Introduction
Lung cancer, including small cell lung cancer (SCLC)
and non-SCLC (NSCLC), is the leading cause of cancer-related deaths
worldwide (1,2). Among the risk factors associated with
lung cancer, the role of asthma, a chronic inflammatory disease
affecting the airways, has become a topic of marked interest in
recent years. Certain evidence points to a positive correlation
between an asthma diagnosis and an elevated risk of lung cancer
(3,4). The chronic inflammation induced by
asthma could potentially promote carcinogenic processes within lung
tissues, which lead to malignant transformation and subsequent
cancer development (5). This
emerging perspective offers novel insights into the role of chronic
inflammatory respiratory diseases in lung cancer etiology.
Nevertheless, some studies provide a contrasting viewpoint. In a
study on contemporaneous chronic obstructive pulmonary disease,
Rosenberger et al (6)
discovered a negative correlation between asthma and lung cancer.
Additionally, studies that account for co-occurring allergic
diseases found a weakened positive connection between asthma and
lung cancer (7,8).
Asthma is a chronic inflammatory disorder impacting
the respiratory airways, driven by a complex interplay between
inflammatory and structural airway cells, and cytokines (9,10).
Asthma is primarily triggered by allergic reactions, often caused
by environmental and dietary factors, which culminate in bronchial
asthma (11). Although there are
various treatments, such as nebulized therapy and topical
corticosteroids, to treat asthma (12), its molecular mechanism is still
unclear due to its complexity. A previous study reported that
asthma is a risk factor for lung cancer (4), shedding light on a possible
intersection between these respiratory conditions. However,
exploring the underlying mechanisms of lung cancer and asthma
remains a challenge.
To explore the specific mechanisms underlying the
pathogenesis of asthma and lung cancer, the present study conducted
a comprehensive bioinformatics analysis and identified triggering
receptor expressed on myeloid cells 1 (TREM1) as a key gene
associated with both diseases. The results of expression
validation, immunoassays and in vitro cell assays suggest
that TREM1 may serve as a novel and effective biomarker for lung
cancer. This finding could potentially inform more targeted
strategies for the prevention and treatment of asthma in patients
who are at a heightened risk of developing lung cancer.
Materials and methods
Gene expression files
The Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/) is
the world's biggest and most comprehensive repository of gene
expression data (13). The keyword
was set as ‘asthma’, the organism as ‘Homo sapiens’, the
experiment type as ‘expression analysis by array’, and the data set
GSE165934 (14) was selected, which
included 10 patients with asthma and 9 healthy controls.
Differentially expressed genes (DEGs) were screened for using the
‘limma’ software package, with P<0.05, log2(fold
change)>1 for upregulation and log2(fold
change)<-1 for downregulation. The STRING database (https://string-db.org/) was used to build a
protein-protein interaction (PPI) network of aggregated DEGs
(15), and Cytoscape software
(v3.8.2) was used for visualization (16). Gene Ontology (GO) analysis
(http://geneontology.org/) was performed,
and terms with P<0.01 were selected. Pathway analysis of node
genes was also performed in WebGestalt (http://www.webgestalt.org/) using the WikiPathway
functional database (https://www.wikipathways.org/).
Immune Cell Abundance Identifier
(ImmuCellAI)
ImmuCellAI was used to predict the abundance of 24
immune cell types in a sample. Differences in immune cell
infiltration in different groups were analyzed by examining immune
cell abundance in the groups. The abundance of 24 immune cells in
19 samples from the GSE165934 database was first analyzed. Next,
the abundance of 24 immune cells in asthma and normal groups were
investigated.
The Cancer Genome Atlas (TCGA)
dataset
RNAseq and relevant clinical data of NSCLC were
obtained from the TCGA dataset (https://portal.gdc.com) to screen TCGA-DEGs. The
common genes of regulation of Toll-like receptor signaling pathway
genes and TCGA-DEGs were targeted using the online Venn tool
(http://bioinformatics.psb.ugent.be/webtools/Venn/).
Kaplan-Meier (KM) curves were generated by Kaplan-Meier Plotter
(https://kmplot.com/analysis/). Cut-off
values and other parameters were chosen as default to assess
differences in overall survival (OS) across overlapping genes in
NSCLC. Survival results were visualized using KM plots and
statistically significance was assessed using the log-rank
test.
Gene Set Cancer Analysis (GSCA)
database
The transcriptomic data, gene mutation data and
clinical data of the TCGA-Lung Adenocarcinoma (LUAD) and TCGA-Lung
Squamous Cell Carcinoma (LUSC) datasets were obtained from the TCGA
database. The GSCA database (http://bioinfo.life.hust.edu.cn/GSCA/#/expression) was
used to assess the copy number variation (CNV) and single
nucleotide variation (SNV) of the survival-associated mutant genes
(17). Furthermore, to
comprehensively study somatic mutations in patients with LUAD and
LUSC, mutation data were acquired and processed by the ‘Maftools’ R
package (version 4.10) (18).
Prognostic nomogram construction using
independent parameters
Univariate Cox analysis was used to evaluate the
prognostic power of 6 survival-associated mutant genes and a number
of clinical parameters, including patient age, tumor grade, and pT,
pN and pTNM stages (19).
Subsequently, multivariate Cox analysis was used to determine
whether these genes and clinical parameters could serve as
independent indicators for patients. According to the results of
multivariate Cox analysis, a composite nomogram was designed by the
‘rms’ R software package (version 4.3.1; http://www.r-project.org/) to evaluate the impact of
independent indicators on the probability of 1-, 3- and 5-year OS.
The 45° line represents a perfect match between predictions and
observations, and the closer the nomogram model to the calibration
curve, the better the prediction result of the model.
University of Alabama at Birmingham
cancer data analysis portal (UALCAN)
UALCAN is a smart web application for the deep
analysis of TCGA and the retrieval of cancer data (20); it allows users to find potential
genes of interest between biomarker or computer approvals and
assess gene expression across different clinical factors such as
sex, ethnicity and tumor grade (21). In the present study, TREM1
expression in patients with LUSC and LUAD with different sample
types, tumor grades, smoking and lymph node metastasis statuses
were assessed.
Tumor Immune Estimation Resource
(TIMER)
TIMER (https://cistrome.shinyapps.io/timer/) enables
systematic analysis of immune infiltrate abundance in different
cancer types (22). Correlations
between the hub gene (TREM1) and immune cells (CD4+ T
cells, B cells, CD8+ T cells, macrophages, neutrophils,
and dendritic cells) in the TCGA-LUAD dataset and the TCGA-LUSC
dataset were analyzed by Spearman correlation analysis. P<0.05
was selected as the cut-off value. For reliable immune score
evaluation, the ‘immunedeconv’ TIMER algorithm in the R software
package was adopted to evaluate the immune scores. The samples were
divided into TREM1 high-expression and low-expression groups based
on the median expression value of TREM1. Those with TREM1
expression above the median were classified as the high-expression
group, while those with expression below the median were classified
as the low-expression group. Subsequently, the expression of 8
immune checkpoints in the TREM1 high-expression group and
low-expression group was analyzed by the R software package. The
relationship between TREM1 expression and the tumor mutational
burden (TMB) was examined. P<0.05 was regarded as statistically
significant when using Spearman's correlation analysis.
Cell culture
Human bronchial epithelioid cells (16HBE) and lung
cancer cell lines (H292, A549 and H1299) were purchased from the
American Type Culture Collection. Cells were cultured in DMEM
(Thermo Fisher Scientific, Inc.) at 37°C in a humidified atmosphere
with 5% CO2, supplemented with 10% FBS (Thermo Fisher
Scientific, Inc.) and 1% penicillin/streptomycin.
Reverse transcription-quantitative PCR
(RT-qPCR)
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract RNA from lysed cells
of the lung cancer cell lines, and Prime script RT Master mix
(Takara Biotechnology Co., Ltd.) used to reverse transcribe the
extracted RNA into cDNA according to the manufacturer's
instructions. Reactions were performed in duplicate using an ABI
7500 Fast Real-Time PCR system (Applied Biosystems). The reaction
conditions were 95°C for 10 min, followed by 40 cycles at 95°C for
15 sec and 60°C for 35 sec. Subsequently, qPCR detection was
performed using the SYBR-Green I (cat. no. S7563; Thermo Fisher
Scientific, Inc.) fluorescence technique computed using the
2−ΔΔCq method (23). The
primer sequences were as follows: β-actin forward,
5′-TGGATCAGCAAGCAGGAGTATG-3′ and reverse, 5′-GCATTTGCGGTGGACGAT-3′;
TREM1 forward, 5′-TCCGAATGGTCAACCTTCAAGTGG-3′ and reverse,
5′-GAACAGCATGTGAGGCTCCTTGG-3′; MyD88 forward,
5′-GGCTGCTCTCAACATGCGA-3′ and reverse, 5′-CTGTGTCCGCACGTTCAAGA-3′;
TLR2 forward, 5′-CTTCACTCAGGAGCAGCAAGCA-3′ and reverse,
5′-ACACCAGTGCTGTCCTGTGACA-3′; and TLR4 forward,
5′-CCAGCCTCCTCAGAAACA-3′and reverse, 5′-TCCAGCAGTGAAGAAGGG-3′.
Cell transfection
The lung cancer cells were cultured in 6-well plates
at 37°C overnight to reach 70% confluence, and the overexpression
plasmid (pcDNA3.1-TREM1; cat. no. V79020; Invitrogen; Thermo Fisher
Scientific, Inc.) and negative control (pcDNA3.1 empty vector; cat.
no. V79020; Invitrogen; Thermo Fisher Scientific, Inc.) were
constructed. When the cells were at ~90% confluency, 1 µg plasmid
was transfected into the cells using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C in 5%
CO2 for 4–6 h, according to the manufacturer's
instructions. After 48 h, subsequent experiments were
performed.
Cell Counting Kit-8 (CCK-8)
A total of 2×103 lung cancer cells were
added to a 96-well plate, and then treated with 10 µl CCK-8
solution (Dojindo Molecular Technologies, Inc.), and incubated at
37°C for 2 h. The optical density (OD) value at 450 nm was recorded
at 1, 2, 3 and 4 days using a microplate reader to generate a
proliferation curve. The analysis was performed in triplicate.
Cell invasion and migration assay
In the migration experiment, 4×104 lung
cancer cells, in serum-free DMEM, were seeded into the upper
chamber of a Transwell insert and a medium with 20% FBS was added
to the lower chamber as a chemoattractant. For the invasion
experiment, Matrigel (BD Biosciences) was coated on the upper
chamber at 37°C for 2 h prior to being seeded with 9×104
cells, and the lower chamber contained medium with 20% FBS. After
incubation at 37°C, in 5% CO2, for 48 h, the Transwell
chamber was removed and the medium in the well was discarded and
washed with calcium-free PBS. The cells were fixed with methanol
for 15 min at room temperature and then stained with DAPI for 10
min at room temperature. The upper unmigrated cells were gently
removed with a cotton swab, and cells in the lower chamber were
counted under a fluorescence microscope (×200 magnification).
Western blotting assay
The lung cancer cells were lysed by RIPA lysis
buffer (Thermo Fisher Scientific, Inc.) with 1% PMSF. The cell
lysates were then centrifuged at 14,000 × g for 15 min at 4°C to
separate the soluble proteins. Proteins were extracted from the
cell's lysates and supernatants. The concentration of proteins was
determined using a Pierce® BCA Protein Assay Kit (Thermo
Fisher Scientific, Inc.). The quantified proteins (50 µg/lane) were
separated by SDS-PAGE on 10% gels and then transferred onto a PVDF
membrane. The membrane was blocked with 5% non-fat dry milk at room
temperature for 3 h. Membranes were probed at 4°C overnight with
primary antibodies against TREM1 (1:5,000; cat. no. ab90808;
Abcam), MyD88 (1:5,000; cat. no. ab133739; Abcam), TLR2 (1:5,000;
cat. no. ab9100; Abcam), TLR4 (1:5,000; cat. no. ab13556; Abcam)
and GAPDH (1:5,000; cat. no. ab9485; Abcam). The following day, the
blot was probed using an HRP-conjugated secondary antibody
(1:5,000; cat. no. ab205718; Abcam) and a Goat Anti-Mouse IgG
H&L (HRP) secondary antibody (1:5,000; cat. no. ab97023; Abcam)
for 1 h at room temperature. Finally, protein bands were visualized
using an ECL Plus kit (Cytiva) and the band density was
semi-quantified using ImageJ software (version 1.52; National
Institutes of Health).
Statistical analysis
All study data were processed by SPSS 22.0 software
(IBM Corp), and each experiment was performed in triplicate. All
quantitative data are expressed as the mean ± SD. Comparison
between groups was performed using one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Identification and analysis of
GSE165934-DEGs
Through analysis of the dataset and comparison
between asthmatic and control groups, 235 upregulated DEGs and
1,040 downregulated DEGs from GSE165934 were acquired and presented
on a volcano plot and heat map (Fig. 1A
and B). Subsequently, the PPI network of all DEGs was
constructed, and the interconnectedness between genes was shown
(Fig. 1C), with 460 nodes and 1,590
edges. The 460 node genes were used for functional enrichment
analysis. Node genes were enriched in ‘neutrophil mediated
immunity’, ‘intracellular membrane-bounded organelle’, ‘RNA
binding’ and others in the GO analysis (Fig. 1D). WikiPathway analysis showed that
node genes were mainly associated with Toll-like receptor signaling
related to MyD88, regulation of the Toll-like receptor signaling
pathway and the glucocorticoid receptor pathway (Fig. 1E).
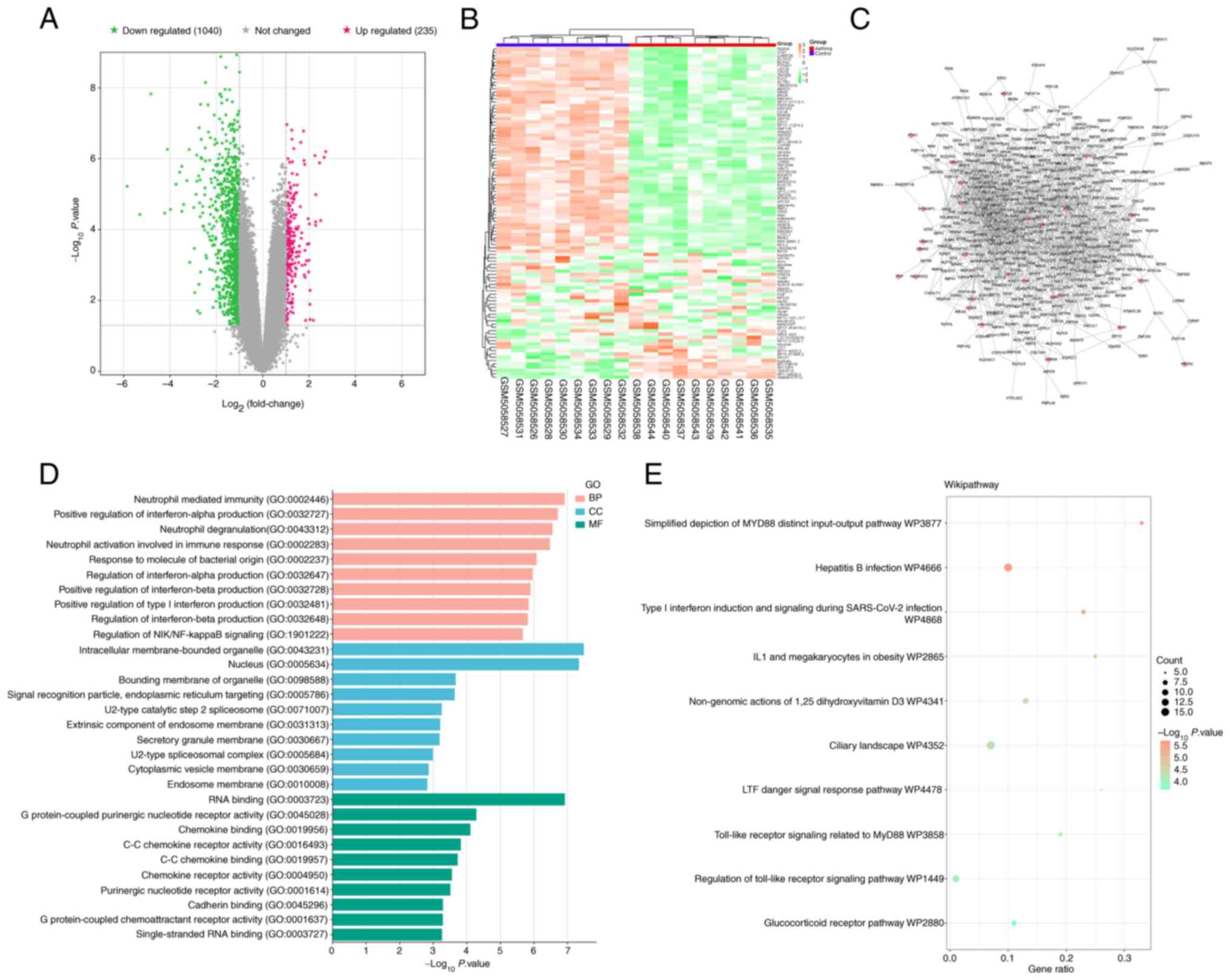 | Figure 1.Identification and bioinformatics
analysis of DEGs. (A) Volcano plot of 235 upregulated DEGs and
1,040 downregulated DEGs, with the gray area in the middle
representing unchanged genes. (B) Cluster heatmap of DEGs in
GSE165934. In the heatmap, the color gradient from green to orange
represents the expression levels of DEGs, with green indicating
lower expression levels and orange indicating higher expression
levels. Blue denotes the control group and red denotes the
asthmatic group. (C) Protein-protein interaction network of DEGs,
where nodes represent genes (pink diamonds represent upregulated
DEGs, gray arrowheads represent downregulated DEGs) and edges
represent interconnectedness between genes. (D) Bar graph of GO
analysis. BP enrichment result (pink), CC enrichment result (blue),
and MF enrichment result (green). (E) WikiPathway analysis on 460
node genes. The larger the node, the more genes are enriched on
this pathway. DEGs, differentially expressed genes; GO, Gene
Ontology; BP, biological process; CC, cellular component; MF,
molecular function. |
Genes in GSE165934 are associated with
immune cells in asthma
The present study explored immune cells in 19
samples of GSE165934 by ImmuCellAI. Fig. 2A shows the proportion of immune
cells in control samples (GSM5058526-GSM5058534) and asthma samples
(GSM5058535-GSM5058544) marked with different colors, and the
length of the bars in the bar graph represent the level of immune
cell populations. The percentages of NK cells and Tc cells in the
samples were significantly reduced. As shown in Fig. 2B, only nTreg, Th17, CD8 naive, NKT
and Tex expression was significantly reduced in the asthma group.
While other cells, such as B cells, macrophages and neutrophils,
increased significantly.
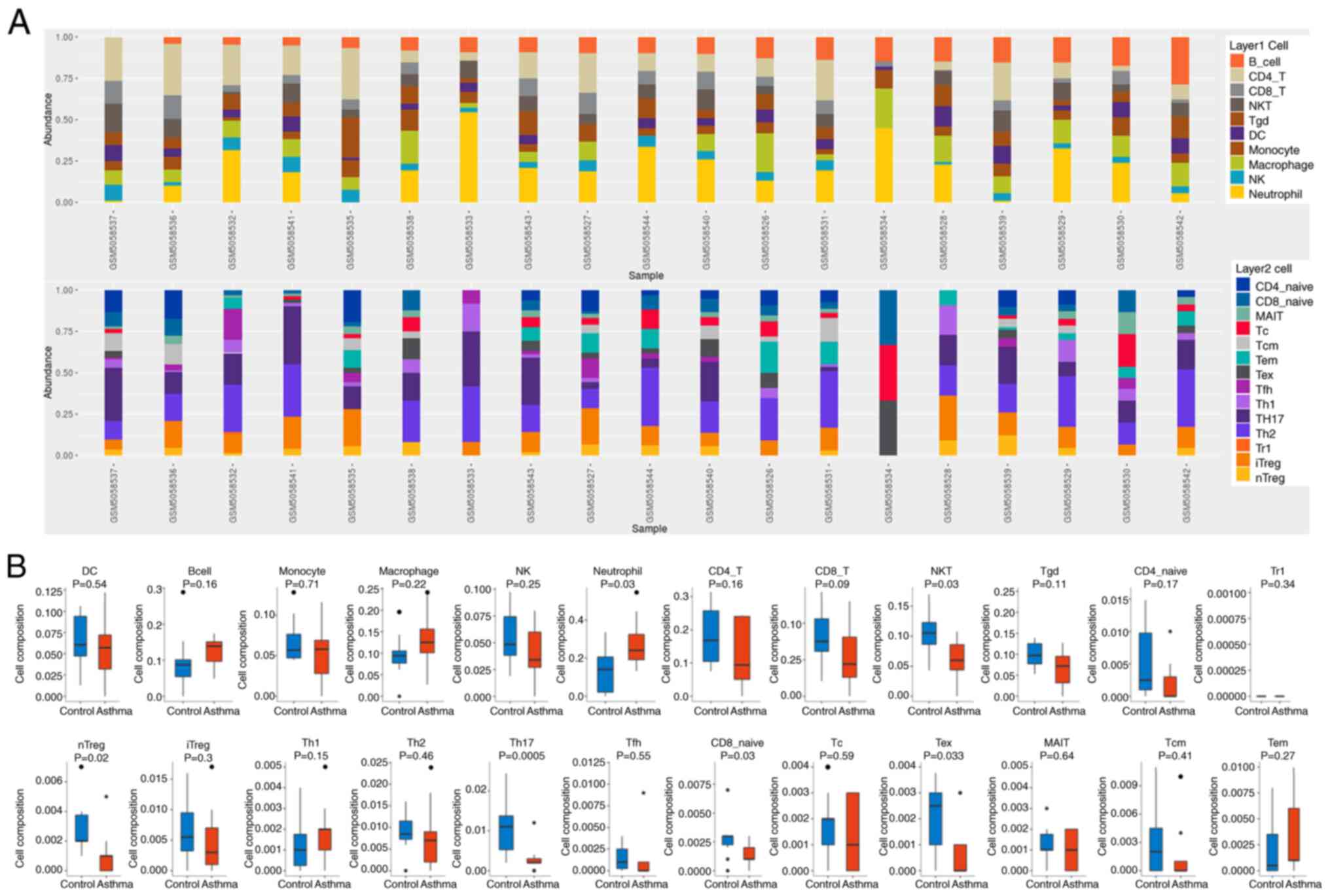 | Figure 2.ImmuCellAI analysis of 19 samples
from GSE165934. (A) The proportions of 24 immune cells in control
samples (GSM5058526-GSM5058534) and asthma samples
(GSM5058535-GSM5058544), with colored squares representing
different types of immune cells. (B) Immune cell abundance was
analyzed and examined between asthma (red) and normal (blue)
tissues by ImmuCellAI. The black dots represent outliers. DC,
dendritic cells; NK, natural killer cells; NKT, natural killer T
cells; Tr1, Type 1 regulatory T cells; nTreg, natural regulatory T
cells; iTreg, induced regulatory T cells; Th1, T helper 1 cells;
Tfh, T follicular helper cell; Tc, cytotoxic T cells; Tex,
exhausted T cells; MAIT, mucosal-associated invariant T cells; Tcm,
central memory T cells; Tem, effector memory T cells. |
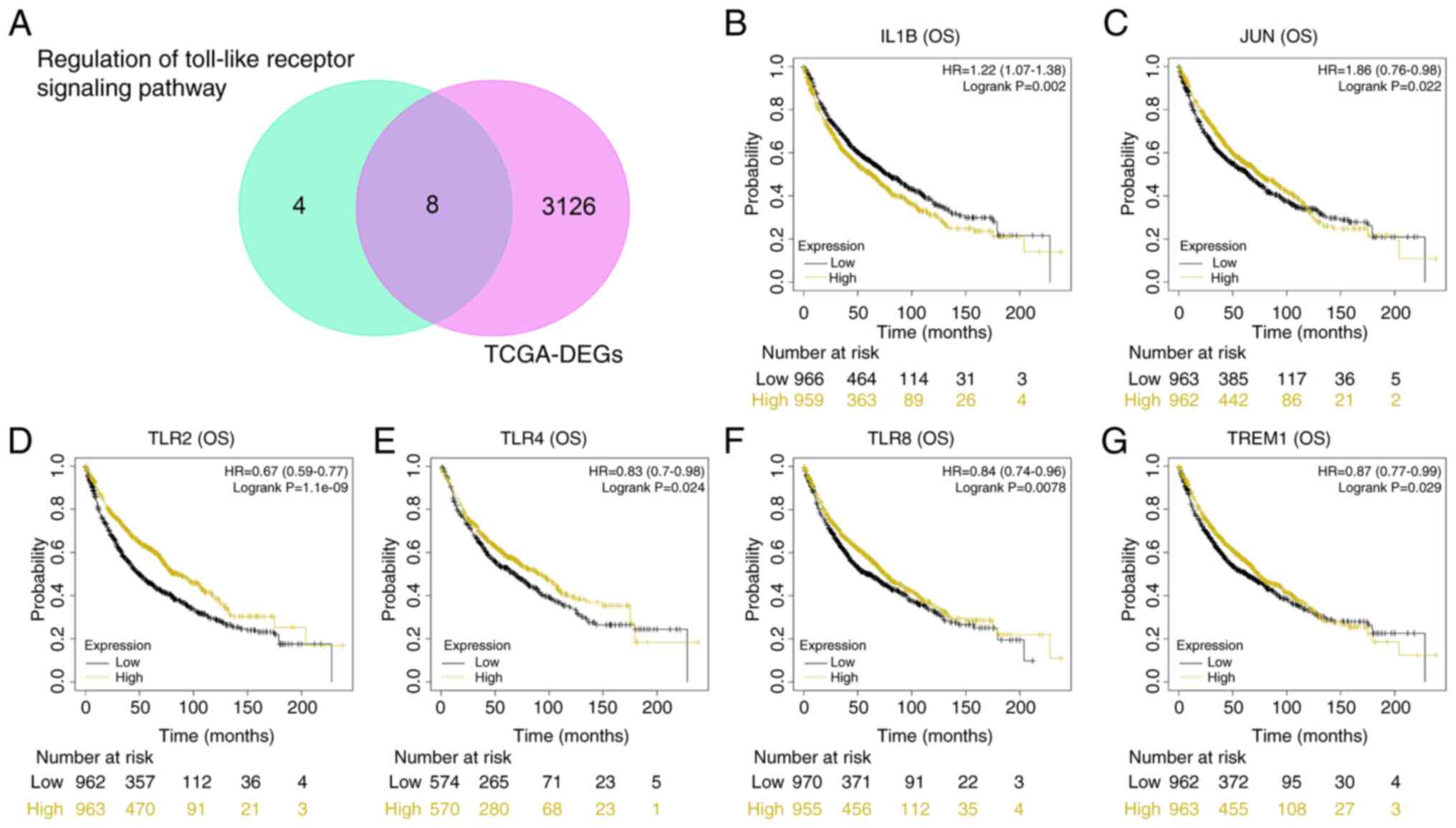
Survival analysis of gene overlap in
the Toll-like receptor signaling pathway and TCGA-DEG
Fig. 3A shows a Venn
diagram of 8 overlapping genes from genes regulated by ‘regulation
of Toll-like receptor signaling pathway’ and TCGA-DEGs. Using batch
survival analysis, 6 genes with significant P-values for OS
analysis were retained (Fig. 3B-G;
P<0.05). High expression of IL1B resulted in poor OS
probability, while low expression of JUN, TLR4, TREM1, TLR2 and
TLR8 indicated poor OS.
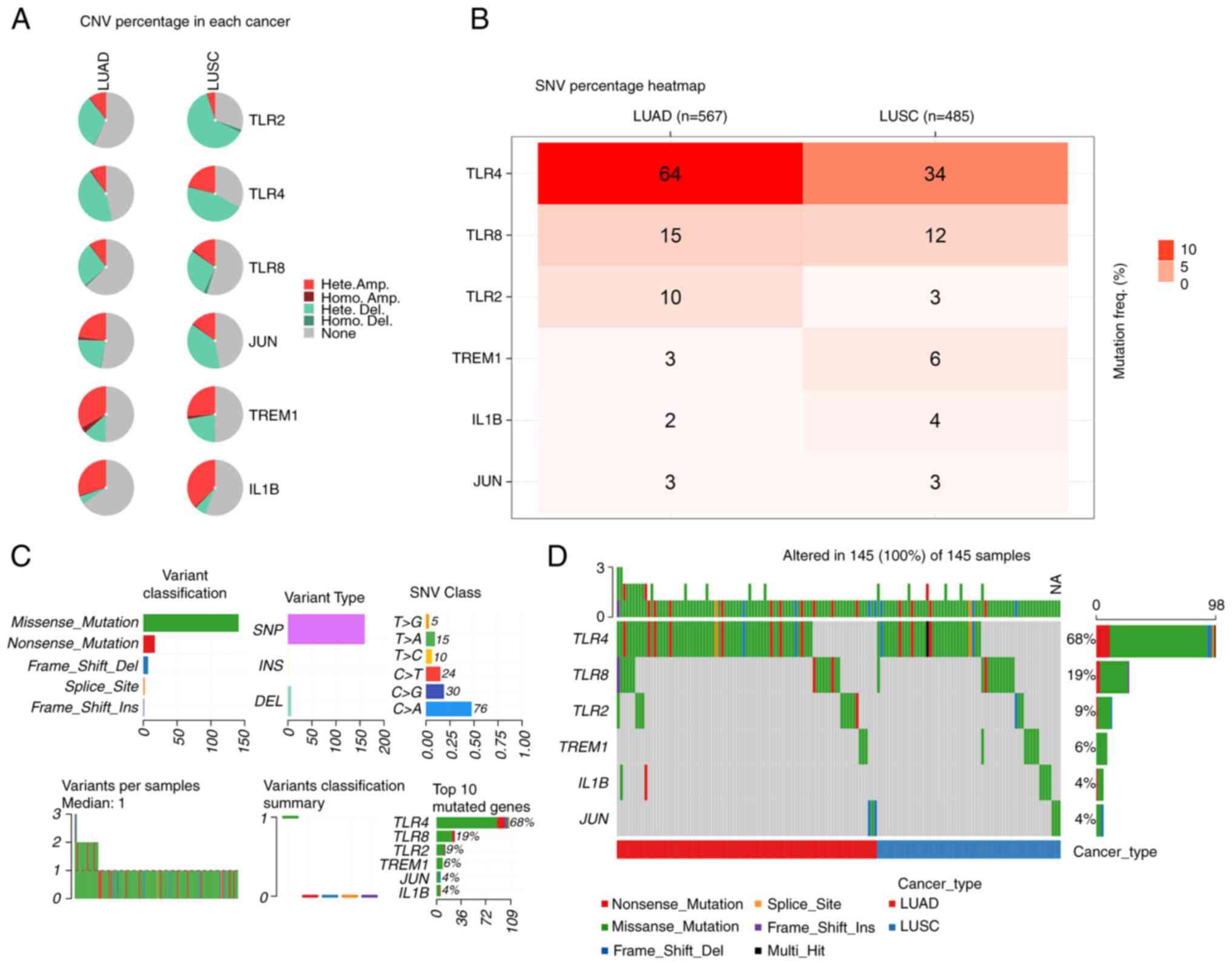
Mutational landscape analysis of CNVs,
SNVs and cellular mutations for 6 identified genes
A genetic variation analysis for 6 genes with
significant P-values in survival analysis, including CNVs and SNVs,
was performed using the GSCA database in LUAD and LUSC. In LUAD,
TLR4 had the highest percentage of CNVs whereas in LUSC, TLR2 had
the highest percentage of CNVs (Fig.
4A). The percentage of SNVs for the 6 genes in LUAD and LUSC
were also explored, and it was found that in LUAD, TLR4 exhibited
the most SNVs, followed by TLR8, TLR2, TREM1, IL1B and JUN. In
LUSC, TLR4 exhibited the most SNVs, followed by TLR8, TREM1, IL1B,
TLR2 and JUN (Fig. 4B). The most
common type of mutation in patients with LUAD and LUSC was a
missense mutation. Single nucleotide polymorphisms (SNPs) were the
main type of mutational variation, with C>A being dominant over
other SNV categories. As shown in Fig.
4C, the median mutation variation per sample was 1, and each
color box represented a mutation. Fig.
4C displays the six most frequently mutated genes in the
present study, including TLR4 (68%), TLR8 (19%), TLR2 (9%), TREM1
(6%), JUN (4%) and IL1B (4%). Histograms in Fig. 4D show the mutation frequencies of
the 6 genes in the patients with LUAD and LUSC (n=145).
TREM1 is a potential prognostic
indicator of lung cancer
After univariate and multivariate Cox analyses were
performed, it was apparent that TREM1, pT-stage and pTNM-stage were
independent prognostic variables to construct the predictive
nomogram, and that TREM1 was considered as the hub gene (Fig. 5A and B). A composite nomogram was
designed with TREM1, pT-stage and pTNM-stage to predict 1-, 3- and
5-year OS rates in patients with NSCLC (Fig. 5C). The presentation of the
calibration plot for patient survival prediction demonstrated the
predicted results of the prognostic nomogram matched with the
observed results, suggesting that the model had good prognostic
prediction for patients (Fig.
5D).
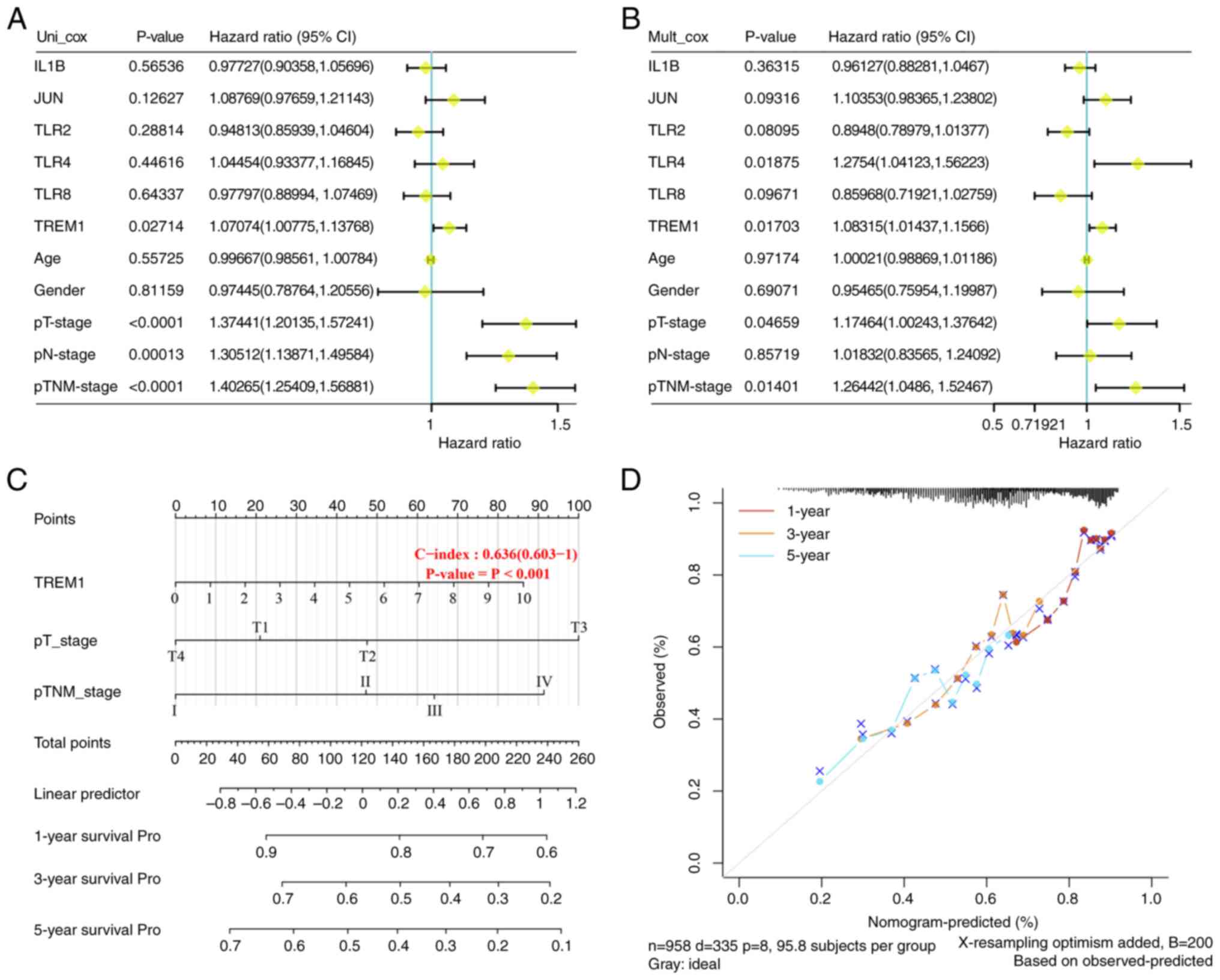 | Figure 5.TREM1 could be a prognostic biomarker
in lung cancer. (A) Univariate Cox regression analysis showed that
TREM1, pT-stage, pN-stage and pTNM-stage were significant
prognostic variables. (B) Multivariate Cox regression analysis
showed that the significant prognostic variables were TLR4, TREM1,
pT-stage, and pTNM-stage. (C) Nomogram with independent indicators,
with a scale marked on the corresponding line segment of each
variable, representing the range of possible values of the
variable. The longer the line segment, the greater its contribution
to the prognosis. (D) Calibration curve of the nomogram model, and
the diagonal gray line is the ideal nomogram. TREM1, triggering
receptor expressed on myeloid cells 1. Pro, prognosis. |
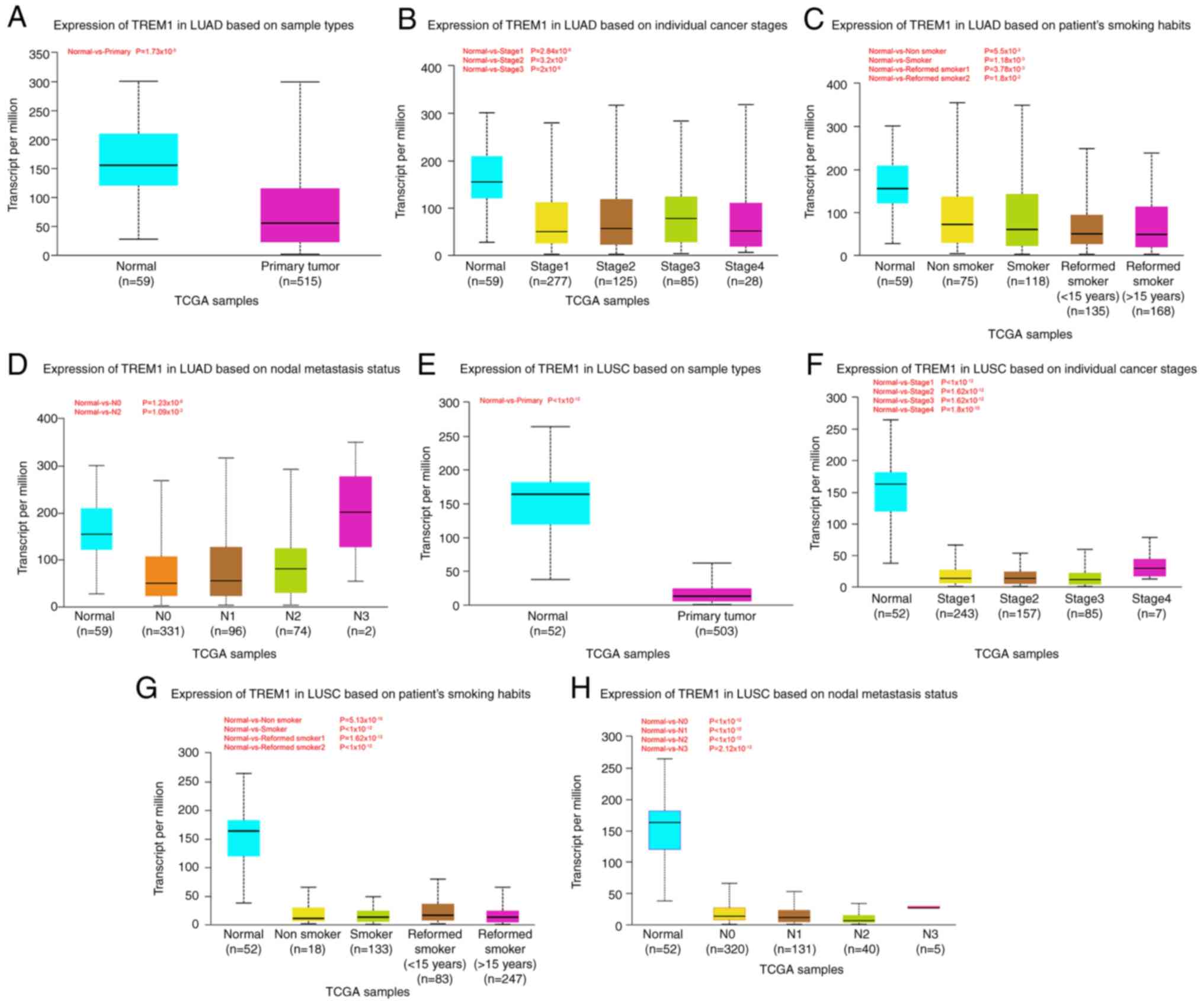
Association between TREM1 expression
and clinical factors in LUSC and LUAD
Comparing different patient samples, including both
LUAD and LUSC tissue samples, TREM1 expression levels were found to
be lower in primary tumor compared with those in normal tissues
(Fig. 6A and E). However, among
different clinical factors of LUAD and LUSC, TREM1 expression had
no significant association with individual cancer stages, patient's
smoking habits or nodal metastasis status (Fig. 6B-D and F-H).
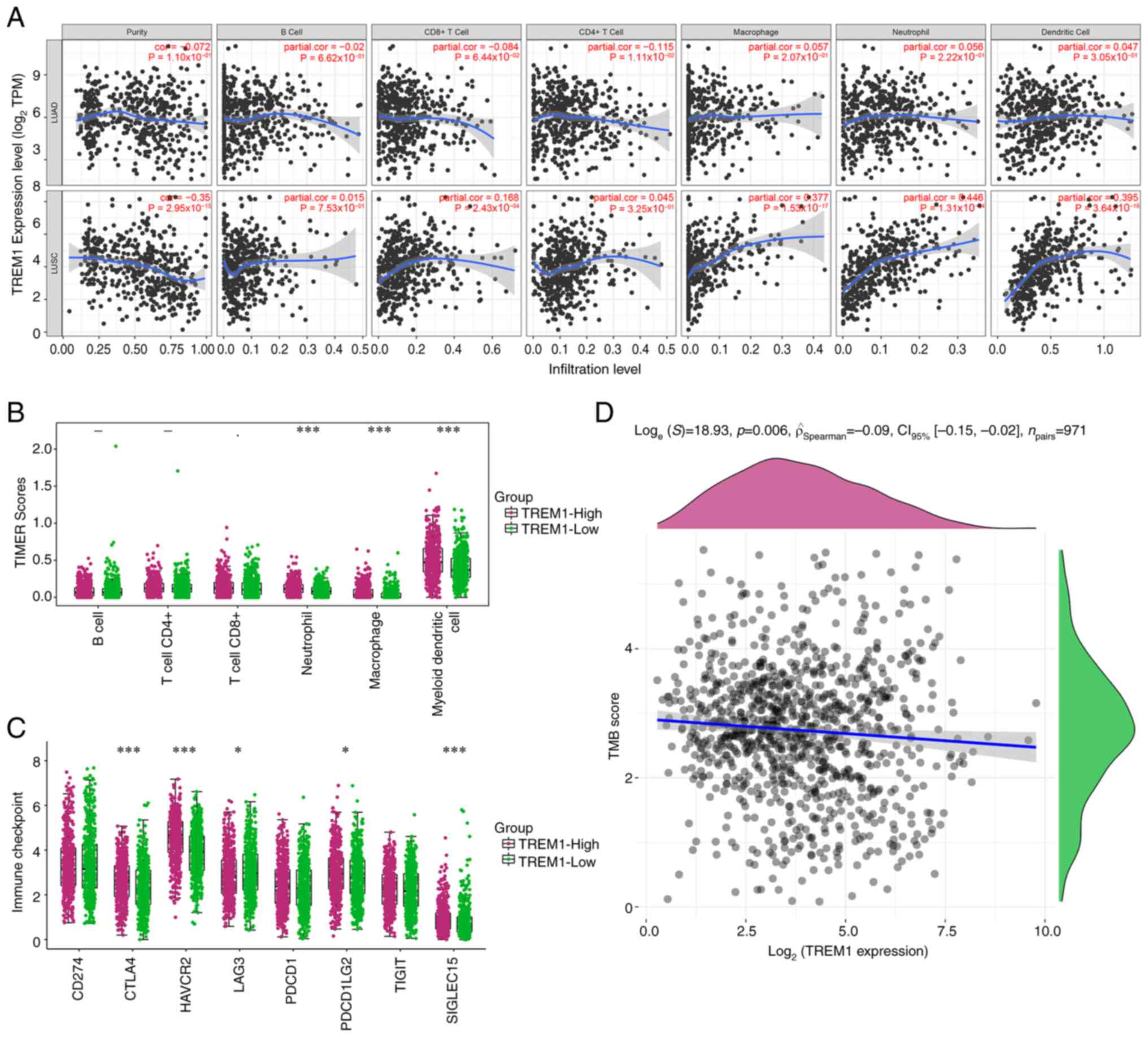
Immunoassay on TREM1 in lung
cancer
Fig. 7A shows the
association between TREM1 and 6 immune cell types in LUSC and LUAD.
A positive correlation was denoted by Cor>0, and a negative
correlation by Cor<0. In LUAD, TREM1 expression was negatively
correlated with tumor purity, B cells, CD8+ T cells and
CD4+ T cells. The expression of TREM1 was positively
correlated with the expression of macrophages, neutrophils and
dendritic cells. In LUSC, TREM1 expression was inversely correlated
with tumor purity. The expression of TREM1 was positively
correlated with the expression of B cells, CD8+ T cells,
CD4+ T cells, macrophages, neutrophils and dendritic
cells. Fig. 7B shows the fraction
of immune cells that infiltrated tumors in samples with high and
low TREM1 expression. Myeloid dendritic cells had higher TIMER
scores in TREM1-expressing samples. In addition, the TIMER score
was higher in the high TREM1 expression group. The differences in
expression of the 8 immune checkpoint molecules between the two
groups were not obvious. However, it was observed that the level of
immune checkpoint molecules was higher in samples with high TREM1
expression (Fig. 7C). Over the past
few years, TMB has been a significant prognostic biomarker;
however, its prognostic value in NSCLC has remained unclear.
Through TCGA, the correlation between TMB and the levels of TREM1
in NSCLC was comprehensively analyzed to determine the impact of
TREM1 in the development of NSCLC. The present results suggested
that TREM1 had a negative association with the TMB score in NSCLC
(Fig. 7D).
TREM1 inhibits cell proliferation,
invasion and migration via the Toll-like receptor pathway in lung
cancer
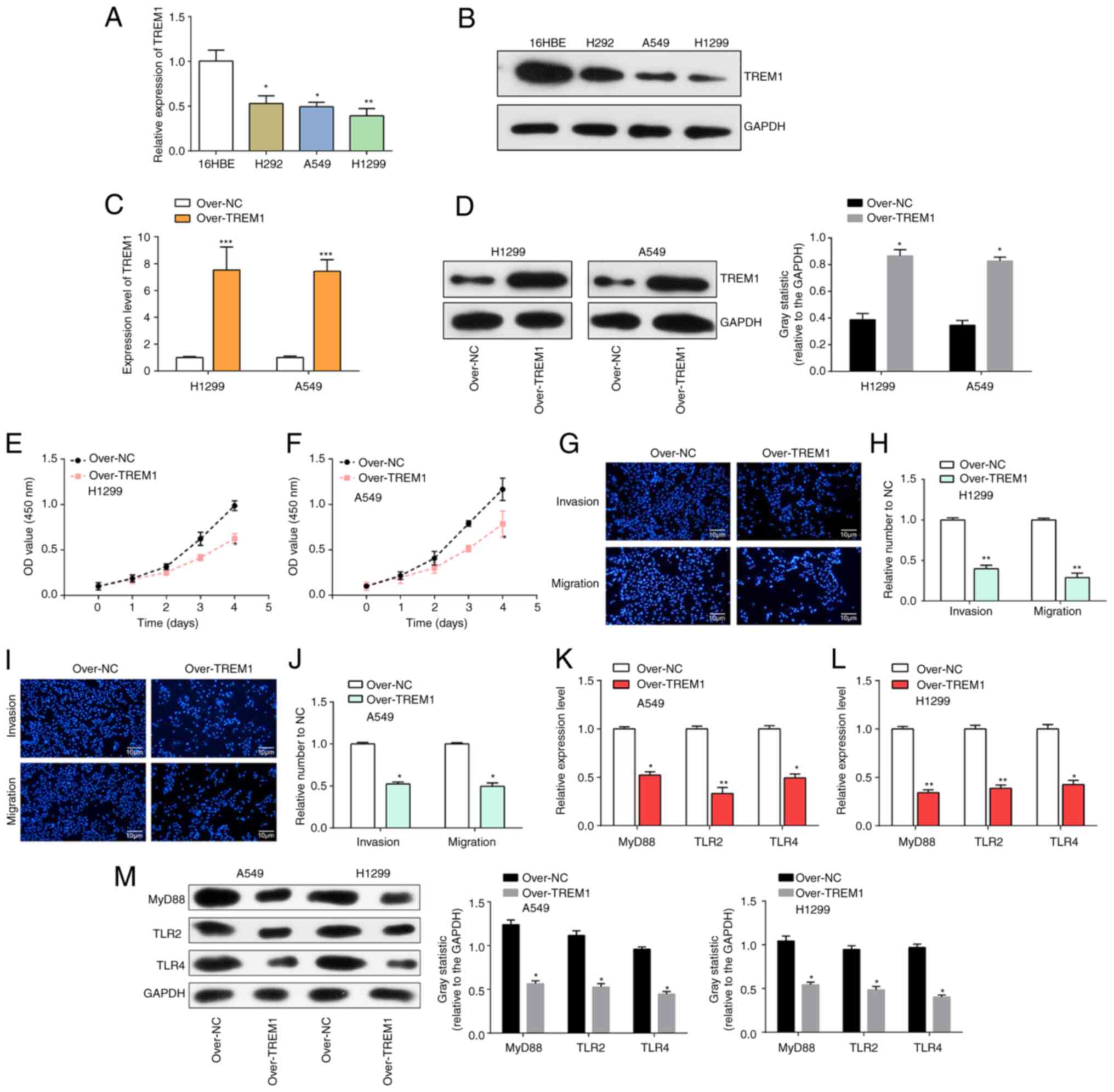
Results of RT-qPCR and western blotting experiments
showed downregulation of TREM1 in lung cancer cells (H292, A549 and
H1299) compared with human bronchial epithelial-like cells (16HBE),
especially in A549 and H1299 cells (Fig. 8A and B). Subsequently, TREM1 was
overexpressed in A549 and H1299 cells, and the overexpression
efficiency was examined by RT-qPCR and western blotting. The
findings indicated a marked increase in TREM1 expression within
A549 and H1299 cells (Fig. 8C and
D). CCK-8 experiments were performed in H1299 and A549 cell
lines, and it was observed that overexpressed TREM1 significantly
inhibited the proliferation of A549 and H1299 cells (Fig. 8E and F). Since TREM1 was most
significantly expressed in H1299 cells and A549 cells, a Transwell
assay using these two cell lines was performed, which showed that
overexpression of TREM1 significantly inhibited the invasion and
migration of H1299 as well as A549 cells (Fig. 8G-J). Furthermore, the effect of
overexpressed TREM1 on the related proteins of the Toll-like
receptor pathway, including TLR2, TLR4 and MyD88, was explored.
RT-qPCR results suggested that the mRNA levels of MyD88, TLR2 and
TLR4 were reduced in lung cancer cells after overexpression of
TREM1 (Fig. 8K and L). Western
blotting results suggest the protein levels of MyD88, TLR2 and TLR4
were also reduced (P<0.05) in lung cancer cells after
overexpression of TREM1, indicating that TREM1 negatively regulated
MyD88, TLR2 and TLR4 (Fig. 8M).
Discussion
Asthma can cause a series of reactions such as
wheezing and chest tightness, which usually occur at night or in
the early morning and can seriously affect the quality of life of
patients (24). According to the
number of exacerbations during oral systemic corticosteroid
therapy, asthma can be classified into four categories:
Intermittent, mild, moderate and severe persistent asthma (25). The chronic inflammatory state in the
lungs of asthmatics is considered to cause oxidative damage that
may contribute to the development of lung cancer (26). Jiang et al (7) suggested that proper control of asthma
symptoms not only reduces asthma attacks but also helps reduce the
incidence of lung cancer. Therefore, identifying the key genes
related to both lung cancer and asthma could reveal the molecular
mechanism behind their connection.
In the present study, 1,275 DEGs were extracted from
the GSE165934 database, and the PPI network of DEGs yielded 460
nodes. The enriched pathways of node genes included the
‘glucocorticoid receptor pathway’ and the ‘Toll-like receptor
signaling pathway’. Glucocorticoids are a common therapy for
controlling airway inflammation in asthma and work by their
attachment to intracellular glucocorticoid receptors, thereby
promoting enhanced production of anti-inflammatory genes and
blocking the activation of pro-inflammatory genes in asthmatic
airways (27). Inhaled
corticosteroids have been successful in treating the majority of
asthmatic patients, improving lung function and reducing
exacerbations (28,29). Toll-like receptors are essential for
identifying invading pathogens and activating the immune system
(30,31). Wu et al (32) showed that a combination of Toll-like
receptor-related genes could be a promising indicator for asthma
prognosis. In addition, a study by Pandey et al (33) found genetic variants in the
Toll-like receptor signaling pathway related to childhood asthma.
Further research on the molecular mechanisms of these pathways
could be crucial for developing asthma therapies.
Combining genes regulated by the regulation of
Toll-like receptor signaling pathway, TCGA-DEGs and OS analysis, 6
genes (IL1B, JUN, TLR2, TLR4, TLR8 and TREM1) were identified for
further analysis. Mutation profiling showed that patients with LUAD
and LUSC exhibited different types of mutations. TLR4 had the
highest mutation frequency in patients with LUAD and LUSC (68%),
followed by TLR8 (19%), TLR2 (9%), TREM1 (6%), JUN (4%), and IL1B
(4%). Poltorak et al (34)
reported that disruptive mutations in TLR4 are associated with the
emergence of gram-negative sepsis while maintaining the majority of
immune system components. Additionally, it was discovered that the
majority of the missense mutations of these altered genes were
found in patients with LUAD and LUSC, and missense mutations with a
high frequency might alter the structure and function of proteins
(35), which suggested a possible
role in the pathogenesis of LUAD and LUSC. In the present study, in
patients with LUAD and LUSC, SNPs were the primary mutation variant
type, and C>T was the most common DNA nucleotide substitution
compared with other SNV classes.
Through a series of bioinformatics analyses, TREM1
was discovered as the hub gene related to both asthma and lung
cancer, suggesting it could serve as a prognostic indicator of lung
cancer. Five members of the immunoglobulin superfamily make up the
TREM family, including TREM1, TREM2, TREM3, and TREM-like
transcripts-1 (TLT1) and −2 (TLT2) (36). TREM1 and TREM2 are immunoglobulin
superfamily receptors that typically regulate innate immunity
through inflammatory responses (37). Liu et al (38) showed that peripheral TREM1 induction
amplified pro-inflammatory responses to the brain- and gut-derived
immunogenic components after a stroke. Bernal-Martínez et al
(39) suggested that TREM1
performed a significant role in the pathophysiology of acute
inflammatory disorders with various etiologies, including acute
myocardial infarction, atherosclerosis and viral illnesses. Chen
et al (40) showed that
TREM1/Dap12-based chimeric antigen receptor-T cells exhibited
powerful anticancer activity both in vitro and in
vivo by designing a chimeric immune receptor. Furthermore,
TREM1 was differentially related to the clinical features of
patients with LUSC and LUAD. In the present study, it was found
that overexpression of TREM1 could block cell migration, invasion
and proliferation in lung cancer, and reduce the expression of
proteins related to the Toll-like receptor signaling pathway,
suggesting that TREM1 is a lung cancer suppressor gene involved in
the Toll-like receptor signaling pathway.
As immune cells are crucial for the development,
metastasis, prognosis and treatment of tumors (41), immune infiltration assays should be
performed to investigate how immune cells and tumors interact.
Immune checkpoint molecules expressed on immune cells can inhibit
immune cell activity and prevent the body from mounting successful
antitumor immune responses, leading to the development of tumor
immune escape (42). The present
study found that the expression levels of immune checkpoint genes
were higher in the high TREM1 expression group. TMB, which includes
the total amount of base substitution, insertion and deletion
mutations in somatic proteins, is also a critical prognostic
biomarker for immune checkpoint inhibitors in a number of cancer
types, such as lung cancer, melanoma and colorectal cancer
(43). Increased somatic mutation
can lead to neoantigen expression and tumorigenesis, which
activates CD8+ cytotoxic T cells and triggers the
antitumor effect of the T cell-dependent immune response (44). TMB has been recognized as a novel
biomarker of immunotherapy response and a candidate for the
prediction of response to immune checkpoint inhibitors (45). Cheng et al (46) reported that the degree of TREM1
expression was significantly inversely linked with TMB in NSCLC. A
high TMB score is considered to increase the number of neoantigens
that are present on the surface of tumor cells, enhancing
immunogenicity and improving the response of malignancies to immune
checkpoint inhibitor therapy. Therefore, the suppressor gene,
TREM1, may be used in a treatment for lung cancer.
The present bioinformatics-based approach identified
a hub gene, TREM1, involved in the molecular mechanism underlying
asthma and lung cancer. Further analysis showed that TREM1 was
downregulated in lung cancer cells as a tumor suppressor gene,
however, its overexpression could significantly reduce the
proliferation of lung cancer cells by regulating the Toll-like
receptor pathway. The present study reveals the pathogenesis
between asthma and lung cancer, and provides a new potential
biomarker for the treatment and prognosis of lung cancer. However,
the present study has certain limitations, as the results of the
current study have not been validated in samples from patients with
asthma and lung cancer. Additionally, the mechanism of action of
TREM1 downstream genes targeting the Toll-like receptor pathway in
lung cancer remains unclear and requires further analysis.
Acknowledgements
Not applicable.
Funding
Funding was provided by the Shanghai Medical Key Specialty
Project (grant. no. ZK2019B08).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZY conceived the idea for and designed the present
study. WZ acquired the data, and KS was responsible for data
analysis and interpretation. Statistical analysis was performed by
WZ and ZY. KS drafted the manuscript, and WZ, ZY and KS were
responsible for the revision of the manuscript for intellectual
content. WZ and KS confirm the authenticity of all the raw data.
All authors have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests
References
|
1
|
Dorantes-Heredia R, Ruiz-Morales JM and
Cano-Garcia F: Histopathological transformation to small-cell lung
carcinoma in non-small cell lung carcinoma tumors. Transl Lung
Cancer Res. 5:401–412. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Buttery R, Monaghan H, Salter DM and Sethi
T: Galectin-3: Differential expression between small-cell and
non-small-cell lung cancer. Histopathology. 44:339–344. 2010.
View Article : Google Scholar
|
|
3
|
Santillan AA, Camargo CA Jr and Colditz
GA: A meta-analysis of asthma and risk of lung cancer (United
States). Cancer Causes Control. 14:327–334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qu YL, Liu J, Zhang LX, Wu CM, Chu AJ, Wen
BL, Ma C, Yan XY, Zhang X, Wang DM, et al: Asthma and the risk of
lung cancer: A meta-analysis. Oncotarget. 8:11614–11620. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stading R, Gastelum G, Chu C, Jiang W and
Moorthy B: Molecular mechanisms of pulmonary carcinogenesis by
polycyclic aromatic hydrocarbons (PAHs): Implications for human
lung cancer. Semin Cancer Biol. 76:3–16. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rosenberger A, Bickeböller H, McCormack V,
Brenner DR, Duell EJ, Tjønneland A, Friis S, Muscat JE, Yang P,
Wichmann HE, et al: Asthma and lung cancer risk: A systematic
investigation by the international lung cancer consortium.
Carcinogenesis. 33:587–597. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang L, Sun YQ, Langhammer A, Brumpton
BM, Chen Y, Nilsen T, Leivseth L, Wahl SGF and Mai XM: Asthma and
asthma symptom control in relation to incidence of lung cancer in
the HUNT study. Sci Rep. 11:45392021. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Denholm R, Schüz J, Straif K, Stücker I,
Jöckel KH, Brenner DR, De Matteis S, Boffetta P, Guida F, Brüske I,
et al: Is previous respiratory disease a risk factor for lung
cancer? Am J Respir Crit Care Med. 190:549–559. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Garth J, Barnes J and Krick S: Targeting
cytokines as evolving treatment strategies in chronic inflammatory
airway diseases. Int J Mol Sci. 19:34022018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Toskala E and Kennedy DW: Asthma risk
factors. Int Forum Allergy Rhinol. 5:S11–S16. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bjorksten B: Genetic and environmental
risk factors for the development of food allergy. Curr Opin Allergy
Clin Immunol. 5:249–253. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hunt LW, Frigas E, Butterfield JH, Kita H,
Blomgren J, Dunnette SL, Offord KP and Gleich GJ: Treatment of
asthma with nebulized lidocaine: A randomized, placebo-controlled
study. J Allergy Clin Immunol. 113:853–859. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhu Y, Davis S, Stephens R, Meltzer PS and
Chen Y: GEOmetadb: Powerful alternative search engine for the Gene
Expression Omnibus. Bioinformatics. 24:2798–2800. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wen S, Li F, Tang Y, Dong L, He Y, Deng Y
and Tao Z: MIR222HG attenuates macrophage M2 polarization and
allergic inflammation in allergic rhinitis by targeting the
miR146a-5p/TRAF6/NF-κB axis. Front Immunol. 14:11689202023.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Simonson MA, Mcqueen MB and Keller MC:
Plot generated using STRING 9.0 (Search Tool for the Retrieval of
Interacting Genes). 2014.
|
|
16
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xu Y, Shu D, Shen M, Wu Q, Peng Y, Liu L,
Tang Z, Gao S, Wang Y and Liu S: Development and validation of a
novel PPAR signaling pathway-related predictive model to predict
prognosis in breast cancer. J Immunol Res. 2022:94121192022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeng Z, Yu J, Yang Z, Du K, Chen Y and
Zhou L: Investigation of M2 macrophage-related gene affecting
patients prognosis and drug sensitivity in non-small cell lung
cancer: Evidence from bioinformatic and experiments. Front Oncol.
12:10964492022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang W, Hong HJ and Chen YL:
Establishment of a gallbladder cancer-specific survival model to
predict prognosis in non-metastatic gallbladder cancer patients
after surgical resection. Dig Dis Sci. 63:2251–2258. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu ZH and Yang DL: High CENPM mRNA
expression and its prognostic significance in hepatocellular
carcinoma: A study based on data mining. Cancer Cell Int.
20:4062020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Q, Xia T, Qi C, Du J and Ye C: High
expression of S100A2 predicts poor prognosis in patients with
endometrial carcinoma. BMC Cancer. 22:772022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li B, Li T, Liu JS and Liu XS:
Computational deconvolution of tumor-infiltrating immune components
with bulk tumor gene expression data. Methods Mol Biol.
2120:249–262. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sistek D, Tschopp JM, Schindler C,
Brutsche M, Ackermann-Liebrich U, Perruchoud AP and Leuenberger P:
Clinical diagnosis of current asthma: Predictive value of
respiratory symptoms in the SAPALDIA study. Swiss study on air
pollution and lung diseases in adults. Eur Respir J. 17:214–219.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hom S and Pisano M: Reslizumab (Cinqair):
An interleukin-5 antagonist for severe asthma of the eosinophilic
phenotype. P T. 42:564–568. 2017.PubMed/NCBI
|
|
26
|
van der Vliet A, Janssen-Heininger YMW and
Anathy V: Oxidative stress in chronic lung disease: From
mitochondrial dysfunction to dysregulated redox signaling. Mol
Aspects Med. 63:59–69. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Henderson I, Caiazzo E, McSharry C, Guzik
TJ and Maffia P: Why do some asthma patients respond poorly to
glucocorticoid therapy? Pharmacol Res. 160:1051892020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Barnes PJ: Efficacy of inhaled
corticosteroids in asthma. J Allergy Clin Immunol. 102:531–538.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Raissy HH, Kelly HW, Harkins M and Szefler
SJ: Inhaled corticosteroids in lung diseases. Am J Respir Crit Care
Med. 187:798–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Miller LS: Toll-like receptors in skin.
Adv Dermatol. 24:71–87. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kaisho T and Akira S: Toll-like receptor
function and signaling. J Allergy Clin Immunol. 117:979–987. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu X, Wang P, Zhang Y, Gao L, Zheng B, Xu
Y and Mo J: Toll-like receptor characterization correlates with
asthma and is predictive of diagnosis. DNA Cell Biol. 39:1313–1321.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pandey RC, Michel S, Tesse R, Binia A,
Schedel M, Liang L, Klopp N, Franke A, von Berg A, Bufe A, et al:
Genetic variation in the toll-like receptor signaling pathway is
associated with childhood asthma. J Allergy Clin Immunol.
131:602–605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Poltorak A, He X, Smirnova I, Liu MY,
Huffel CV, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al:
Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations
in Tlr4 gene. Science. 282:2085–2088. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gnad F, Baucom A, Mukhyala K, Manning G
and Zhang Z: Assessment of computational methods for predicting the
effects of missense mutations in human cancers. BMC Genomics.
14:1–13. 2013. View Article : Google Scholar
|
|
36
|
Pelham CJ, Pandya AN and Agrawal DK:
Triggering receptor expressed on myeloid cells receptor family
modulators: A patent review. Exp Opin Ther Pat. 24:1383–1395. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun H, Feng J and Tang L: Function of
TREM1 and TREM2 in liver-related diseases. Cells. 9:26262020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Q, Johnson EM, Lam RK, Wang Q, Ye HB,
Wilson EN, Minhas PS, Liu L, Swarovski MS, Tran S, et al:
Peripheral TREM1 responses to brain and intestinal immunogens
amplify stroke severity. Nat Immunol. 20:1023–1034. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bernal-Martínez L, Gonçalves SM, de Andres
B, Cunha C, Jimenez IG, Lagrou K, Mellado E, Gaspar ML, Maertens
JA, Carvalho A and Alcazar-Fuoli L: TREM1 regulates antifungal
immune responses in invasive pulmonary aspergillosis. Virulence.
12:570–583. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Chen B, Zhou M, Zhang H, Wang C, Hu X,
Wang B and Wang E: TREM1/Dap12-based CAR-T cells show potent
antitumor activity. Immunotherapy. 11:1043–1055. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mlecnik B, Bindea G, Pagès F and Galon J:
Tumor immunosurveillance in human cancers. Cancer Metastasis Rev.
30:5–12. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ribas A: Adaptive immune resistance: How
cancer protects from immune attackadaptive immune resistance.
Cancer Discov. 5:915–919. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chan TA, Yarchoan M, Jaffee E, Swanton C,
Quezada SA, Stenzinger A and Peters S: Development of tumor
mutation burden as an immunotherapy biomarker: Utility for the
oncology clinic. Ann Oncol. 30:44–56. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Palucka AK and Coussens LM: The basis of
oncoimmunology. Cell. 164:1233–1247. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Klempner SJ, Fabrizio D, Bane S, Reinhart
M, Peoples T, Ali SM, Sokol ES, Frampton G, Schrock AB, Anhorn R
and Reddy P: Tumor mutational burden as a predictive biomarker for
response to immune checkpoint inhibitors: A review of current
evidence. Oncologist. 25:e147–e159. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cheng X, Wang X, Nie K, Cheng L, Zhang Z,
Hu Y and Peng W: Systematic pan-cancer analysis identifies TREM2 as
an immunological and prognostic biomarker. Front Immunol.
12:6465232021. View Article : Google Scholar : PubMed/NCBI
|