, Yin M, Liu Y, Wu S
and Li H: SUMOylation alterations are associated with multidrug
resistance in hepatocellular carcinoma. Mol Med Rep. 9:877–881.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:(Database Issue). D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hu Y and Smyth GK: ELDA: Extreme limiting
dilution analysis for comparing depleted and enriched populations
in stem cell and other assays. J Immunol Methods. 347:70–78. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tomasi ML and Ramani K: SUMOylation and
phosphorylation cross-talk in hepatocellular carcinoma. Transl
Gastroenterol Hepatol. 3:202018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuan H, Lu Y, Chan YT, Zhang C, Wang N and
Feng Y: The role of protein SUMOylation in human hepatocellular
carcinoma: A potential target of new drug discovery and
development. Cancers (Basel). 13:57002021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mohammed MK, Shao C, Wang J, Wei Q, Wang
X, Collier Z, Tang S, Liu H, Zhang F, Huang J, et al: Wnt/β-catenin
signaling plays an ever-expanding role in stem cell self-renewal,
tumorigenesis and cancer chemoresistance. Genes Dis. 3:11–40. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cao W, Chen HD, Yu YW, Li N and Chen WQ:
Changing profiles of cancer burden worldwide and in China: A
secondary analysis of the global cancer statistics 2020. Chin Med J
(Engl). 134:783–791. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen Z, Xie H, Hu M, Huang T, Hu Y, Sang N
and Zhao Y: Recent progress in treatment of hepatocellular
carcinoma. Am J Cancer Res. 10:2993–3036. 2020.PubMed/NCBI
|
|
31
|
Gramantieri L, Fornari F, Callegari E,
Sabbioni S, Lanza G, Croce CM, Bolondi L and Negrini M: MicroRNA
involvement in hepatocellular carcinoma. J Cell Mol Med.
12:2189–2204. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: A decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yeh ETH: SUMOylation and De-SUMOylation:
Wrestling with life's processes. J Biol Chem. 284:8223–8227. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Q, Zhou B, Liao R, Zhou X and Yan X:
PIAS4, upregulated in hepatocellular carcinoma, promotes
tumorigenicity and metastasis. J Cell Biochem. 121:3372–3381. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ong JR, Bamodu OA, Khang NV, Lin YK, Yeh
CT, Lee WH and Cherng YG: SUMO-activating enzyme subunit 1 (SAE1)
is a promising diagnostic cancer metabolism biomarker of
hepatocellular carcinoma. Cells. 10:1782021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tsui YM, Chan LK and Ng IO: Cancer
stemness in hepatocellular carcinoma: Mechanisms and translational
potential. Br J Cancer. 122:1428–1440. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cui CP, Wong CC, Kai AK, Ho DW, Lau EY,
Tsui YM, Chan LK, Cheung TT, Chok KS, Chan ACY, et al: SENP1
promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation
and SENP1/HIF-1α positive feedback loop. Gut. 66:2149–2159. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang N, Wang Q, Shen D, Sun X, Cao X and
Wu D: Downregulation of microRNA-122 promotes proliferation,
migration, and invasion of human hepatocellular carcinoma cells by
activating epithelial-mesenchymal transition. Onco Targets Ther.
9:2035–2047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Turato C, Fornari F, Pollutri D, Fassan M,
Quarta S, Villano G, Ruvoletto M, Bolondi L, Gramantieri L and
Pontisso P: MiR-122 targets SerpinB3 and is involved in sorafenib
resistance in hepatocellular carcinoma. J Clin Med. 8:1712019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fornari F, Gramantieri L, Giovannini C,
Veronese A, Ferracin M, Sabbioni S, Calin GA, Grazi GL, Croce CM,
Tavolari S, et al: MiR-122/cyclin G1 interaction modulates p53
activity and affects doxorubicin sensitivity of human
hepatocarcinoma cells. Cancer Res. 69:5761–5767. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vilchez V, Turcios L, Marti F and Gedaly
R: Targeting Wnt/β-catenin pathway in hepatocellular carcinoma
treatment. World J Gastroenterol. 22:823–832. 2016. View Article : Google Scholar : PubMed/NCBI
|