Introduction
Currently, most end-stage disorders can be
successfully treated by organ transplantation, including heart
transplantation, liver transplantation and kidney transplantation
(1–3). However, recipients of organ
transplants require a lengthy course of immunosuppressive therapy,
which may increase the frequency of malignancies (1,4).
Notably, previous studies have shown that patients who have
undergone organ transplantation have a 2–8 times higher incidence
of malignant melanoma than those who are immunocompetent (5–8).
Post-transplantation malignancy seriously affects the quality of
life of patients, and it is one of the main causes of death among
transplant recipients (9). A
previous study showed that of the 126,474 deaths in the United
States after solid organ transplants between 1987 and 2018, 13%
were due to cancer (10).
Therefore, the search for drugs that have both anti-rejection and
antitumor properties is essential.
Rapamycin, a macrolide antibiotic, exerts its
immunosuppressive function mainly by acting on mammalian target of
rapamycin (mTOR) (11). Unlike
calcineurin inhibitors, which may increase tumor incidence after
transplantation (12), rapamycin
can be applied to inhibit tumor spread and recurrence following
organ transplantation (13–15). Rapamycin has been shown to inhibit
the growth of a mouse hepatocellular carcinoma xenograft model by
targeting STAT3 and affecting c-Myc, in addition to inhibiting
angiogenesis in an in vivo CT-26 cell model of liver
metastasis (16,17). In addition, rapamycin has been
reported to inhibit tumor growth in a mouse model of breast cancer
established using the MC4-L2 cell line, and to inhibit the
viability of kaposiform hemangioendothelioma primary cells and
human osteosarcoma MG-63 cells by inducing autophagy and apoptosis
(18–20). For different tumor types, rapamycin
may act in diverse ways. Malignant melanoma is easily metastasized
and has a poor prognosis once it has progressed to an advanced
stage (21). Research has found
that the median duration from transplant to melanoma diagnosis in
the individuals who have undergone lung and heart transplants is
2.5 years (22). Some studies have
shown that rapamycin inhibits the growth of A375 malignant melanoma
in an in vitro model (23,24).
However, the mechanism of action of the effects of rapamycin on
malignant melanoma remains to be explored.
The present study aimed to examine the effects of
rapamycin on cell viability, cell apoptosis, cell cycle
progression, and cellular autophagy and related signaling pathways
in B16-F10 (B16) murine melanoma cells. Additionally, in
vivo experiments were performed to verify the anti-melanoma
effect of rapamycin.
Materials and methods
Cell culture
Mouse B16 melanoma cells purchased from EK
Biosciences GmbH were cultured in Gibco Dulbecco's modified Eagle's
medium supplemented (cat. no. C1995500BT) with 1%
penicillin-streptomycin and 10% gibco fetal bovine serum (cat. no.
10099-141) (Thermo Fisher Scientific, Inc.) in an incubator at 37°C
and 5% CO2. The medium was changed every 1–2 days, and
when the cells covered 90% of the culture flasks, they were
trypsinized and passaged. All cell lines tested negative for
Mycoplasma contamination.
Reagents and drugs
The Annexin V-FITC/PI double staining cell apoptosis
detection kit (cat. no. BD556547) was purchased from BD
Biosciences. Mouse GAPDH monoclonal antibody (cat. no. 60004-1-Ig),
mouse Beclin-1 monoclonal antibody (cat. no. 66665-1-Ig), mouse
caspase 3/p17/p19 monoclonal antibody (cat. no. 66470-2-Ig), mouse
Bax monoclonal antibody (cat. no. 60267-1-Ig), mouse Bcl2
monoclonal antibody (cat. no. 68103-1-Ig), mouse cyclin D1
monoclonal antibody (cat. no. 60186-1-Ig), mouse CDK4 monoclonal
antibody (cat. no. 66950-1-Ig), mouse CDK6 monoclonal antibody
(cat. no. 66278-1-Ig), mouse CDK2 monoclonal antibody (cat. no.
60312-1-Ig), rabbit cyclin E1 polyclonal antibody (cat. no.
11554-1-AP), rabbit microtubule-associated protein light chain 3
(LC3) polyclonal antibody (cat. no. 14600-1-AP), goat anti-rabbit
IgG-HRP (cat. no. SA00001-2) and goat anti-mouse IgG-HRP (cat. no.
SA00001-1) antibodies were purchased from ProteinTech Group, Inc.
Rabbit sequestosome 1/p62 (cat. no. AF5384), mTOR (cat. no.
AF6308), p70 ribosomal S6 kinase (p70-S6k; cat. no. AF6226),
eukaryotic initiation factor 4E-binding protein (4E-BP1; cat. no.
AF6432), phosphorylated (p)-mTOR (cat. no. AF3308), p-p70-S6k (cat.
no. AF3228) and p-4E-BP1 (cat. no. AF3830) polyclonal antibodies
were purchased from Affinity Biosciences. Thiazolyl Blue (MTT; cat.
no. HY-15924), rapamycin (cat. no. HY-10219) and chloroquine (CQ;
cat. no. HY-17589A), an autophagy inhibitor that can block the
degradation of LC3 and p62 by inhibiting the fusion of
autophagosomes and lysosomes, were purchased from MedChemExpress.
Rapamycin was configured with DMSO into 10°, 101,
102, 103, 104, 105,
106, 107 and 108 nM concentrated
reservoirs (later diluted), and stored frozen at −20 or −80°C for
in vitro cell experiments. For in vivo animal
studies, the drug solution was prepared in anhydrous ethanol and
phosphate-buffered saline (PBS) at a concentration of 1 mg/ml and
stored at −20°C, ready to use.
In vitro cell viability assay
Cell suspensions made from logarithmic cells were
inoculated into 96-well culture plates (20,000 cells/well).
Solubilized rapamycin solution (final drug concentrations,
10−3, 10−2, 10−1, 10°,
101, 102, 103, 104 and
105 nM) was added. After 48 h at 37°C, 20 µl MTT working
solution (5 g/l) was added to each well and the cells were
incubated at 37°C for 4 h. After discarding the supernatant, the
purple formazan crystals were solubilized with 150 µl DMSO.
Finally, the optical density was measured at 492 nm using a plate
reader (Multiskan FC; Thermo Fisher Scientific, Inc.).
Cell apoptosis assay
B16 cells pretreated with different concentrations
(0, 0.1, 1, 10 and 100 nM) of rapamycin for 48 h at 37°C were
collected. The supernatant was collected from the 6-well plate, and
~1×105 cells/well were digested with 0.25% EDTA-free
trypsin after washing with PBS. After termination of digestion, the
cells were centrifuged at 500 × g for 5 min at 4°C. The cells were
then washed twice with PBS, 500 µl binding buffer was added, and
the cells were incubated with Annexin V-FITC (5 µl) and PI (5 µl)
at room temperature for 15 min in the dark. Finally, the stained
cells were detected by flow cytometry (NovoCyte 2040R with FlowJo
v10.9; ACEA Bioscience, Inc.) within 2 h (25).
Cell cycle analysis
The culture fluid was collected from the cells into
a centrifuge tube and set aside for termination of digestion, and
1×105 cells/well were digested with 0.25% trypsin after
PBS rinsing. After the cells were collected into a centrifuge tube,
they were centrifuged at 4°C for 3 min at 1,000 × g. The cells were
washed twice with pre-cooled PBS and added to 1 ml pre-cooled 70%
ethanol. The cells were gently aspirated and blown with a spiking
gun to prevent clumps, then mixed and fixed at 4°C for 12 h. The
cells were centrifuged for a further 3 min at 1,000 × g and 4°C.
The supernatant was then aspirated and washed once with PBS.
Subsequently, 500 µl staining buffer (Cell Cycle Kit; cat. no.
C1052; Beyotime Institute of Biotechnology), 25 µl PI staining
solution (20 X) and 10 µl RNase A (50X) was added to each sample.
After incubation at room temperature for 30 min, PI staining was
examined by flow cytometry (NovoCyte 2040R with FlowJo v10.9; ACEA
Bioscience, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from the B16 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), and the A260/A280 of total RNA in each group was determined.
A PrimeScript® RT Reagent Kit (cat. no. RR047A; Takara
Bio, Inc.) was used to reverse transcribe RNA into cDNA. qPCR was
performed on a real-time PCR detection instrument (Light Cycler
480II; Roche Diagnostics) with a TB Green® Premix Ex
Taq™ II (Tli RNaseH Plus) cat. no. RR820A; Takara Bio, Inc.)
following standard procedures. The thermal cycling conditions were
as follows: 95°C for 30 sec of pre-denaturation; followed by 40
cycles at 95°C for 5 sec and 60°C for 30 sec; and a final
incubation at 95°C for 5 sec and 60°C for 1 min. Finally, the
reaction was cooled to 50°C in 30 sec. The relative expression
levels of autophagy-related genes were quantified using the
2−ΔΔCq (26) method
using GAPDH as a control. The mRNA expression levels were
normalized to those of GAPDH. The primer sequences are shown in
Table I.
 | Table I.Sequences of primers used in the
present study. |
Table I.
Sequences of primers used in the
present study.
| Gene | Primer sequence
(5′-3′) | Accession no. |
|---|
| Mouse GAPDH | F:
GGTTGTCTCCTGCGACTTCA | NM_008084 |
|
| R:
TGGTCCAGGGTTTCTTACTCC |
|
| Mouse LC3 II | F:
CAAGCCTTCTTCCTCCTGGTGAATG | NM_025735.3 |
|
| R:
CCATTGCTGTCCCGAATGTCTCC |
|
| Mouse Beclin-1 | F:
GACGAACTCAAGAGTGTGGAGAACC | NM_001359819.1 |
|
| R:
AGATGTGGAAGGTGGCATTGAAGAC |
|
| Mouse p62 | F:
TTCCAGCACAGGCACAGAAGAC | NM_001290769.1 |
|
| R:
TCCCACCGACTCCAAGGCTATC |
|
Transmission electron microscopy
(TEM)
A total of 200,000 B16 cells per well were
inoculated in 6-well culture plates and allowed to grow for 48 h.
TEM was used to examine the morphology of B16 cells under 0, 1 and
10 nM rapamycin treatment for 48 h at 37°C. Briefly, cells were
immediately fixed in electron microscopy fixative (3% osmium
tetroxide) for 2 h at 4°C. After low-speed centrifugation for 3 min
at 1,000 × g and 4°C, they were rinsed three times with PBS (pH
7.4). Subsequently, the cells were dehydrated in a graded ethanol
series, then permeabilized and embedded in acetone, and cut into
60–80 nm ultrathin sections. Sections were double-stained with
uranium-lead (2% uranyl acetate saturated alcohol solution and 3%
lead citrate, each for 15 min at room temperature 25°C), and
sections were dried at room temperature overnight. The sections
were finally observed under TEM (HT7700; Hitachi, Ltd.) and images
were collected for analysis.
Western blotting
Lysis buffer (cat. no. P0013B; Beyotime Institute of
Biotechnology) containing 1 mM phenylmethanesulfonyl fluoride was
used to extract total cellular or mouse tumor tissue proteins after
treatment, and the bicinchoninic acid protein assay kit (cat. no.
P0010; Beyotime Institute of Biotechnology) was used to measure
protein concentration. SDS-PAGE at a 12% concentration was used to
separate equal amounts of 0.8 µg/µl of protein samples, which were
then transferred to PVDF membranes. The membranes were blocked for
15 min at room temperature in Rapid Blocking Solution (cat. no.
P0252; Beyotime Institute of Biotechnology) and incubated for 2 h
at room temperature with different primary antibodies against
Beclin-1, LC3, p62, CDK1, caspase 3, Bax, Bcl2, cyclin D1, CDK4,
CDK6, CDK2, cyclin E1, mTOR, p-mTOR, p70-S6k, p-p70S6k, 4E-BP1,
p-4E-BP1 (dilution for all, 1:1,500) and GAPDH (dilution,
1:10,000). Membranes were then washed three times with PBS-0.05%
Tween 20 and then incubated with HRP-coupled anti-rabbit or
anti-mouse secondary antibodies (dilution of both, 1:10,000) for 2
h at room temperature. After PBS washing three times, protein bands
were observed using a protein blotting assay kit (Applygen
Technologies Inc.) with ultrasensitive enhanced chemiluminescence,
and ImageJ software (National Institutes of Health) was used for
semi-quantitative analysis of protein bands (27).
Animals
A total of 32 male C57BL/6 mice (age, 8 weeks;
weight, 18 g) were purchased from the Guangdong Medical Laboratory
Animal Centre. The mice were housed in an environment of 50–60%
relative humidity and 21–25°C, with an alternating light and dark
cycle of 10/14 h. The mice had free access to clean water and food.
The present study was performed in strict accordance with the
recommendations in the Guide for the Care and Use of Laboratory
Animals of the National Institutes of Health, Eighth Edition, 2010,
and also complied with the ARRIVE guidelines and the AVMA
euthanasia guidelines 2020 (28,29).
The animal experiments were approved by the Ethics Committee of
Hainan Medical University (Haikou, China; approval no. HYLL:
2022-227). All protocols were in accordance with the approved
guidelines and regulations.
Model building and treatment
The abdominal hair of the mice was first shaved and
sterilized, and then B16 cells (1×106 in 100 µl) were
injected subcutaneously into the right abdomen of 8-week-old male
C57BL/6 mice to establish a subcutaneous tumor transplantation
model. These mice were randomly and evenly grouped into four groups
(n=8 mice/group). To ensure successful inoculation, tumor length
was regularly observed and recorded daily. To assess the inhibitory
effect of rapamycin on B16 melanoma cells in vivo, the mice
were treated with different doses of rapamycin (1, 1.5 and 2
mg/kg/day) for 12 days, and the control group was injected
intraperitoneally with an equal amount of PBS. When the tumor
diameter of control mice reached the execution criteria (15 mm),
all mice were euthanized by intravenous injection with an overdose
of anesthetic (sodium pentobarbital, 150 mg/kg). Humane endpoints
included malignancy, ulceration and necrosis. Tumors were removed
and weighed. Tumor volume was measured as follows: Tumor volume
(mm3)=length × width × width/2.
Immunohistochemistry
The tumor tissues were fixed with 4%
paraformaldehyde at room temperature for 24 h, paraffin embedded
and cut into 4-µm sections. The sections were deparaffinized with
xylene for 15 min twice and subsequently placed in 100, 95 and 80%
ethanol, and sequentially rehydrated for 10 min. PBS-rinsed
sections were immersed in 0.1 mol/l citrate (pH 6.0) and incubated
for 15 min in a microwave oven for antigen repair. After the
sections were cooled at room temperature for 10 min, incubation
with 3% H2O2-PBS for 10 min at room
temperature was used to eliminate endogenous peroxidase activity.
Next, 5% BSA (cat. no. ST023; Beyotime Institute of Biotechnology)
was applied at 37°C to close the sections for 1 h. Subsequently,
the sections were incubated with primary antibodies against the
autophagy marker proteins LC3 (dilution, 1:200) and p62 (dilution,
1:200) at 37°C for 1 h. PBS was used to replace the first
antibodies in the blank control. After incubating with
HRP-conjugated goat anti-rabbit (dilution, 1:300) at 37°C for 20
min, tissue sections were stained with 3,3-diaminobenzidine and
hematoxylin at room temperature for 5 min in turn. After being
dehydrated and mounted, the tissues were observed under a
fluorescence inverted microscope. Relative expression was assessed
by measuring the average optical density of positive responses in
each group using ImageJ software.
TUNEL fluorescence staining of
paraffin-embedded tissue sections
The paraffin-embedded sections were deparaffinized
and washed twice with PBS (5 min/wash). After shaking the sections
dry, proteinase K working solution (cat. no. 11684795910;
MilliporeSigma) was applied dropwise and the sections were
incubated for 25 min at 37°C before being washed twice with PBS.
The sections underwent two PBS washes after being dried once more
and treated with a membrane-breaking working solution for 10 min at
room temperature. The sections were treated with a 50-µl 1:9
mixture of reagents TdT and Cy3-dUTP from the TUNEL kit (cat. no.
11684817910; Roche Diagnostics) at 37°C for 1 h. After adding the
DAPI staining solution and washing the sections three times in PBS
(5 min/wash), they were maintained for 10 min at room temperature
in the dark. After submerging the slides in PBS and shaking them
three times (5 min each), the sections were blocked with an
anti-fluorescence quenching sealing agent at 37°C for 2 min once
the sections had somewhat dried. Using a fluorescence microscope
(DS-Fi3; Nikon Corporation), sections were examined and images were
captured.
Statistical analysis
All experiments were conducted at least three times
and data are presented as the mean ± standard deviation. GraphPad
9.5 software (Dotmatics) was used for statistical analysis.
Multiple groups were compared using one-way ANOVA followed by
Tukey's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Rapamycin reduces the viability of B16
cells in vitro
To investigate the effects of rapamycin on B16
melanoma cell viability, the MTT assay was performed. B16 melanoma
cells were treated with a range of rapamycin concentrations (0,
10−3, 10−2, 10−1, 10°,
101, 102, 103, 104 and
105 nM) for 48 h. As shown in Fig. 1A, rapamycin at a concentration of
10−1 nM significantly reduced the viability of B16
melanoma cells compared with that in the control group. In
addition, the half-maximal inhibitory concentration of rapamycin in
B16 cells was 84.14 nM (Fig.
1B).
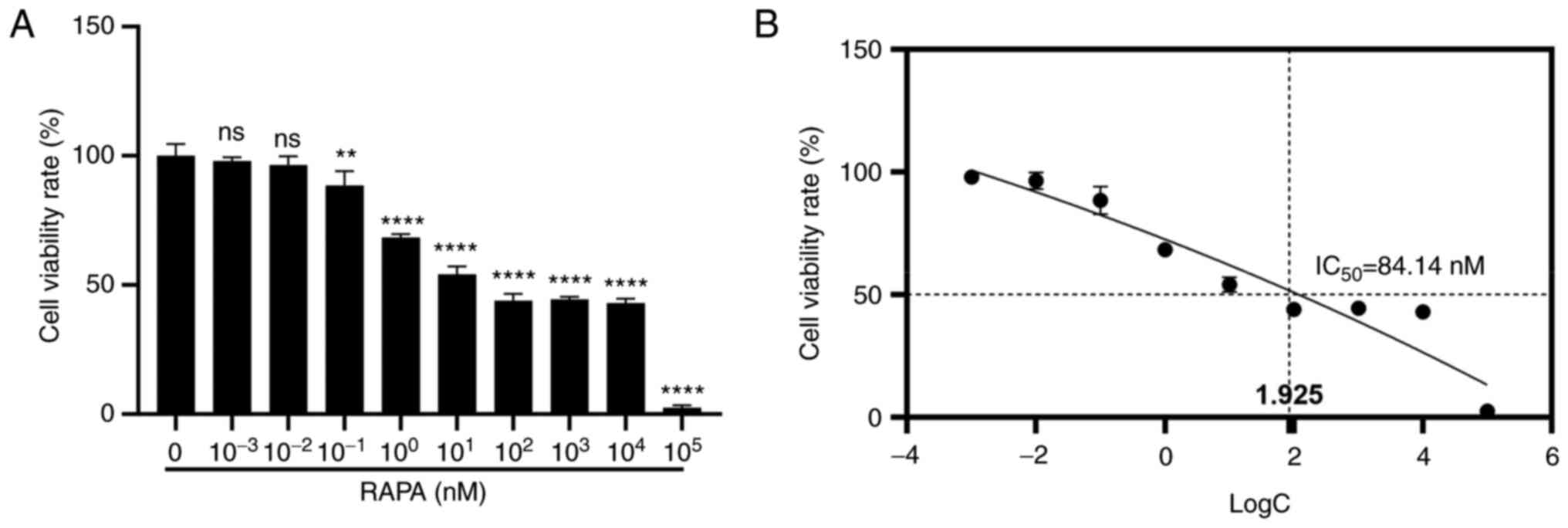 | Figure 1.RAPA inhibits B16 cell viability
in vitro. Cells were treated with RAPA for 48 h. (A) Cell
viability was detected by MTT assay. (B) IC50 value of
RAPA in B16 cells. Data are presented as the mean ± SD logC: for
10−3, 10−2, 10−1, 10°,
101, 102, 103, 104 and
105 nM take the logarithm of the base 10. (n=3).
**P<0.01 and ****P<0.0001 vs. RAPA 0 nM group. B16, B16-F10;
IC50, half-maximal inhibitor concentration; ns, not
significant; RAPA, rapamycin. |
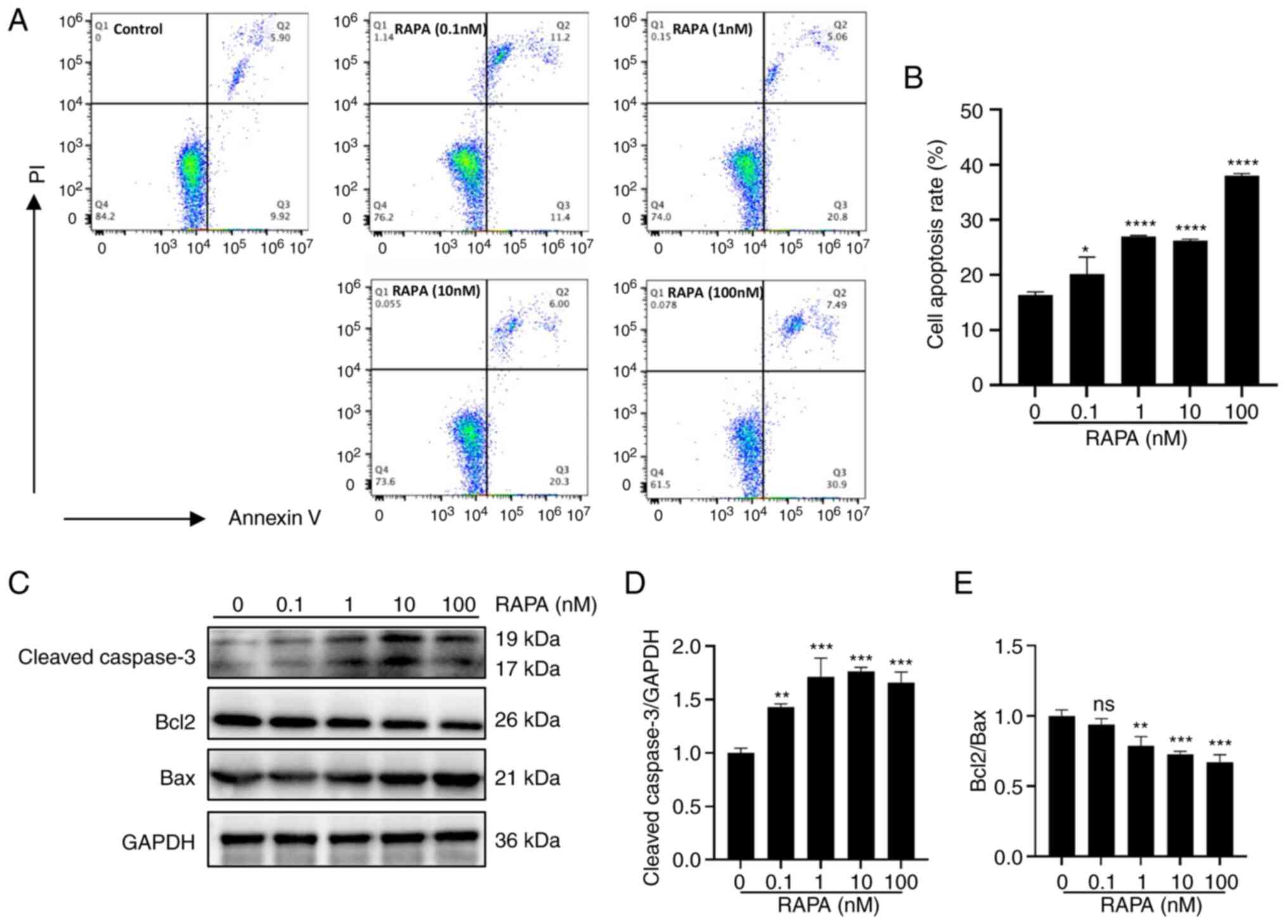
Rapamycin induces the apoptosis of B16
cells in vitro
To investigate the possible mechanism by which
rapamycin inhibits the viability of B16 cells, the present study
examined the apoptosis of B16 cells following rapamycin treatment
using flow cytometry. As shown in Fig.
2A and B, cell apoptosis was increased by rapamycin in the
concentration range of 0.1–100 nM (10−7−10−4
mM). In addition, rapamycin increased the protein expression levels
of cleaved caspase 3 and Bax, and decreased the protein expression
levels of Bcl2 compared with those in the control group (Fig. 2C-E). These results indicated that
rapamycin can induce the apoptosis of melanoma cells in
vitro.
Rapamycin induces cell cycle arrest in
B16 cells in vitro
After treating B16 cells with different
concentrations of rapamycin, the cell cycle distribution of B16
cells was assessed using flow cytometry. As shown in Fig. 3A and B, rapamycin induced cell cycle
arrest in B16 cells, with an increased proportion of cells in
G1 phase and a decreased proportion of cells in
G2/M phase compared with that in the control group.
Furthermore, rapamycin reduced the protein expression levels of
CDK1, cyclin D1 and CDK4, whereas it did not affect CDK6, cyclin E1
and CDK2 expression, compared with in the control group (Fig. 3C-I). These results suggested that
rapamycin may induce B16 cell cycle arrest at
G0/G1 phase.
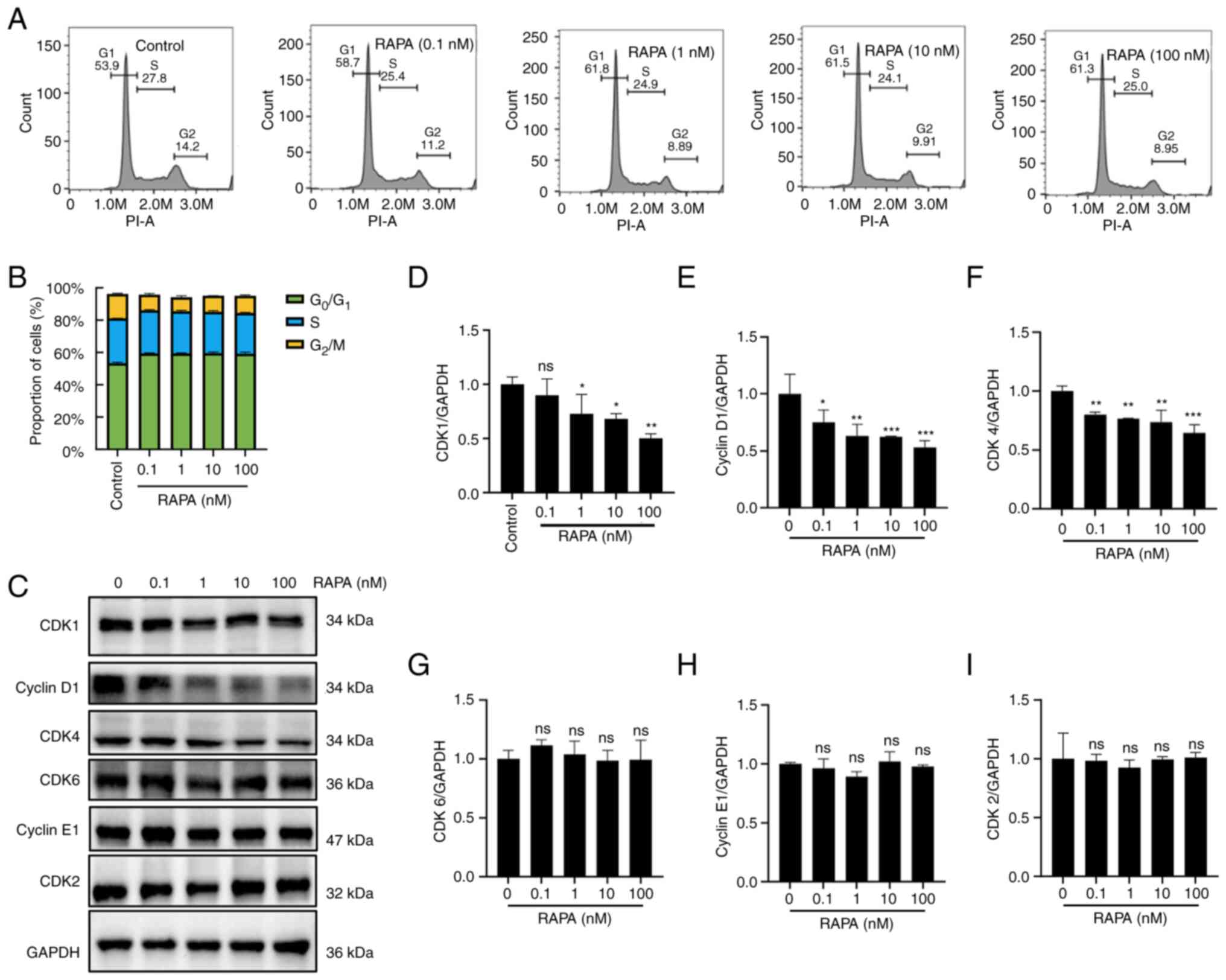 | Figure 3.Effects of RAPA on the cell cycle
progress of B16-F10 cells. (A) Cell cycle distribution was assessed
by flow cytometry. (B) Results of statistical analysis of cell
rates at different time periods for each group in (A). (C) CDK1,
cyclin D1, CDK4, CDK6, cyclin E1 and CDK2 protein expression levels
were detected by western blotting. Relative expression of (D) CDK1,
(E) Cyclin D1, (F) CDK4, (G) CDK6, (H) Cyclin E1 and (I) CDK2
proteins. Data are presented as the mean ± SD (n=3). *P<0.05,
**P<0.01 and ***P<0.001 vs. RAPA 0 nM group. ns, not
significant; RAPA, rapamycin. |
Rapamycin induces autophagy in B16
cells
Autophagic vesicles observed by TEM are the gold
standard for autophagy detection. In order to understand the effect
of rapamycin on cellular autophagy, TEM was performed to observe
the structure of the cells. The results revealed that the control
cells had intact cell membranes, intact nuclear membranes, and a
small number of autophagic vesicles and autophagic lysosomal
structures (Fig. 4A). By contrast,
B16 cells in the prominent 1 and 10 nM rapamycin groups had notably
more autophagic vesicles and autophagic lysosomes than the cells in
the control group. Moreover, cells in the 1 and 10 nM rapamycin
groups exhibited marked cell membrane breaks and mitochondrial
ridge breaks. Furthermore, at the gene and protein levels,
rapamycin increased LC3 and Beclin-1 expression, and decreased p62
expression compared with that in the control group (Fig. 4B-H). To determine whether rapamycin
enhanced autophagy, western blotting was used to detect the changes
in LC3 and p62 expression following treatment with CQ (an autophagy
inhibitor that blocks the degradation of LC3 and p62 by inhibiting
the fusion of autophagosomes and lysosomes). Rapamycin in
combination with CQ further increased LC3 and p62 expression
compared with that in the CQ group, suggesting that rapamycin
indeed induced an increase in autophagy in B16 cells (Fig. 4I-K).
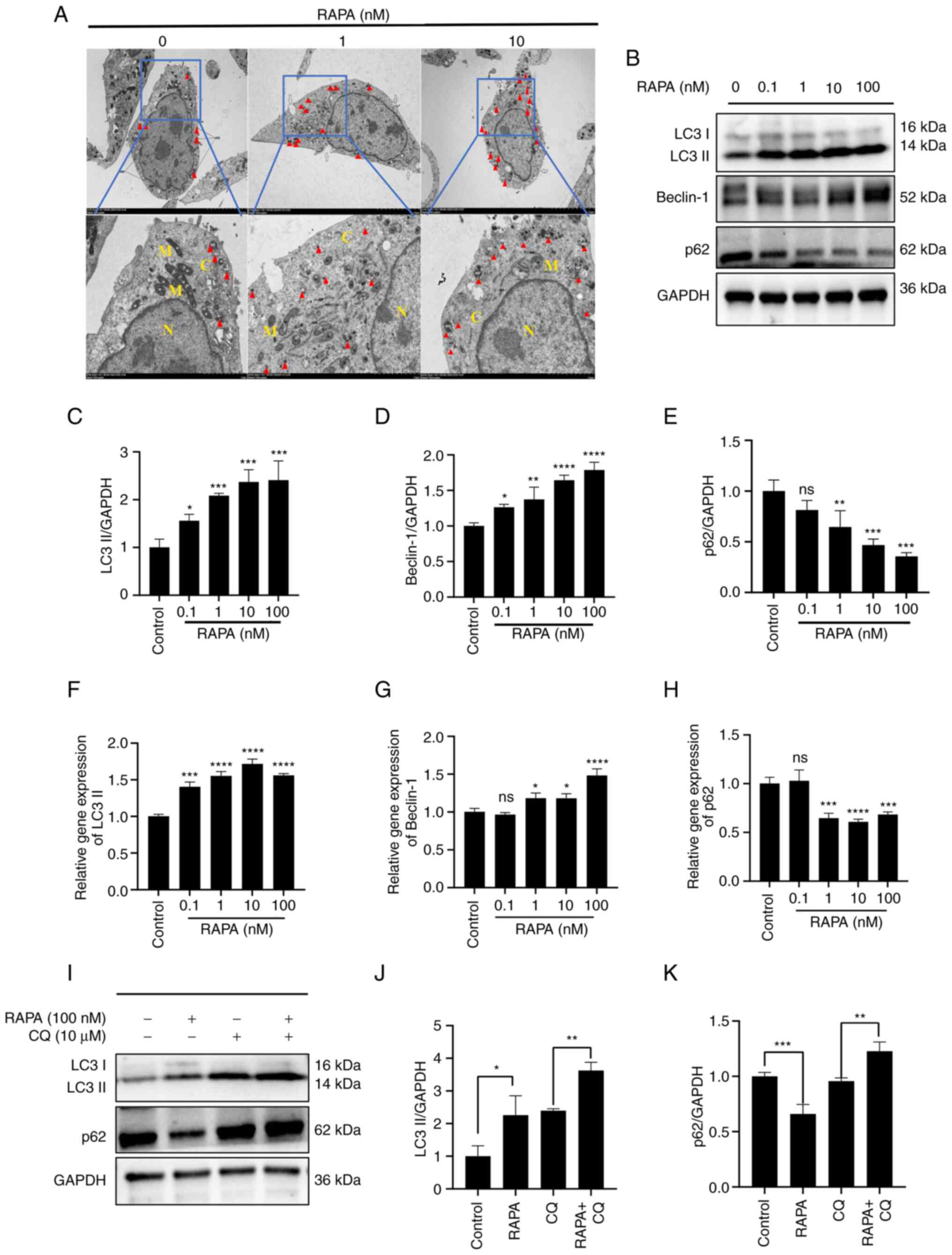 | Figure 4.RAPA induces autophagy in B16 cells.
(A) B16 cells were treated with 0, 1 and 10 nM RAPA for 48 h. The
formation of autophagic vesicles was observed by transmission
electron microscopy. M indicates mitochondrial structure, N the
nucleus, C the cytoplasm, and red triangles indicate autophagic
lysosomes. Magnification, ×8,000; scale bar, 2.0 nm. (B) Expression
levels of autophagy-related LC3, p62 and Beclin-1 in B16 cells were
detected by western blotting. Relative expression of (C) LC3II, (D)
Beclin-1 and (E) p62 proteins. Relative mRNA expression levels of
(F) LC3 II, (G) Beclin-1 and (H) p62. (I) B16 cells were treated
with rapamycin (100 nM) and/or CQ (10 µM) for 48 h. The protein
expression levels of LC3 and p62 were detected by western blotting.
(J) Relative expression of LC3II protein. (K) Relative expression
of LC3II protein. Data are presented as the mean ± SD (n=3).
*P<0.05, **P<0.01, ***P<0.001 and ****P<0.0001 vs. RAPA
0 nM group or as indicated. B16, B16-F10; CQ, chloroquine; LC3,
microtubule-associated protein light chain 3; ns, not significant;
RAPA, rapamycin. |
Rapamycin inhibits the mTOR/p70-S6k
signaling pathway in B16 cells
The mTOR/p70-S6k signaling pathway serves an
important role in the regulation of autophagy, cell proliferation
and cell survival in eukaryotic cells (30). To investigate the molecular
mechanism underlying rapamycin-induced cellular autophagy in B16
cells, the expression and activation of important components of the
mTOR signaling pathway, including p-mTOR, p-p70-S6k and p-4EBP1,
were assessed after 48 h of rapamycin treatment. As shown in
Fig. 5, the phosphorylation of mTOR
and p70-S6k was downregulated by rapamycin compared with that in
the control group, whereas there was no change in 4EBP1
phosphorylation. This result suggested that the mTOR/p70-S6k
signaling pathway may be inhibited by rapamycin.
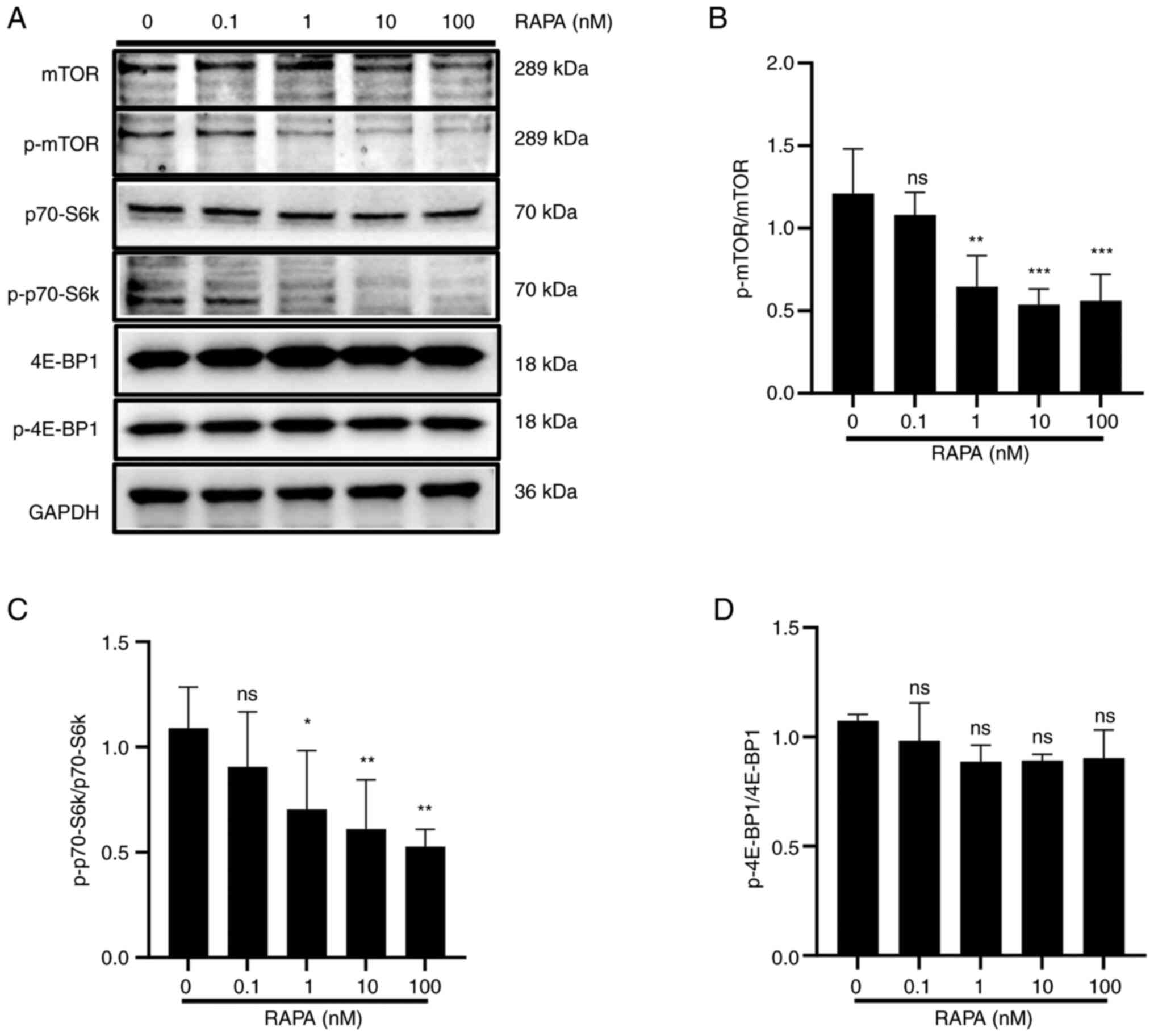 | Figure 5.RAPA inhibits the mTOR/p70-S6k
signaling pathway in B16 cells. B16 cells were treated with 0, 1,
10 and 100 nM RAPA for 48 h. (A) Western blotting results showed
that RAPA inhibited the expression of p-mTOR and p-p70-S6k compared
with in the control group. Statistical analysis of the (B)
p-mTOR/mTOR ratio, (C) the p-p70-S6k/p70-S6k ratio and (D) the
p-4E-BP1/4E-BP1 ratio at the protein level. Data are presented as
the mean ± SD (n=3). *P<0.05, **P<0.01 and ***P<0.001 vs.
RAPA 0 nM group. 4E-BP1, eukaryotic translation initiation factor
4e-binding protein 1; B16, B16-F10; mTOR, mammalian target of
rapamycin; ns, not significant; p70-S6k, p70 ribosomal S6 kinase;
p-, phosphorylated; RAPA, rapamycin. |
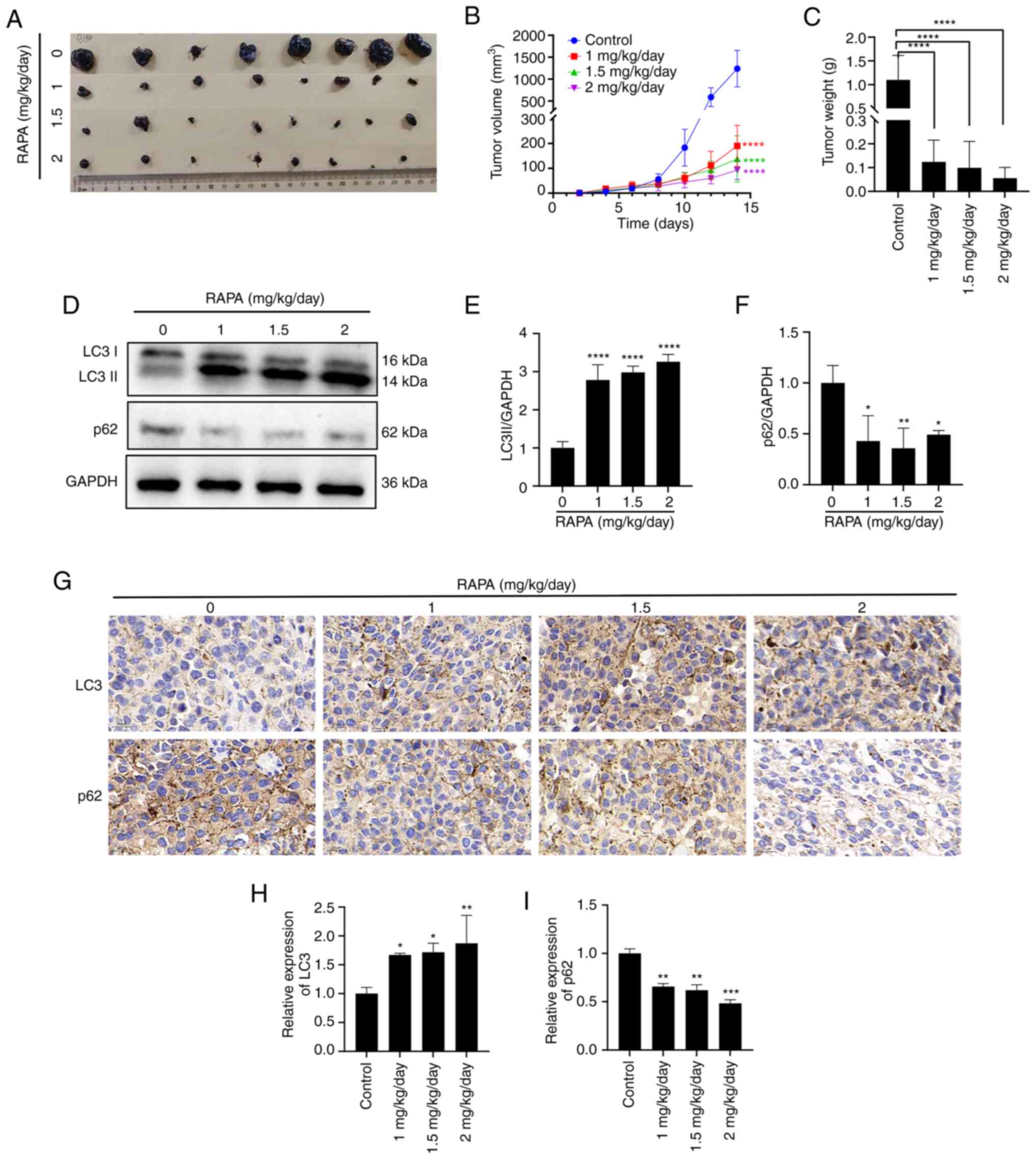
Rapamycin induces autophagy and
inhibits the growth of melanoma B16 cells in the C57BL/6 mouse
model
To determine whether rapamycin inhibits melanoma
growth in vivo, B16 cells were subcutaneously injected into
the abdomen of 8-week-old male mice, and rapamycin was administered
at 1, 1.5 or 2 mg/kg/day. As shown in Fig. 6A-C, rapamycin at 1, 1.5 and 2
mg/kg/day effectively inhibited B16 melanoma growth compared with
that in the control group. Western blotting results showed that the
protein expression levels of LC3 II were increased, whereas the
protein expression levels of p62 were decreased in
rapamycin-treated tumors compared with those in the control group
(Fig. 6D-F). Immunohistochemistry
results showed an increase in the area of LC3 II-positive regions
in rapamycin-treated tumors, along with a decrease in the area of
p62-positive regions compared with that in the control group
(Fig. 6G-I), which was consistent
with the western blotting results. These findings suggested that
rapamycin may induce B16 cell autophagy and suppress B16 melanoma
growth in vivo.
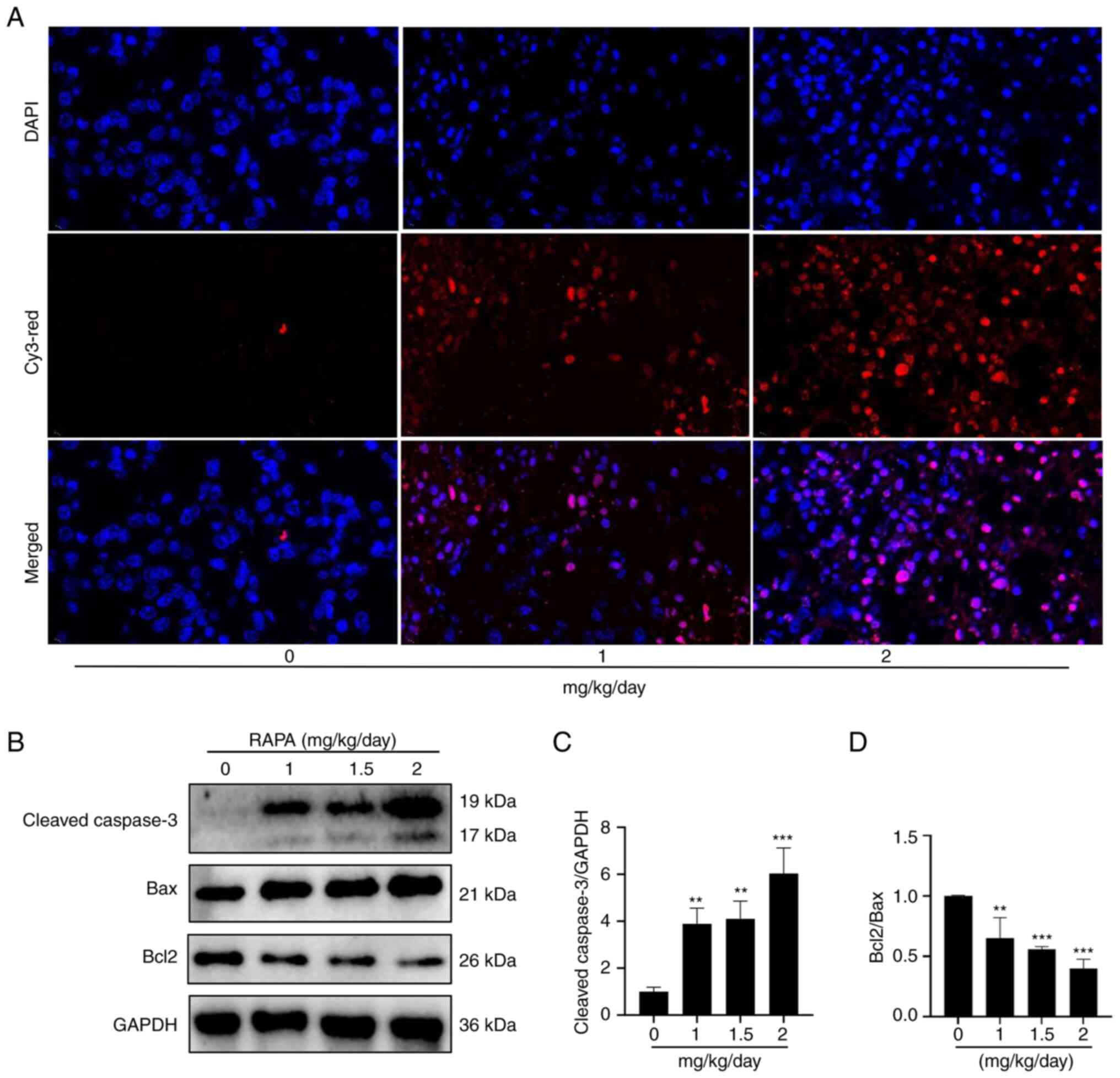
Rapamycin induces apoptosis in mouse
melanoma tumors
TUNEL fluorescent labeling and western blotting were
performed to detect apoptosis in tumor tissues to examine whether
rapamycin may trigger apoptosis in mice with melanoma in
vivo. TUNEL immunofluorescence results showed that rapamycin
promoted apoptosis of tumor cells in tumor tissues compared with
that in the control group (Fig.
7A). The results of western blotting demonstrated that
rapamycin induced a reduction in Bcl2 expression, and an increase
in the expression levels of cleaved caspase 3 and Bax compared with
those in the control group (Fig.
7B-D). These findings indicated that rapamycin may cause mouse
melanoma tumor cells to undergo apoptosis.
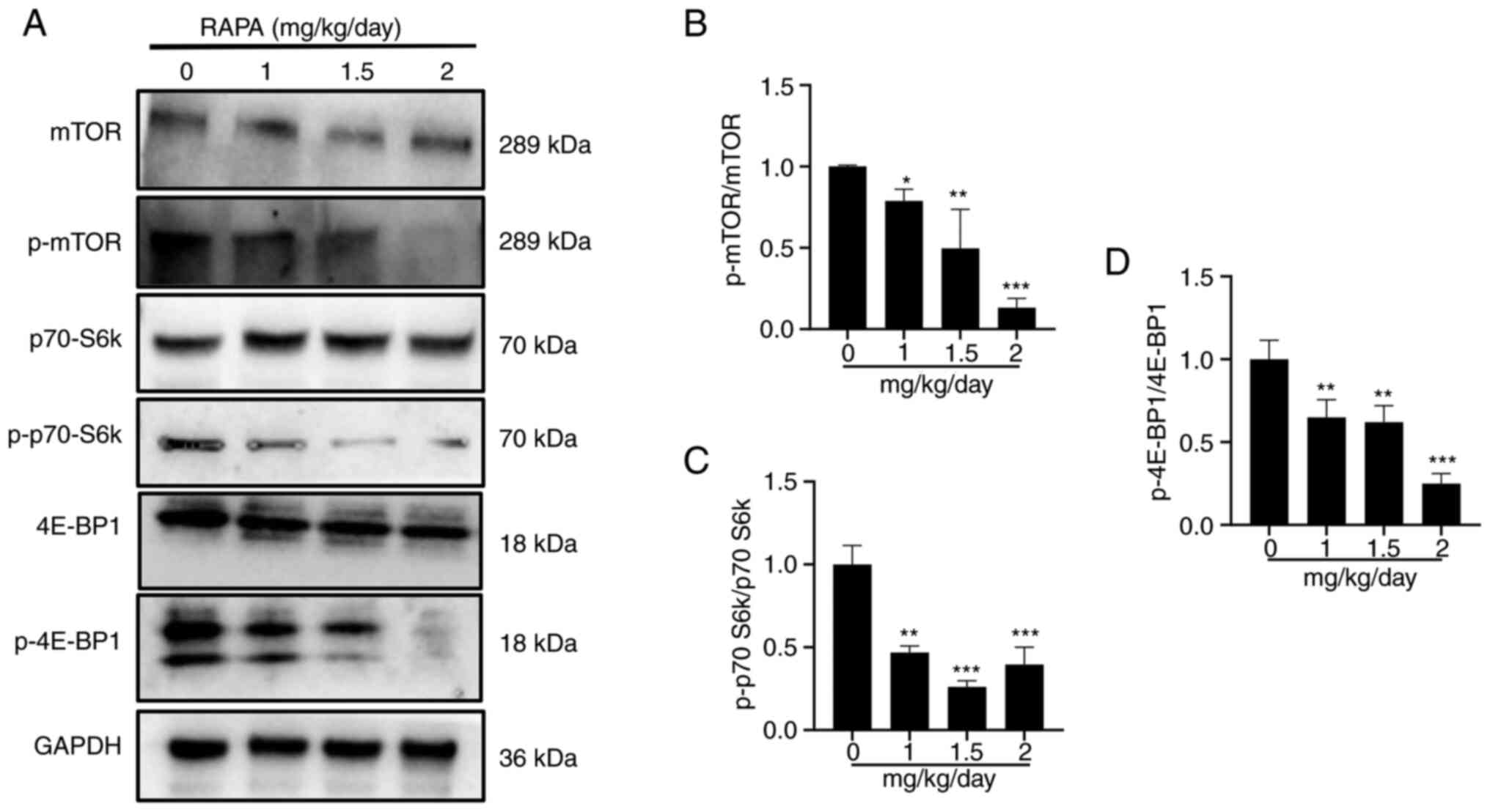
Rapamycin inhibits the
mTOR/p70-S6k/4E-BP1 signaling pathway in a mouse model
To determine if proteins associated with the
mTOR/p70-S6k signaling pathway were altered in tumors, western
blotting was performed. The administration of 1, 1.5 and 2
mg/kg/day rapamycin resulted in a decrease in the protein
expression levels of p-mTOR, p-P70 S6k and p-4E-BP1 in comparison
to those in the control group (Fig.
8A-D). These findings suggested that rapamycin can inhibit the
mTOR/p70-S6k/4E-BP1 signaling pathway in tumor tissues.
Discussion
Rapamycin, a powerful and specific mTOR inhibitor,
can specifically activate autophagy. Data from clinical studies
have indicated that rapamycin has promising outcomes in the
prevention or treatment of post-transplant tumor recurrence
(13–15,31).
However, the mechanism underlying the antitumor action of rapamycin
has not yet been fully elucidated. The present study demonstrated
that rapamycin may inhibit B16 melanoma cell growth by inducing
autophagy in a C57BL/6 mouse model.
Melanoma is a malignant tumor, the frequency of
which has quickly increased in recent decades (32). Melanoma is difficult to diagnose in
the early stages and is often fatal once it has progressed to an
advanced stage (33). Wang et
al (24) reported that
rapamycin can inhibit A375 melanoma cell growth. In the current
study, it was confirmed that rapamycin could inhibit the viability
of B16 melanoma cells both in vivo and in vitro.
The mechanism underlying the suppressive effects of
rapamycin on B16 melanoma cells is not well understood. Cell cycle
dysregulation is one of the key hallmarks of cancer cells. Numerous
chemotherapeutic agents cause cell cycle arrest and promote
apoptosis to stop the growth of human glioblastoma and other types
of cancer cells (34,35). Furthermore, rapamycin has been shown
to induce cell cycle arrest in the G1 phase and promote
apoptosis in several cancer types, including renal cancer cells and
glioma cells (36,37). The present results revealed that
rapamycin treatment increased the proportion of B16 cells in the
G1 phase, and decreased the protein expression levels of
CDK1, cyclin D1 and CDK4. We therefore hypothesized that
rapamycin-induced cell cycle arrest may be one of the mechanisms
that inhibits the viability of B16 cells.
Induction of apoptosis is key in the suppression of
tumors. Rapamycin is a classic autophagy inducer, which is able to
induce autophagy in most vital cells (38–41).
However, some studies have also found that it can induce the
apoptosis of certain types of cells, including Kaposiform
hemangioendothelioma primary cells, human MG-63 osteosarcoma cells
and SHSY5Y neuroblastoma cells (19,20,42,43).
It has been shown that rapamycin induces B16 cell apoptosis in a
lung metastasis model (44). The
present study showed that rapamycin can cause B16 cells to undergo
apoptosis both in vivo and in vitro by increasing the
expression levels of the apoptotic proteins cleaved caspase 3 and
Bax. This may be one of the underlying mechanisms by which
rapamycin prevents B16 cell viability.
The relationship between autophagy and cancer has
long been discussed, and there exists an ‘autophagy paradox’. On
the one hand, autophagy breaks down cellular components that may
supply substrates for biogenesis, including cancer cells. On the
other hand, if this process is overactive, cells may excessively
degrade and eventually die, including cancer cells (45). Studies have shown that in breast
cancer, osteosarcoma and pancreatic cancer, inducing autophagy can
significantly inhibit growth and proliferation (46–48).
The present results also showed that rapamycin induced B16 melanoma
cell autophagy and suppressed cell viability in a dose-dependent
manner. LC3 and p62 proteins are two important indicators of
autophagy. LC3 is the only mammalian autophagy-related protein
known to be uniquely associated with autophagosomes and is
positively correlated with the amount of autophagosomes. By
contrast, the p62 protein is degraded after the formation of
autophagic lysosomes and therefore its concentration is generally
considered to be inversely correlated with autophagic activity
(49). p62 has been used as a
marker for the inhibition of autophagy, with its reduction
indicating the activation of autophagy (50). The present study demonstrated that
rapamycin promoted autophagy in mouse B16 melanoma cells in
vivo and in vitro. We therefore hypothesized that
cellular autophagy may be one of the main mechanisms by which
rapamycin inhibits B16 cell viability.
The fact that autophagy is a highly dynamic,
multi-step process that can be regulated at various levels. The
ratio of the rate of autophagosome formation to the rate of
destruction by fusion with lysosomes determines the number of
autophagosomes that may be detected at any given time point
(51). Therefore, an increase in
LC3 II may indicate either an increase in autophagosome production
brought on by the induction of autophagy or an obstruction of the
subsequent autophagic stages, such as ineffective fusion or
decreased autophagosome destruction (52). To confirm that rapamycin induced
autophagy in B16 cells, changes in LC3 II expression were observed
in response to CQ intervention. The results showed that rapamycin
combined with CQ induced a further increase in LC3 II expression,
which indicated that autophagy was further induced by rapamycin at
the level of basal autophagy. This finding suggested that induction
of autophagic kinetics is an important mechanism by which rapamycin
inhibits the viability of B16 cells.
Dysregulated mTOR signaling is linked to cancer,
metabolic dysregulation and aging. The mTOR and PI3K/AKT/mTOR
complex 1 (mTORC1) signaling pathway are essential for the
regulation of numerous fundamental cell processes, including
protein synthesis, cell viability, metabolism, survival, catabolism
and autophagy (53). Two essential
molecules, p70-S6k and 4E-BP1, that support translation and protein
synthesis are phosphorylated by mTORC1 to control protein synthesis
(54). It has been reported that
inhibition of the mTOR/p70-S6k signaling pathway suppresses cell
viability, and induces apoptosis and autophagy (30). Rapamycin differentially inhibits
S6Ks and 4E-BP1 to mediate cell type-specific repression of mRNA
translation. (55,56). The present study revealed that
rapamycin inhibited the phosphorylation of mTOR and p70-S6k in B16
cells in a dose-dependent manner both in vivo and in
vitro. These findings indicated that rapamycin may slow down
the viability of B16 cells by inhibiting the mTOR/p70-S6k pathway,
inducing autophagy and/or inhibiting protein synthesis in B16
cells.
Overall, the present study demonstrated that
rapamycin inhibited the viability of B16 cells in vitro and
in vivo by inducing autophagy and apoptosis, and that
inducing cell cycle arrest in the G1 phase may also be
one of the underlying mechanisms. In addition, rapamycin inhibited
activation of the mTOR/p70-S6k signaling pathway, which controls
protein synthesis and negatively regulates autophagy. Therefore, it
was hypothesized that rapamycin may inhibit B16 cell viability by
inducing autophagy and/or inhibiting protein synthesis through
inactivation of the mTOR/p70-S6k signaling pathway; however, this
hypothesis requires further study. In conclusion, the present study
identified the possible mechanisms underlying rapamycin-induced
tumorigenesis inhibition and provides a theoretical basis for
rapamycin therapy of tumors in organ transplant recipients.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the National Natural
Science Foundation of China (grant no. 81660270), Hainan Provincial
Natural Science Foundation of China (grant nos. 823RC497, 823RC602
and 2019RC211), the Project of Nanhai Series of Talent Cultivation
Program (grant no. 20192031), the Youth Science and Technology
Talent Innovation Program of the Hainan Association for Science and
Technology (grant no. QCXM201920), and the Key Discipline Project
of Pathophysiology at Hainan Medical College (grant no. 04).
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
PW performed experiments, provided all figures and
wrote the article. HZ analyzed data and designed experiments. KG,
CL, BP and TF performed experiments and provided data for the
paper. ShC and SiC analyzed the data and interpreted the results.
HJ and CG contributed the central idea, designed the research, and
provided reagents and funding. ShC and SiC confirm the authenticity
of all the raw data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The animal experiments were approved by the Ethics
Committee of Hainan Medical College (approval no. HYLL: −2022-227).
All protocols were in accordance with the approved guidelines and
regulations.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Awad MA, Shah A and Griffith BP: Current
status and outcomes in heart transplantation: A narrative review.
Rev Cardiovasc Med. 23:112022. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hahn D, Hodson EM, Hamiwka LA, Lee VW,
Chapman JR, Craig JC and Webster AC: Target of rapamycin inhibitors
(TOR-I; sirolimus and everolimus) for primary immunosuppression in
kidney transplant recipients. Cochrane Database System Rev.
12:CD0042902019.PubMed/NCBI
|
|
3
|
Yang LS, Shan LL, Saxena A and Morris DL:
Liver transplantation: A systematic review of long-term quality of
life. Liver Int. 34:1298–1313. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Geissler EK: Post-transplantation
malignancies: Here today, gone tomorrow? Nat Rev Clin Oncol.
12:705–717. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dzambova M, Secnikova Z, Jirakova A,
Jůzlová K, Viklický O, Hošková L, Göpfertovà D and Hercogová J:
Malignant melanoma in organ transplant recipients: Incidence,
outcomes, and management strategies: A review of literature.
Dermatol Ther. 29:64–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Park CK, Dahlke EJ, Fung K, Kitchen J,
Austin PC, Rochon PA and Chan AW: Melanoma incidence, stage, and
survival after solid organ transplant: A population-based cohort
study in Ontario, Canada. J Am Acad Dermatol. 83:754–761. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Russo I, Piaserico S, Belloni-Fortina A
and Alaibac M: Cutaneous melanoma in solid organ transplant
patients. G Ital Dermatol Venereol. 149:389–394. 2014.PubMed/NCBI
|
|
8
|
Zwald F, Carvajal RD, Walker J, Sawinski D
and Al-Adra D: Analysis of malignant melanoma risk and outcomes in
solid organ transplant recipients: Assessment of transplant
candidacy and the potential role of checkpoint inhibitors. Clin
Transplant. 35:e142642021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Masuda Y, Mita A, Ohno Y, Kubota K, Notake
T, Shimizu A and Soejima Y: De novo malignancy after adult-to-adult
living donor liver transplantation: A single-center long-term
experience. Transplant Proc. 55:952–955. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang JH, Pfeiffer RM, Musgrove D,
Castenson D, Fredrickson M, Miller J, Gonsalves L, Hsieh MC, Lynch
CF, Zeng Y, et al: Cancer mortality among solid organ transplant
recipients in the United States during 1987–2018. Transplantation.
107:2433–2442. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Naik MG, Arns W, Budde K, Diekmann F,
Eitner F, Gwinner W, Heyne N, Jürgensen JS, Morath C, Riester U, et
al: Sirolimus in renal transplant recipients with malignancies in
Germany. Clin Kidney J. 14:2047–2058. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kulbat A, Richter K, Stefura T,
Kołodziej-Rzepa M, Kisielewski M, Wojewoda T and Wysocki WM:
Systematic review of calcineurin inhibitors and incidence of skin
malignancies after kidney transplantation in adult patients: A
study of 309,551 cases. Curr Oncol. 30:5727–5737. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Berenguer M, Burra P, Ghobrial M, Hibi T,
Metselaar H, Sapisochin G, Bhoori S, Man NK, Mas V, Ohira M, et al:
Posttransplant management of recipients undergoing liver
transplantation for hepatocellular carcinoma. Working group report
from the ILTS transplant oncology consensus conference.
Transplantation. 104:1143–1149. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim M, Rhu J, Choi GS, Kim JM and Joh JW:
Risk factors for poor survival after recurrence of hepatocellular
carcinoma after liver transplantation. Ann Surg Treat Res.
101:28–36. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rajendran L, Ivanics T, Claasen MP, Muaddi
H and Sapisochin G: The management of post-transplantation
recurrence of hepatocellular carcinoma. Clin Mol Hepatol. 28:1–16.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guba M, Graeb C, Jauch KW and Geissler EK:
Pro- and anti-cancer effects of immunosuppressive agents used in
organ transplantation. Transplantation. 77:1777–1782. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sun L, Yan Y, Lv H, Li J, Wang Z, Wang K,
Wang L, Li Y, Jiang H and Zhang Y: Rapamycin targets STAT3 and
impacts c-Myc to suppress tumor growth. Cell Chem Biol.
29:373–385.e376. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rostamzadeh D, Haghshenas MR, Samadi M,
Mojtahedi Z, Babaloo Z and Ghaderi A: Immunosuppressive effects and
potent anti-tumor efficacy of mTOR inhibitor everolimus in breast
tumor-bearing mice. Iran J Allergy Asthma Immunol. 21:287–299.
2022.PubMed/NCBI
|
|
19
|
Wang Z, Han Q, Wang J, Yao W, Wang L and
Li K: Rapamycin induces autophagy and apoptosis in Kaposiform
hemangioendothelioma primary cells in vitro. J Pediatr Surg.
57:1274–1280. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu WX, Lu C, Wang B, Ren XY and Xu K:
Effects of rapamycin on osteosarcoma cell proliferation and
apoptosis by inducing autophagy. Eur Rev Med Pharmacol Sci.
24:915–921. 2020.PubMed/NCBI
|
|
21
|
Cust AE and Scolyer RA: Melanoma in
situ-getting the diagnosis and prognosis right. JAMA Dermatol.
159:699–701. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Puza CJ, Cardones AR and Mosca PJ:
Examining the incidence and presentation of melanoma in the
cardiothoracic transplant population. JAMA Dermatol. 154:589–591.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bundscherer A, Hafner C, Maisch T, Becker
B, Landthaler M and Vogt T: Antiproliferative and proapoptotic
effects of rapamycin and celecoxib in malignant melanoma cell
lines. Oncol Rep. 19:547–553. 2008.PubMed/NCBI
|
|
24
|
Wang M, Xu Y, Wen GZ, Wang Q and Yuan SM:
Rapamycin suppresses angiogenesis and lymphangiogenesis in melanoma
by downregulating VEGF-A/VEGFR-2 and VEGF-C/VEGFR-3 expression.
Onco Targets Ther. 12:4643–4654. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jurisic V, Srdic-Rajic T, Konjevic G,
Bogdanovic G and Colic M: TNF-α induced apoptosis is accompanied
with rapid CD30 and slower CD45 shedding from K-562 cells. J Membr
Biol. 239:115–122. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scherbakov AM, Vorontsova SK, Khamidullina
AI, Mrdjanovic J, Andreeva OE, Bogdanov FB, Salnikova DI, Jurisic
V, Zavarzin IV and Shirinian VZ: Novel pentacyclic derivatives and
benzylidenes of the progesterone series cause anti-estrogenic and
antiproliferative effects and induce apoptosis in breast cancer
cells. Invest New Drugs. 41:142–152. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Leary S, Underwood W and Anthony R: AVMA
guidelines for the euthanasia of animals: 2020 edition. AVMA;
Schaumburg, IL: 2020
|
|
29
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press (US); Washington, DC: 2011
|
|
30
|
Yang B and Zhao S: Polydatin regulates
proliferation, apoptosis and autophagy in multiple myeloma cells
through mTOR/p70s6k pathway. Onco Targets Ther. 10:935–944. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Saber-Moghaddam N, Nomani H, Sahebkar A,
Johnston TP and Mohammadpour AH: The change of immunosuppressive
regimen from calcineurin inhibitors to mammalian target of
rapamycin (mTOR) inhibitors and its effect on malignancy following
heart transplantation. Int Immunopharmacol. 69:150–158. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gruber P and Zito PM: Skin Cancer.
StatPearls. StatPearls Publishing LLC; Treasure Island, FL:
2023
|
|
33
|
Rubatto M, Sciamarrelli N, Borriello S,
Pala V, Mastorino L, Tonella L, Ribero S and Quaglino P: Classic
and new strategies for the treatment of advanced melanoma and
non-melanoma skin cancer. Front Med (Lausanne). 9:9592892022.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tsai TH, Su YF, Tsai CY, Wu CH, Lee KT and
Hsu YC: RTA dh404 induces cell cycle arrest, apoptosis, and
autophagy in glioblastoma cells. Int J Mol Sci. 24:40062023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao W, Zhang L, Zhang Y, Jiang Z, Lu H,
Xie Y, Han W, Zhao W, He J, Shi Z, et al: The CDK inhibitor AT7519
inhibits human glioblastoma cell growth by inducing apoptosis,
pyroptosis and cell cycle arrest. Cell Death Dis. 14:112023.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sabarwal A, Chakraborty S, Mahanta S,
Banerjee S, Balan M and Pal S: A novel combination treatment with
honokiol and rapamycin effectively restricts c-met-induced growth
of renal cancer cells, and also inhibits the expression of tumor
cell PD-L1 involved in immune escape. Cancers (Basel). 12:17822020.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Z, Wang X, Cheng F, Wen X, Feng S, Yu
F, Tang H, Liu Z and Teng X: Rapamycin inhibits glioma cells growth
and promotes autophagy by miR-26a-5p/DAPK1 Axis. Cancer Manag Res.
13:2691–2700. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bjedov I and Rallis C: The target of
rapamycin signalling pathway in ageing and lifespan regulation.
Genes (Basel). 11:10432020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao G, Chen W, Yan M, Liu J, Luo H, Wang C
and Yang P: Rapamycin regulates the balance between cardiomyocyte
apoptosis and autophagy in chronic heart failure by inhibiting mTOR
signaling. Int J Mol Med. 45:195–209. 2020.PubMed/NCBI
|
|
40
|
Masaki N, Aoki Y, Obara K, Kubota Y,
Bouvet M, Miyazaki J and Hoffman RM: Targeting autophagy with the
synergistic combination of chloroquine and rapamycin as a novel
effective treatment for well-differentiated liposarcoma. Cancer
Genomics Proteomics. 20:317–322. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ni Z, Li H, Mu D, Hou J, Liu X, Tang S and
Zheng S: Rapamycin alleviates 2,4,6-trinitrobenzene sulfonic
acid-induced colitis through autophagy induction and NF-κB pathway
inhibition in mice. Mediators Inflamm. 2022:29232162022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ozates NP, Soğutlu F, Lerminoglu F, Demir
B, Gunduz C, Shademan B and Avci CB: Effects of rapamycin and
AZD3463 combination on apoptosis, autophagy, and cell cycle for
resistance control in breast cancer. Life Sci. 264:1186432021.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kocoglu SS, Sunay FB and Akkaya PN:
Effects of monensin and rapamycin combination therapy on tumor
growth and apoptosis in a xenograft mouse model of neuroblastoma.
Antibiotics (Basel). 12:9952023. View Article : Google Scholar
|
|
44
|
Yang Z, Lei Z, Li B, Zhou Y, Zhang GM,
Feng ZH, Zhang B, Shen GX and Huang B: Rapamycin inhibits lung
metastasis of B16 melanoma cells through down-regulating alphav
integrin expression and up-regulating apoptosis signaling. Cancer
Sci. 101:494–500. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo JY, Teng X, Laddha SV, Ma S, Van
Nostrand SC, Yang Y, Khor S, Chan CS, Rabinowitz JD and White E:
Autophagy provides metabolic substrates to maintain energy charge
and nucleotide pools in Ras-driven lung cancer cells. Genes Dev.
30:1704–1717. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hashemi M, Paskeh MDA, Orouei S, Abbasi P,
Khorrami R, Dehghanpour A, Esmaeili N, Ghahremanzade A, Zandieh MA,
Peymani M, et al: Towards dual function of autophagy in breast
cancer: A potent regulator of tumor progression and therapy
response. Biomed Pharmacother. 161:1145462023. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li J, Chen X, Kang R, Zeh H, Klionsky DJ
and Tang D: Regulation and function of autophagy in pancreatic
cancer. Autophagy. 17:3275–3296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Ning B, Liu Y, Huang T and Wei Y:
Autophagy and its role in osteosarcoma. Cancer Med. 12:5676–5687.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang P and Mizushima N: LC3- and
p62-based biochemical methods for the analysis of autophagy
progression in mammalian cells. Methods. 75:13–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zhang Z, Singh R and Aschner M: Methods
for the detection of autophagy in mammalian cells. Curr Protoc
Toxicol. 69:2012212012262016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
du Toit A, Hofmeyr JS, Gniadek TJ and Loos
B: Measuring autophagosome flux. Autophagy. 14:1060–1071.
2018.PubMed/NCBI
|
|
52
|
Klionsky DJ, Abdelmohsen K, Ab A, Abedi
MJ, Abeliovic H, Arozen AA, Adach H, Adams CM, Adams PD, Adeli K,
et al: Guidelines for the use and interpretation of assays for
monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Popova NV and Jucker M: The role of mTOR
signaling as a therapeutic target in cancer. Int J Mol Sci.
22:17432021. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Burnett PE, Barrow RK, Cohen NA, Snyder SH
and Sabatini DM: RAFT1 phosphorylation of the translational
regulators p70 S6 kinase and 4E-BP1. Proc Natl Acad Sci USA.
95:1432–1437. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Choo AY, Yoon SO, Kim SG, Roux PP and
Blenis J: Rapamycin differentially inhibits S6Ks and 4E-BP1 to
mediate cell-type-specific repression of mRNA translation. Proc
Natl Acad Sci USA. 105:17414–17419. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Li B, Zhou C, Yi L, Xu L and Xu M: Effect
and molecular mechanism of mTOR inhibitor rapamycin on
temozolomide-induced autophagic death of U251 glioma cells. Oncol
Lett. 15:2477–2484. 2018.PubMed/NCBI
|