Introduction
As the leading cause of cancer-related death
worldwide, lung cancer ranks first among malignant tumors in terms
of both incidence and mortality rates (1). Non-small cell lung cancer (NSCLC)
makes up 80–85% of all lung cancers and 50–70% of patients have
distant metastasis at the time of diagnosis. The five-year survival
rate of patients with NSCLC is <15–20% (2). A number of molecular targeted
therapies and immunotherapies have markedly improved outcomes in
NSCLC over the past two decades (3–8).
However, the vast majority of advanced NSCLC cases become resistant
to current treatments and eventually progress. Notably, among
these, patients with lung squamous cell carcinoma (LSCC) is a
special group that does not benefit from targeted therapy (9). Although immunotherapy has markedly
improved the prognosis of patients with cancer, relatively low
response rates and serious adverse events have hindered the
clinical use of this promising treatment in LSCC (9). Thus, identifying biomarkers is
essential to screen populations with a dominance of
treatment-responsive individuals for current therapies.
In the past decade, ferroptosis, an iron-dependent
form of regulated cell death driven by excessive lipid peroxidation
(10), has been implicated in the
development and therapeutic responses of several types of cancer,
including LSCC (11). The process
of ferroptosis promotes and suppresses tumor development during
tumorigenesis, which is triggered by the release of
damage-associated molecules and the activation of immune responses
triggered by ferroptotic damage within the tumor microenvironment
(11). As ferroptosis influences
the efficacy of chemotherapy, radiotherapy and immunotherapy, it is
likely that these therapies may be improved by agents targeting
ferroptosis signaling (10–15). It has been reported that numerous
ferroptosis-related genes promote tumor growth and are potential
targets for cancer treatment (11,14,15).
Despite this, the prognostic impact of ferroptosis-related genes on
cancers and the roles of ferroptosis-related genes in patients with
LSCC remain unclear. Therefore, it is necessary to investigate the
expression pattern and prognostic potential of ferroptosis-related
genes in LSCC.
Using the Cancer Genome Atlas (TCGA) database, the
present study assessed the expression levels of 22
ferroptosis-related genes in LSCC and para-cancerous tissues
derived from previous studies. Furthermore, Kaplan-Meier survival
analyses, univariate analyses and multivariate Cox analyses were
performed and a nomogram based on the expression of heat shock
protein (HSP)A5 was generated. In addition, the immune checkpoint
blockade (ICB) responses, stemness features and half-maximal
inhibitory concentration (IC50) scores of chemotherapy
drugs were evaluated. Thereafter, the prognostic significance of
HSPA5 in metastatic LSCC in the study cohort was assessed.
Accordingly, it may be hypothesized that HSPA5 expression can
influence the outcome of LSCC and that higher expression of HSPA5
can be predicted by chemotherapy. In conclusion, the present study
demonstrated that the expression of the ferroptosis-related gene
HSPA5 is a negative prognostic marker for LSCC.
Materials and methods
Analysis of differential
ferroptosis-related gene expression
RNA-sequencing (RNA-Seq) expression profiles for
LSCC were downloaded from the TCGA database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga)
and the full TCGA-LSCC dataset (501 patients with LSCC, phs000178)
was included in the present study. The ferroptosis-related genes
presented in the present study were obtained from Liu et al
(16). All of the analysis methods
were implemented in R (version 4.0.3; R Foundation for Statistical
Computing), as previously described (17–19).
Gene expression datasets and
functional enrichment
The method used was similar to that of previous
studies (17–20). The R software's ‘limma’ package was
used to study differentially expressed mRNA. To identify mRNAs with
differential expression, P<0.05 and log2 (fold
change) >1 or log2 (fold change) <-1 were used as
thresholds. An enrichment analysis of the Kyoto Encyclopedia of
Genes and Genomes (KEGG) can be used to study gene function in
conjunction with high-level genome functional information. KEGG
pathways were enriched using the ‘clusterProfiler’ package (version
3.18.0) in R software to describe the carcinogenesis of mRNA. In
addition, a box plot was created using R software with the
‘ggplot2’ package and a heat map with the ‘pheatmap’ package.
Kaplan-Meier survival analysis
In the present study, Kaplan-Meier survival analysis
was divided into two parts: The first part was a Kaplan-Meier
survival analysis (R software version 4.0.3) of the TCGA data. To
generate Kaplan-Meier curves, log-rank tests were used to calculate
P-values, hazard ratios (HRs) with 95% confidence intervals (CIs)
and survival time between distinct groups. Disease-free survival
(DFS) time was the period between the start of randomization and
the time of the last follow-up visit or the time of death (from any
cause). The progression-free survival (PFS) time was defined as the
time between the start of randomization and the onset of (any
aspect of) tumor progression, death (from any cause) or the last
visit after the last randomization. The overall survival (OS) time
was calculated from the time of randomization until death (from any
cause) or the last follow-up visit. All of the analysis methods and
R packages were implemented in R (version 4.0.3). P<0.05 was
considered to indicate a statistically significant difference
(21). The second part of the
analysis was a Kaplan-Meier survival analysis of the cohort data of
the present study. PFS and OS time for metastatic LSCC were
compared for high and low expression of HSPA5 in the cohort and
calculated using Kaplan-Meier analyses. The final follow-up was
performed on May 1st, 2022.
Univariate analysis, multivariate Cox
analysis and nomogram
As previously described, multivariate and univariate
Cox regression analyses were performed to identify the suitable
terms required for building a nomogram (22). A forest plot was used to show the
P-value, HR and 95% CI of each variable with the ‘forestplot’
package in R software (version 4.0.3), and a nomogram was developed
according to the results of the multivariate Cox proportional
hazards analysis to predict the 1-, 3- and 5-year overall
recurrence. The nomogram was used to provide a graphic
representation of the factors used to calculate the risk of
recurrence for an individual patient using the points associated
with each risk factor through the ‘rms’ package in R software
(version 4.0.3).
Cancer stemness analysis using the
one-class logistic regression machine-learning algorithm
The mRNA stemness index was calculated using the
machine-learning method of one-class logistic regression (OCLR)
developed by Malta et al (23). Based on mRNA expression signatures,
11,774 genes were detected in the gene expression profile.
Spearman's correlation coefficient was utilized to analyze the
relationship between variables, followed by the transformation of
the dryness index to a standardized range of [0,1] through a linear
conversion involving subtraction of the minimum value and division
by the maximum value. The aforementioned analysis methods and R
packages were implemented in R software (version 4.0.3) as
previously described (23).
Immune checkpoint analysis
SIGLEC15, TIGIT, CD274, HAVCR2, PDCD1, CTLA4, LAG3
and PDCD1LG2 were selected as immune checkpoint-relevant
transcripts (24–29), and all the expression values of
these eight genes were extracted. The aforementioned analyses and R
packages were implemented in R software (version 4.0.3) using the
‘ggplot2’ and ‘pheatmap’ packages (version 4.0.3) (30–32).
ICB response analysis by tumor immune
dysfunction and exclusion
First, the potential ICB response was predicted
using a tumor immune dysfunction and exclusion (TIDE) algorithm as
previously described (33). The
TIDE algorithm uses a set of gene expression markers to assess two
different mechanisms of tumor immune escape: Dysfunction of
tumor-infiltrating cytotoxic T lymphocytes (CTLs) and exclusion of
CTLs by immunosuppressive factors. High TIDE scores were associated
with poor efficacy of ICB therapy and short survival time after
administration of ICB therapy.
Signaling pathway analysis
The ‘GSVA’ package in R software (version 4.0.3) was
used for the analysis, selecting the single-sample gene set
enrichment analysis (‘ssGSEA’) method as a parameter. The
correlation between genes and pathway scores was then analyzed
using Spearman's correlation coefficient (34).
Chemotherapeutic sensitivity of
LSCC
The chemotherapeutic response for each sample was
predicted based on the largest publicly available pharmacogenomics
database, the Genomics of Drug Sensitivity in Cancer (https://www.cancerrxgene.org/). The prediction process
was implemented with the R package ‘pRRophetic’ (R Foundation for
Statistical Computing, version 4.0.3). The IC50 of the
samples was estimated by ridge regression. All parameters were set
as the default values using the batch effect of combat and tissue
type of all tissues, and the duplicate gene expression was
summarized as the mean value.
Immunohistochemistry (IHC) and image
analysis
Metastatic LSCC tissues and para-cancerous tissues
(n=100) were obtained from the Fifth Affiliated Hospital of
Zhengzhou University (Zhengzhou, China). A total of 100 patients
were included in the study, which was performed from July 2015 to
December 2018. All patients had undergone surgery and eventually
developed lung metastases. These patients were treated according to
the National Comprehensive Cancer Network guidelines for LSCC
(35). Metastatic patients who have
not undergone surgery are excluded. Formalin-fixed
paraffin-embedding (FFPE) tissues were used in the present study
and the detailed procedure was as described previously (36). In brief, the first step was to dewax
and rehydrate formalin-fixed and paraffin-embedded sections of LSCC
and para-cancerous tissues (5 µm thick). Antigen retrieval was
performed by heating the slides in 10 mM Tris buffer with 1 mM EDTA
(pH 9) in a streamer for 20 min. Samples were immersed in 3%
H2O2 for 5 min to inhibit endogenous
peroxidase activity. Subsequently, after Tris-buffered saline
containing 0.1% Tween had been used for washing, endogenous biotin
was inhibited by sequential incubation with 0.1% anti-biotin
protein and 0.01% biotin (Dako; Agilent Technologies, Inc.) for 10
min at room temperature. Subsequently, 3% skimmed milk powder was
applied for 30 min at room temperature to block unspecific binding.
Thereafter, LSCC and para-cancerous tissue sections were incubated
with HSPA5 monoclonal mouse anti-human antibodies (cat. no.
ab21685; dilution, 1:1,000; Abcam) at 4°C overnight. The sections
were then serially rinsed and incubated with secondary antibody
(cat. no. ab98799; dilution, 1:3,000; Abcam) at room temperature 1
h. Finally, color was developed by incubation with
3,3′-diaminobenzidine (DAB) substrate [ImmPACT DAB Peroxidase (HRP)
substrate; cat. no. ab64238; Abcam], followed by counterstaining
with hematoxylin for 5 min and mounting with Vecmount (cat. no.
H-5000; Vector Laboratories). The IHC staining was evaluated
independently by two experienced pathologists blinded to patient
characteristics and results via microscopy (BX53; Olympus Corp.).
Based on a combination of the % positive stained cells and the
intensity of the staining, a semi-quantitative scoring system
(H-score) was calculated, as previously described (32–34).
The H-score was calculated as follows: [H-score=∑ % (0–100%) ×
intensity (1–3)]=(% weak-intensity cells ×1) + (%
moderate-intensity cells ×2) + (% strong-intensity cells ×3). The
median H-score was selected as the cut-off value for high or low
HSPA5 expression.
Statistical analysis
R software (version 4.0.3) was used for all
statistical analyses of the TCGA data. The statistical details of
all the experiments are reported in the Materials and methods
section, including the statistical analyses performed and
statistical significance. GraphPad Prism 9.0 (Dotmatics) and IBM
SPSS Statistics 24.0 (IBM Corp.) were used to statistically analyze
the cohort data. Quantification of the HSPA5 density was performed
using an unpaired t-test, and PFS and OS were calculated using the
Kaplan-Meier estimator. Univariate and multivariate Cox
proportional hazards regression models were used to estimate the
HRs, along with the associated CIs and P-values. The Student's
unpaired t-test and the χ2 test were used for
inferential statistical analysis. For all data, P<0.05 was used
to indicate a statistically significant difference.
Results
Expression distribution of
ferroptosis-related genes in LSCC
An analysis of RNA-Seq data from the TCGA database
was performed to determine the presence of ferroptosis-related
genes in LSCC. Prior studies have identified 22 genes that have
been reported to serve crucial roles in regulating ferroptosis
(16). These ferroptosis regulator
genes are CDKN1A, HSPA5, TTC35/EMC2, SLC7A11, NFE2L2, MT1G, HSPB1,
GPX4, FANCD2, CISD1, FDFT1, SLC1A5, SAT1, TFRC, RPL8, NCOA4,
LPCAT3, GLS2, DPP4, CS, ALOX15 and ACSL4. Following this, the
expression patterns of ferroptosis-related genes in LSCC and
adjacent tissues were compared. Of these 22 ferroptosis-related
genes, the mRNA expression levels of HSPA5, HSPB1, GPX4, FANCD2,
CISD1, FDFT1, NFE2L2, SLC1A5, RPL8, NCOA4, TFRC and SLC7A11 were
significantly increased in LSCC compared with those in the adjacent
tissues (Fig. 1). However, the
expression levels of CDKN1A, EMC2, SAT1, LPCAT3, GLS2, DPP4, CS and
ACSL4 were significantly decreased in LSCC compared with those in
the adjacent healthy tissues (Fig.
1). However, there was no significant difference in the
expression levels of MT1G and ALOX15 in LSCC compared with those of
the adjacent tissues (Fig. 1).
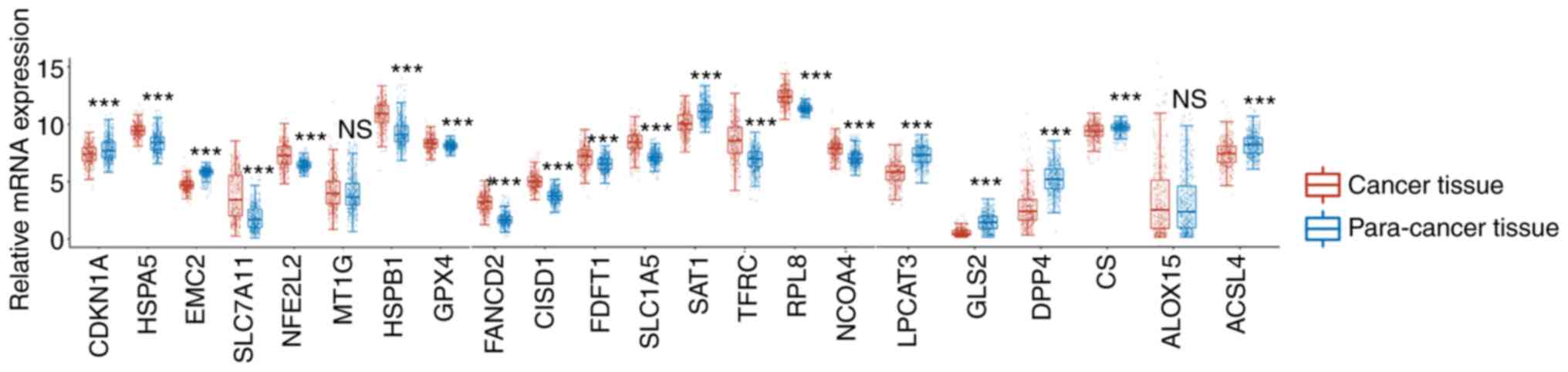 | Figure 1.Relative mRNA expression of CDKN1A,
HSPA5, EMC2, SLC7A11, NFE2L2, MT1G, HSPB1 and GPX4, FANCD2, CISD1,
FDFT1, SLC1A5, SAT1, TFRC, RPL8, NCOA4, and LPCAT3, GLS2, DPP4, CS,
ALOX15 and ACSL4 between lung squamous cell carcinoma and normal
tissue. ***P<0.001. NS, not significant. |
Different prognostic roles of
ferroptosis-related genes in LSCC
Ferroptosis is involved in the metabolism of cells
and the immune system, so it has been suggested that it and its
regulators could be linked to cancer survival (37). In light of this, the prognostic
value of increased ferroptosis-related gene expression was
evaluated in LSCC. To determine whether ferroptosis-related gene
expression is prognostic, the DFS, PFS and OS were evaluated. There
were 12 genes related to ferroptosis with increased expression, and
only the higher expression of HSPA5 was significantly associated
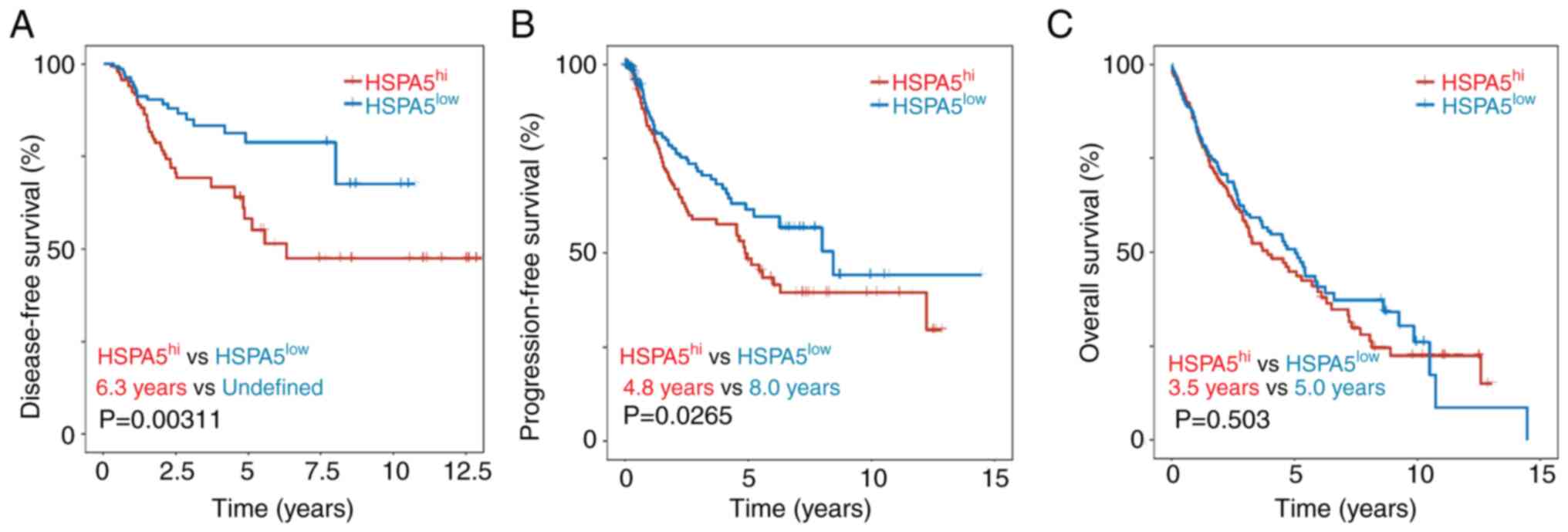
with a shorter DFS (Fig. 2A) and
PFS (Fig. 2B), but not OS (Fig. 2C). There was no significant
difference in DFS, PFS or OS associated with high and low
expression of the other 11 ferroptosis-related genes (Fig. S1). However, HSPA5 was significantly
elevated in lung adenocarcinoma tissue compared with that of normal
tissue (Fig. S2A). However, there
was no significant difference in the DFS, PFS and OS between high
and low expression of HSPA5 (Fig.
S2B-D). Of the 22 ferroptosis-related genes, only HSPA5
predicted DFS, PFS and OS in patients with LSCC, suggesting that it
may be a potential biomarker for predicting LSCC.
Both univariate and multivariate Cox
analyses and the nomogram predict HSPA5 to be a poor prognostic
factor for LSCC
A multivariate and univariate Cox analysis was
performed to evaluate the independent prognostic value of HSPA5 in
terms of DFS, PFS and OS of patients with LSCC. According to the
univariate analysis, the group with high HSPA5 expression had a
significantly worse DFS time (Fig.
3A) and the multivariate analysis demonstrated that high HSPA5
expression was independently associated with a significantly
shorter DFS time (Fig. 3B). This
result indicates that HSPA5 expression may serve as an independent
prognostic factor for LSCC. To develop a clinically applicable
method to predict a patient's survival probability, a nomogram was
generated to construct a predictive model that considered
clinicopathological covariates. With the multivariate and
univariate analysis of DFS rates, a nomogram was created to predict
the 1-, 3- and 5-year DFS rates in the discovery group, which
demonstrates the similar results (Fig.
3C). The probability of the death of patients was also
predicted using generalized linear regression. Among the predictors
under evaluation was when HSPA5hi vs.
HSPA5low met the P<0.05 risk assessment criterion.
According to an ideal model, DFS rates in the entire cohort were
well predicted at 1-, 3- and 5-year intervals (Fig. 3D). The independent prognostic
significance of HSPA5 was then measured in terms of the PFS in
patients with LSCC and the same results were obtained (Fig. 3E-H). By contrast, in terms of the OS
of patients with LSCC, there were no significant differences
between the HSPA5 high and low expression groups (Fig. S3). In summary, in both the
univariate and multivariate Cox analysis, as well as the nomograms,
HSPA5 was predicted to be an unfavorable prognostic factor.
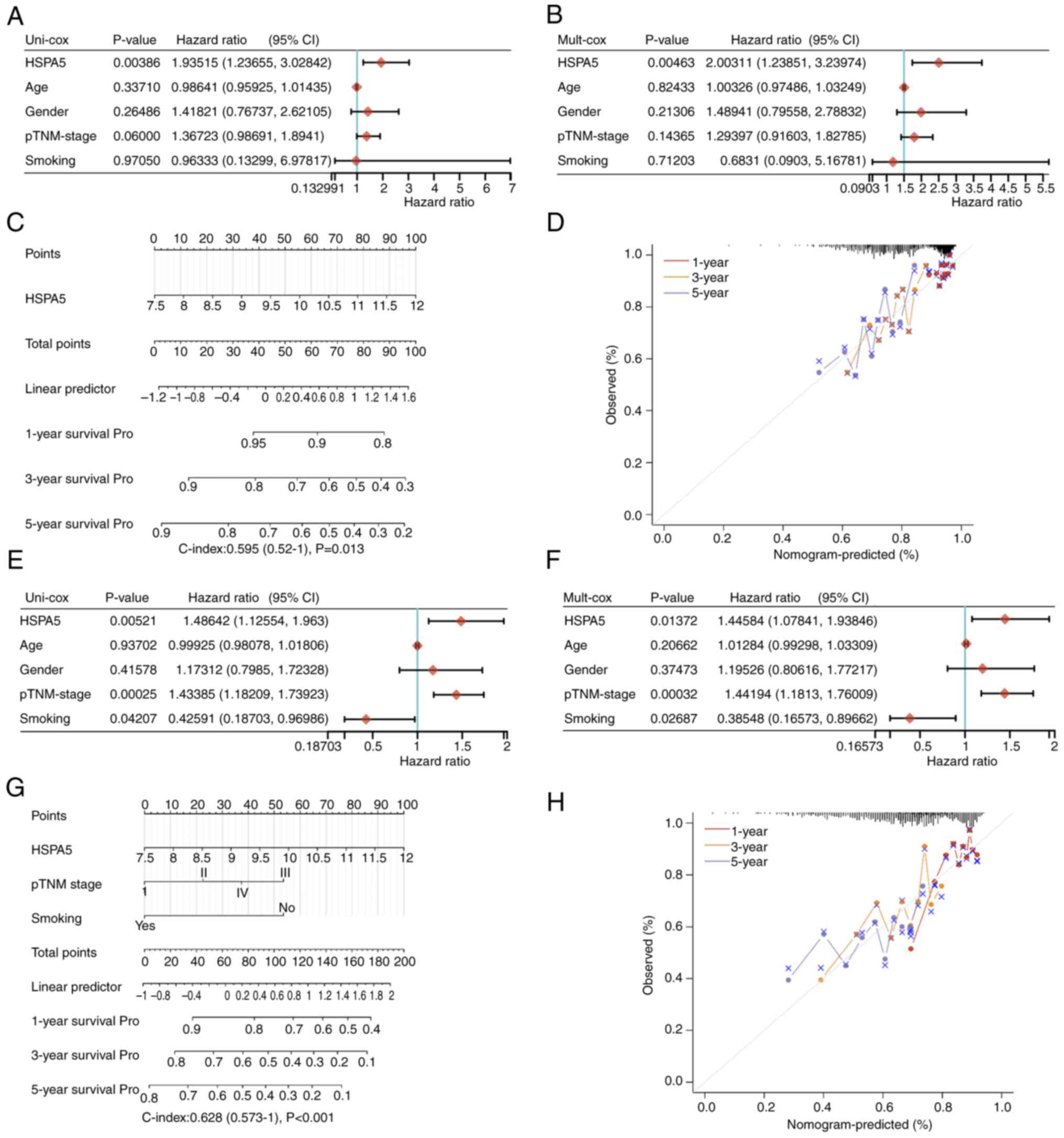 | Figure 3.The (A) univariate and (B)
multivariate Cox regression revealing the HRs and P-values and
certain parameters of the HSPA5, p-TNM stage, age, gender and
smoking of DFS. (C) Nomogram predicting the 1-, 3- and 5-year DFS
of patients with LSCC. (D) Calibration curve for the DFS nomogram
in the discovery group. The dashed diagonal line represents an
ideal nomogram, whereas the red, orange and blue-gray lines
represent the 1-, 3- and 5-year nomograms, respectively. The (E)
univariate and (F) multivariate Cox regression revealing the HRs
and P-values and certain parameters of the HSPA5, p-TNM stage, age,
gender and smoking of PFS. (G) Nomogram predicting the 1-, 3- and
5-year PFS of patients with LSCC. (H) Calibration curve for the PFS
nomogram in the discovery group. The dashed diagonal line
represents an ideal nomogram, whereas the red, orange and blue-gray
lines represent the 1-, 3- and 5-year nomograms, respectively. HR,
hazard ratio; DFS, disease-free survival; HSPA5, heat shock protein
A5; p-TNM, pathological Tumor-Node-Metastasis; LSCC, lung squamous
cell carcinoma; PFS, progression-free survival. |
High HSPA5 expression is associated
with low responses to ICB via increased stemness of LSCC
Immune checkpoint inhibitors have become a common
type of immunotherapy for patients with lung cancer in recent years
(38,39). In the present study, TIDE scores
were used to evaluate the ICB responses of patients with LSCC with
high (HSPA5hi) and low (HSPA5low) expression
of HSPA5. HSPA5hi patients had significantly higher TIDE
scores than HSPA5low patients, indicating lower
responses to ICB (Fig. 4A). Due to
the strong association between immune checkpoint molecules and ICB
responses, the expression of immune checkpoint molecules in
patients with LSCC with HSPA5hi and HSPA5low
was then assessed. PDCD1LG1, CTLA4 and PDCD1LG2 were all
significantly upregulated, but not TIM3, LAG3, PDCD1, TIGIT and
SIGLEC15, in the HSPA5hi group (Fig. 4B). Previous studies have indicated
that cancer progresses through the gradual loss of a differentiated
phenotype and the acquisition of stem-cell-like characteristics
(23). Consequently, numerous
treatment approaches, including immunotherapy, do not kill cancer
stem cells effectively (40,41).
Therefore, the stemness features of patients with LSCC with
HSPA5hi and HSPA5low were tested using an
OCLR machine-learning algorithm, as previously described (23). Patients with LSCC with
HSPA5hi were demonstrated to have similar OCLR scores to
those with HSPA5low (Fig.
4C). In summary, these data indicate that patients with LSCC
with HSPA5hi are not sensitive to ICB.
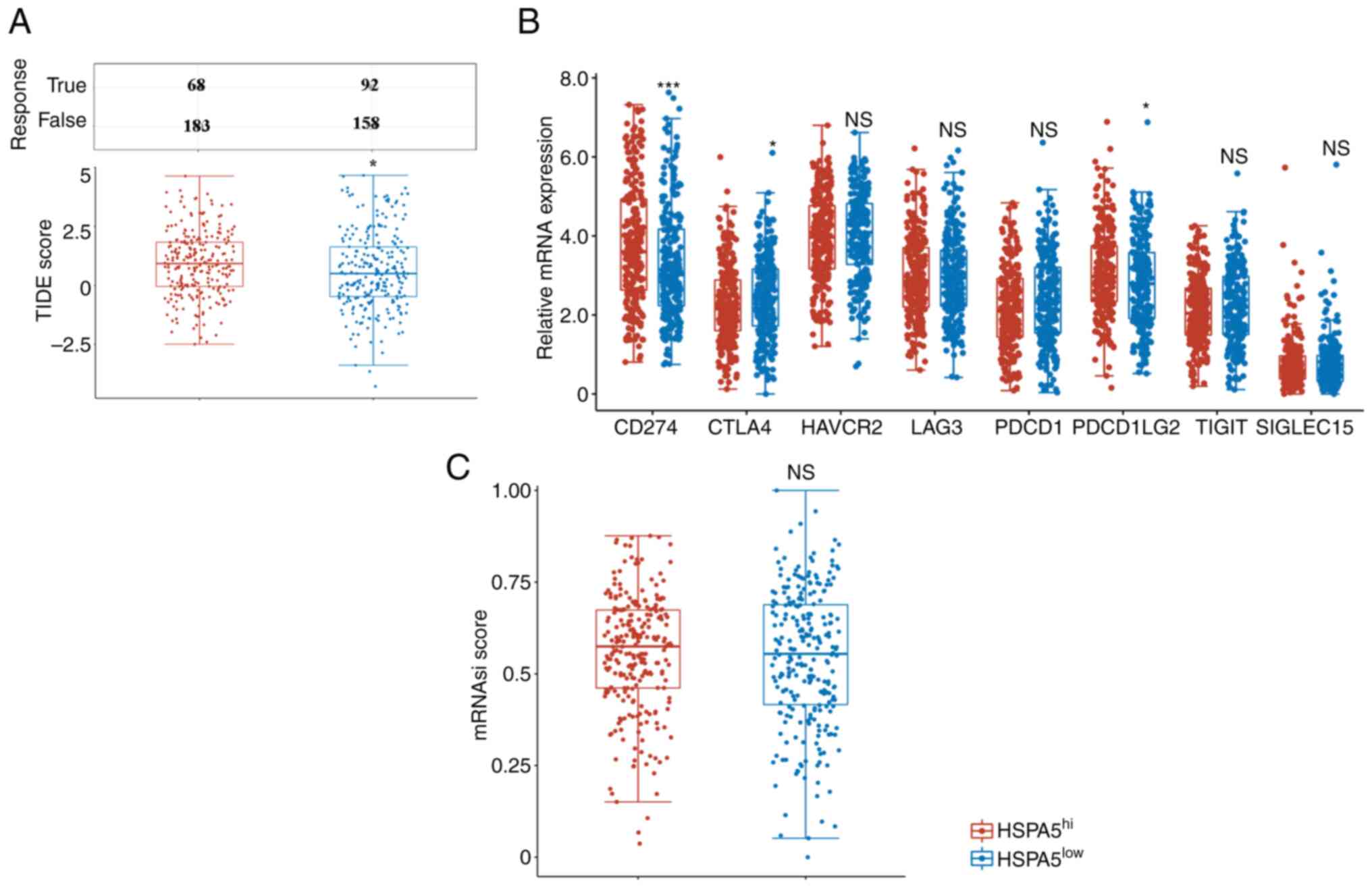 | Figure 4.(A) Distribution of TIDE scores in
the HSPA5hi and HSPA5low groups within the
prediction results. (B) Expression of immune checkpoint molecules
in the HSPA5hi and HSPA5low groups. (C)
Distribution of mRNAsi scores in the HSPA5hi and
HSPA5low groups within the prediction results.
*P<0.05, ***P<0.001. NS, not significant; TIDE, tumor immune
dysfunction and exclusion; HSPA5, heat shock protein A5;
HSPA5hi, high expression of HSPA5; mRNAsi, stemness
index; PD-L1, programmed cell death-ligand 1; CTLA4, cytotoxic T
lymphocyte-associated antigen 4; HAVCR2, hepatitis A virus cellular
receptor 2; LAG3, lymphocyte activating 3; PDCD1, programmed cell
death 1; PDCD1LG2, PDCD1 ligand 2; TIGIT, T-cell immunoreceptor
with Ig and ITIM domains; SIGLEC15, sialic acid binding Ig like
lectin 15. |
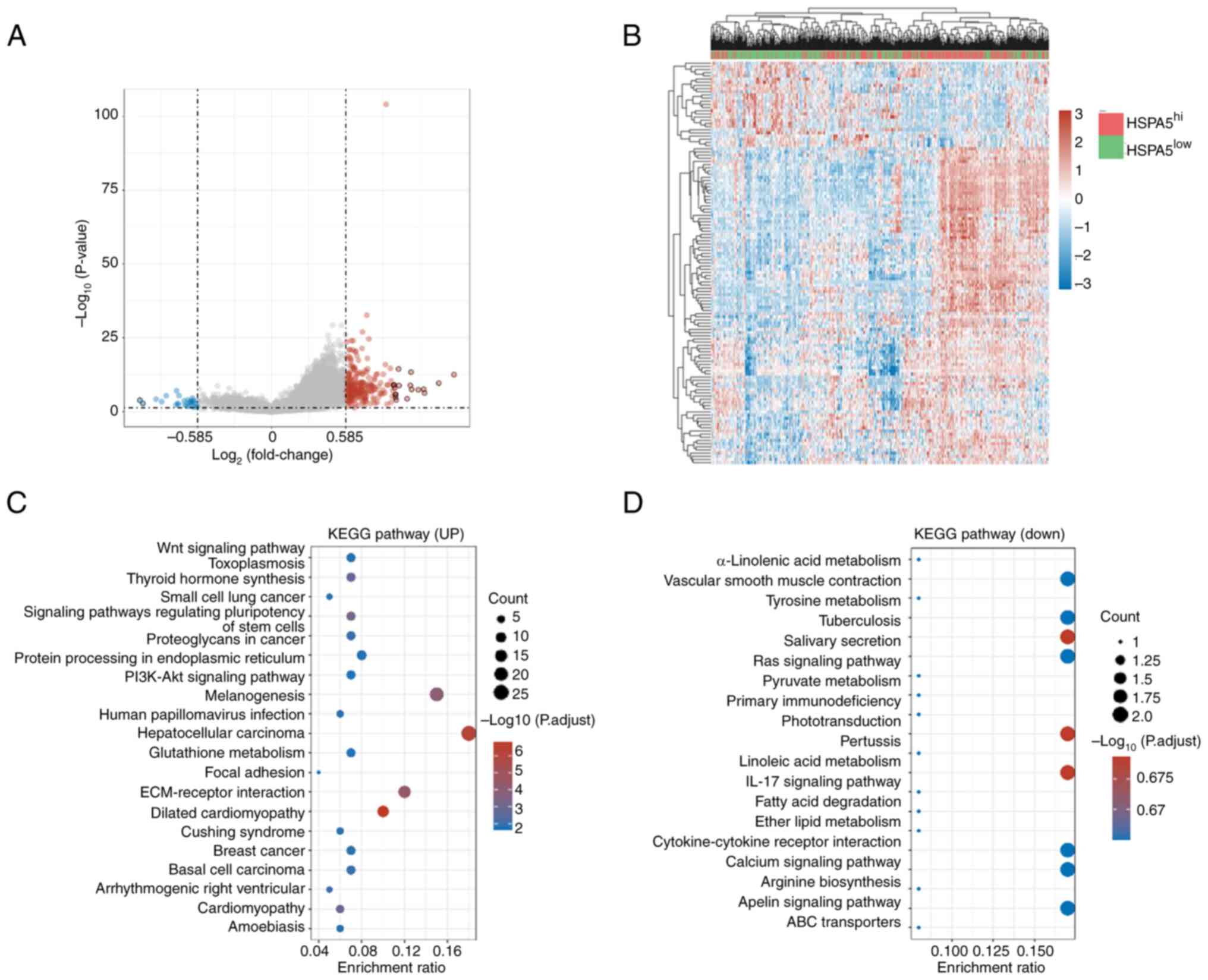
Differentially expressed genes and
KEGG pathway analysis based on high and low expression of
HSPA5
As HSPA5 can be used as a prognostic biomarker for
lung adenocarcinomas, high and low expression levels of HSPA5 of
patients with lung carcinomas were compared. The volcano plot in
Fig. 5A depicts the 237 upregulated
and the 27 downregulated genes in the HSPA5hi vs.
HSPA5low groups (Table
SI) and Fig. 5B presents a heat
map of the differentially expressed genes. The KEGG signaling
pathways enriched with differentially expressed genes were analyzed
to predict their primary biological actions. Fig. 5C shows the KEGG pathways enriched by
the upregulated genes in the HSPA5hi vs.
HSPA5low group, including ‘glutathione metabolism’,
‘focal adhesion’, ‘extracellular matrix (ECM)-receptor interaction’
and numerous cancer pathways. Fig.
5D presents the KEGG pathways enriched by the downregulated
genes in the HSPA5hi vs. HSPA5low group,
including ‘tyrosine metabolism’, ‘fatty acid degradation’ and
‘ATP-binding cassette transporters’. Correlations between the
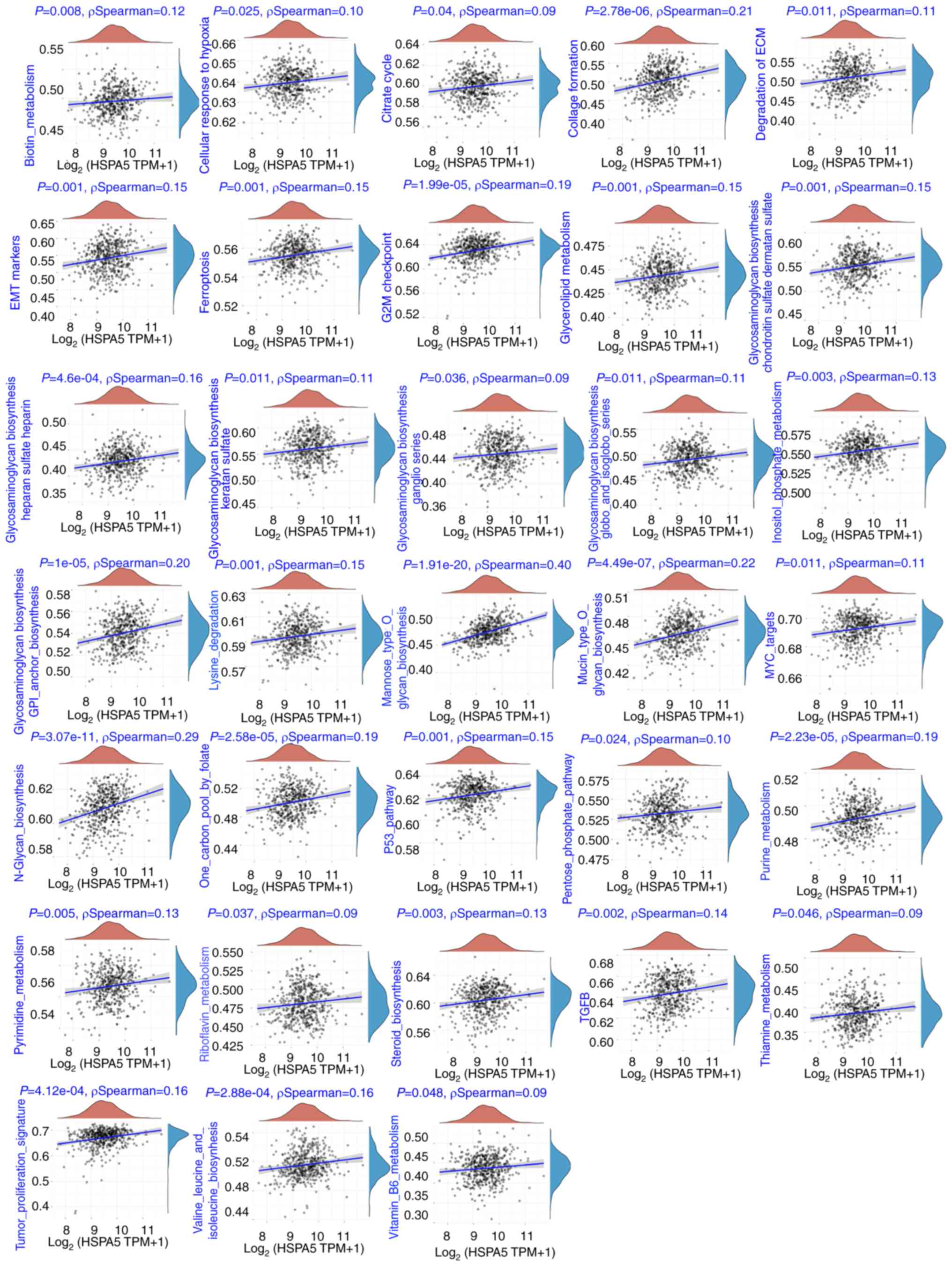
expression of HSPA5 with enriched signaling pathways with the use
of ssGSEA analyses were also assessed, as previously described
(42). Notably, positive
correlations between HSPA5 expression and ‘ferroptosis’, ‘cellular
responses to hypoxia’, ‘tumor proliferation signature’, ‘G2M
checkpoint’, ‘MYC targets’ and ‘TGFB’ were demonstrated (Fig. 6).
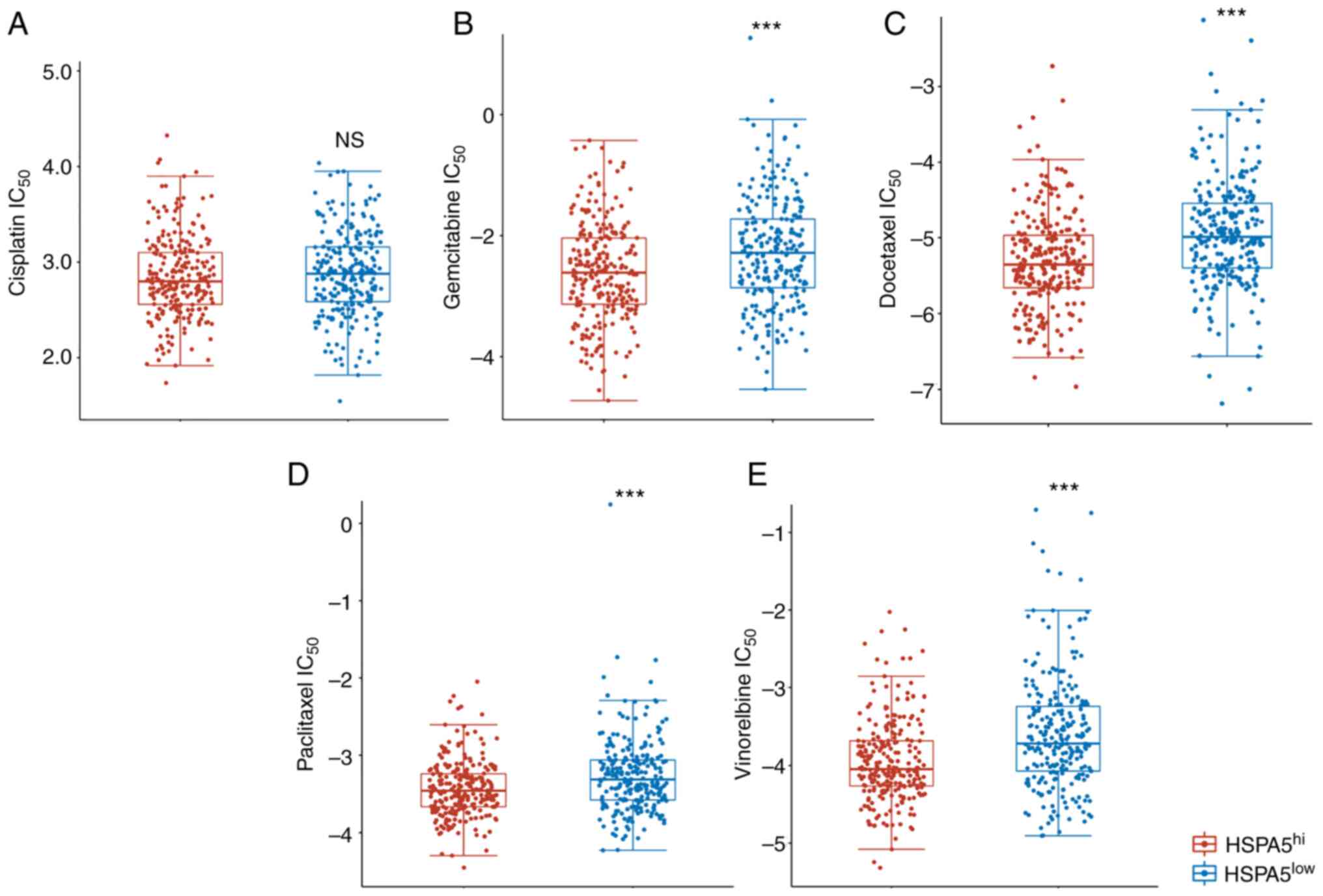
High HSPA5 expression predicts
distinct responses to chemotherapy for patients with LSCC
Cisplatin, gemcitabine, docetaxel, paclitaxel and
vinorelbine are the five most commonly used chemotherapy agents in
the treatment of LSCC (43,44). The present study therefore assessed
whether high or low HSPA5 expression was associated with
sensitivity to treatment with the five different chemotherapeutic
agents commonly used for LSCC by IC50 analysis (Fig. 7). The IC50 scores for
cisplatin, gemcitabine, docetaxel, paclitaxel and vinorelbine were
tested according to previous reports (45). Patients with LSCC in the
HSPA5hi group exhibited significantly lower gemcitabine,
docetaxel, paclitaxel and vinorelbine IC50 scores than
those of the HSPA5low group (Fig. 7B-E); however, the IC50
scores were similar between groups for cisplatin (Fig. 7A). According to these results,
patients with LSCC with HSPA5hi may be more sensitive to
chemotherapy.
High HSPA5 expression predicts poor
prognosis in LSCC in the study cohort
A total of 100 metastatic LSCC tissues and
para-cancerous tissues were obtained from the Fifth Affiliated
Hospital of Zhengzhou University (Zhengzhou, China) to confirm the
expression pattern of HSPA5. A total of 100 patients, of whom 47
(47%) were male and 53 (53%) were female, with a median age of 63
years, were included in the study, which was conducted from July
2015 to December 2018. All patients had undergone surgery and
eventually developed lung metastases. These patients were treated
according to the National Comprehensive Cancer Network guidelines
for LSCC (35). Table I lists the detailed
clinicopathological characteristics of the patients. To detect
HSPA5 expression in each patient, IHC was performed with an
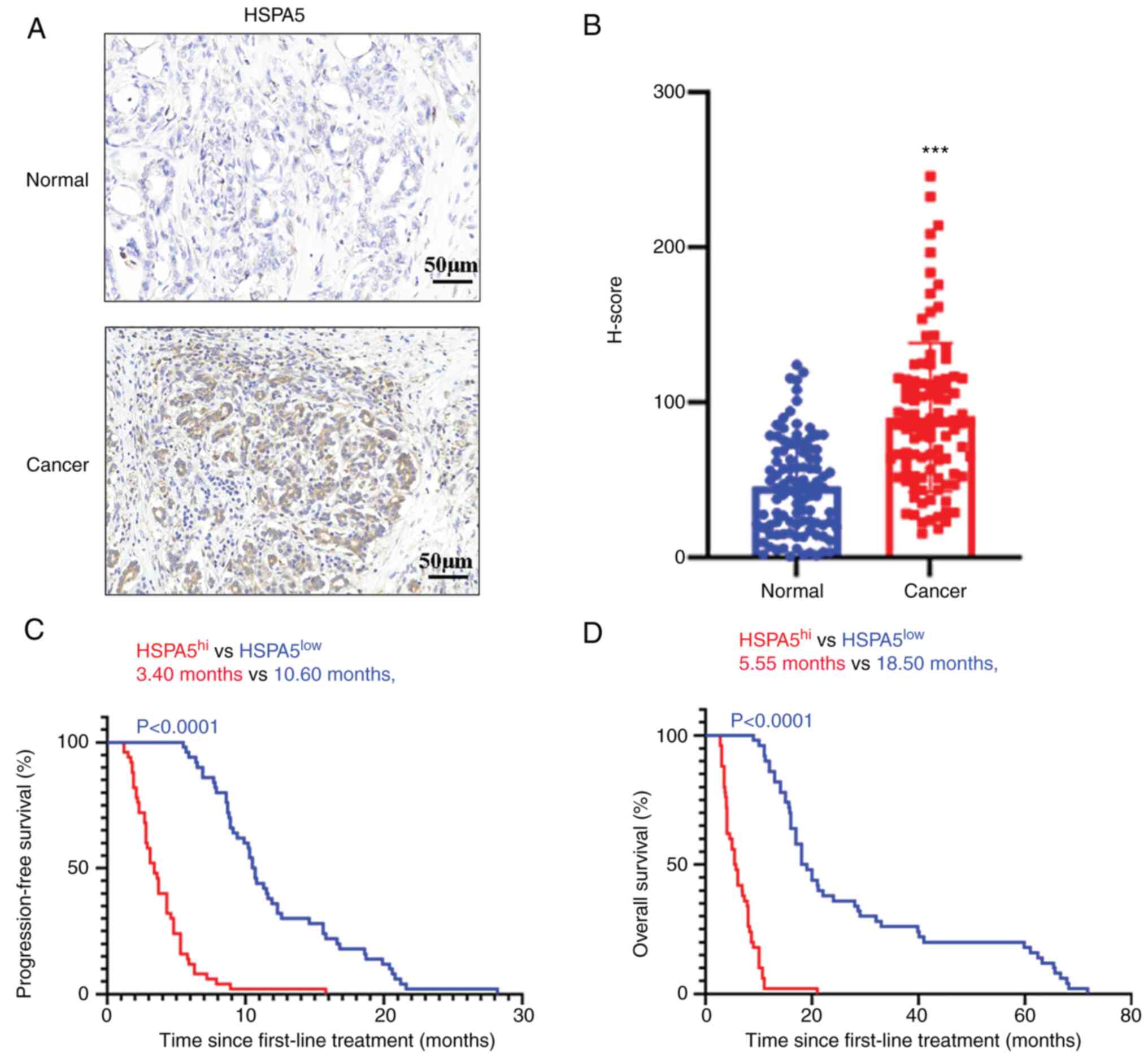
anti-HSPA5 antibody. In Fig. 8A,
representative IHC images of HSPA5 in cancerous and paraneoplastic
tissues are provided. H-scores were used to perform a
semiquantitative analysis of HSPA5 expression, as previously
described (42). According to the
quantitative analysis, the H-scores of HSPA5 in cancerous tissues
were significantly higher than those in paraneoplastic tissues
(Fig. 8B). Furthermore, it was
demonstrated that the HSPA5 expression levels were different in
every LSCC sample (Fig. 8B). The
prognostic value of HSPA5 expression was then evaluated. The median
H-score was used as a cut-off value for determining high and low
expression of HSPA5. On the basis of the cutoff value for LSCC, the
LSCC group of patients was divided into high-expression
(HSPA5hi) and low-expression (HSPA5low)
subgroups. The PFS and OS of the patients with HSPA5hi
and HSPA5low metastatic LSCC was then evaluated. In line
with the results for the TCGA database cohort, higher expression of
HSPA5 in metastatic LSCC was significantly associated with shorter
PFS (Fig. 8C) and OS (Fig. 8D). Furthermore, univariate analysis
revealed that an advanced tumor-node-metastasis (TNM) stage
(46) was a risk factor for PFS and
OS (Table II). These risk factors
from the univariate analysis were adopted as covariates in a
multivariate Cox proportional hazards model, and an advanced TNM
stage and HSPA5 were determined to be independent prognostic
indicators for PFS and OS (Table
III).
 | Table I.Characteristics of patients with lung
squamous cell carcinoma included from the cohort of the present
study. |
Table I.
Characteristics of patients with lung
squamous cell carcinoma included from the cohort of the present
study.
| Characteristic | Patients
(n=100) |
|---|
| Sex |
|
|
Male | 43 (43) |
|
Female | 57 (57) |
| Age, years | 62.8 (40–70) |
| Smoking
history |
|
|
Yes | 45 (45) |
| No | 55 (55) |
| ECOG PS |
|
| 0 | 84 (84) |
| 1 | 16 (16) |
| Size of primary
tumor, cm |
|
| ≥5 | 39 (39) |
|
<5 | 61 (61) |
| Histopathological
grading |
|
|
High | 8 (8) |
|
Intermediate | 43 (43) |
|
Low | 49 (49) |
| TNM stage |
|
|
IVA | 44 (44) |
|
IVB | 56 (56) |
 | Table II.Univariate analysis. |
Table II.
Univariate analysis.
| A, Progression-free
survival |
|---|
|
|---|
| Parameter | Hazard ratio | 95% CI | P-value |
|---|
| TNM stage | 2.56 | (1.53, 3.16) | 0.005 |
| (IVA vs. IVB) |
|
|
|
| HSPA5 (high vs.
low) | 3.48 | (1.87, 4.55) | 0.003 |
|
| B, Overall
survival |
|
|
Parameter | Hazard
ratio | 95% CI | P-value |
|
| TNM stage | 1.89 | (0.99–2.17) | 0.006 |
| (IVA vs. IVB) |
|
|
|
| HSPA5 (high vs.
low) | 2.68 | (1.38–3.24) | 0.005 |
 | Table III.Multivariate analysis. |
Table III.
Multivariate analysis.
| A, Progression-free
survival |
|---|
|
|---|
| Parameter | Hazard ratio | 95% CI | P-value |
|---|
| TNM stage | 2.77 | (1.88, 3.46) | 0.004 |
| (IVA vs. IVB) |
|
|
|
| HSPA5 (high vs.
low) | 2.33 | (1.21, 3.65) | 0.005 |
|
| B, Overall
survival |
|
|
Parameter | Hazard
ratio | 95% CI | P-value |
|
| TNM stage | 1.58 | (0.84–1.96) | 0.008 |
| (IVA vs. IVB) |
|
|
|
| HSPA5 (high vs.
low) | 1.67 | (0.79–2.17) | 0.009 |
Discussion
NSCLC is the most common malignancy in the world,
with the highest incidence and mortality rates (47). In recent years, advances in
immunotherapy and targeted treatments have significantly improved
patient outcomes in lung adenocarcinoma. Despite this, the 5-year
survival rate of these patients remains <20% (1,3–8).
However, patients with LSCC cannot benefit from targeted therapy
(9), and low response rates and
serious adverse events have hindered the clinical use of
immunotherapy in LSCC. Several parameters have been evaluated for
clinical decisions and prognostication of lung cancer (48–50).
However, emerging clinical evidence has shown that patients at the
same TNM stage undergoing the same treatment have different
prognoses, which indicates that prognosis assessments based on the
TNM stage alone may not be adequate in the context of LSCC
(51,52).
Ferroptosis has been associated with cancer
development and represents a promising treatment strategy for
cancer (11,14,15,37).
In this regard, modulating the progress of ferroptosis may affect
the proliferation, colony formation and cell death of lung cancer
cells and improve the therapeutic efficacy of xenografts for lung
cancer (53). Nevertheless,
ferroptosis-related gene profiling remains to be clarified as a
prognostic factor in the context of LSCC. In the present study, the
TCGA database was used to assess the variations in expression
profiling of ferroptosis-related genes in LSCC. Through these
analyses, the signature of 22 ferroptosis-related genes was
identified, and it was demonstrated that, of these genes, the
expression of HSPA5, HSPB1, GPX4, FANCD2, CISD1, FDFT1, NFE2L2,
SLC1A5, RPL8, NCOA4, TFRC and SLC7A11 was significantly increased
in LSCC. Thereafter, the association between ferroptosis-related
genes and the prognosis of patients with LSCC was evaluated. The
survival analysis found that, of the 11 ferroptosis-related genes
with increased expression, only HSPA5 was highly related to the DFS
and PFS of patients with LSCC. The results of the present study on
patients with metastatic LSCC revealed that higher levels of HSPA5
expression predicted poor prognosis and shorter PFS and OS.
Furthermore, the results indicated that increased HSPA5 expression
may also assist with the prediction of metastatic LSCC.
HSPA5 is a member of the HSP 70 superfamily, which
serves as an important regulator in numerous diseases (54). HSPA5 is also related to the
progression of certain cancers, including head and neck cancer,
endometrial cancer, liver cancer, glioblastoma, breast cancer and
osteosarcoma. In addition, it has been reported that HSPA5 is
closely associated with the progression and poor prognosis of
NSCLC, and serves an important role in the treatment of NSCLC
(55). Lung adenocarcinoma and LSCC
are important types of NSCLC; however, they are two different tumor
types with relatively specific aspects in terms of diagnosis,
treatment and prognosis. However, the expression of HSPA5 markedly
increases in both lung adenocarcinoma and LSCC. Using TCGA
datasets, the present study demonstrated that higher expression of
HSPA5 was associated with shorter DFS and PFS of patients with LSCC
but not lung adenocarcinoma. In addition, multivariate and
univariate Cox regression analyses were performed to demonstrate
that the expression of HSPA5 is a prognostic marker for LSCC. In
conclusion, the present study further clarified that HSPA5 may be a
useful prognostic predictor for LSCC.
In general, immune cells regulate tumor ferroptosis
during cancer immunotherapy (56).
Ferroptosis also regulates immune activity within tumor
microenvironments (57). However,
ferroptosis is largely unknown as a predictor of ICB responses in
patients with LSCC. The present study demonstrated that individuals
with higher expression levels of HSPA5 had worse TIDE scores, which
indicates lower ICB responses. Therefore, HSPA5 expression may
offer a novel means of predicting ICB responses. Nevertheless,
further studies are required to identify the role of HSPA5 in
immunotherapy for LSCC.
As a consequence of the aforementioned findings,
differentially expressed genes and KEGG pathways associated with
high and low levels of HSPA5 expression were assessed in patients
with LSCC. Notably, it was demonstrated that high HSPA5 expression
was associated with glutathione metabolism, focal adhesion,
ECM-receptor interaction and numerous cancer pathways, which may
accelerate tumor progression. In addition, ssGSEA signaling pathway
analysis was performed for HSPA5 expression in LSCC, which
demonstrated that the expression of HSPA5 was positively associated
with ‘ferroptosis’, ‘cellular responses to hypoxia’, ‘tumor
proliferation signature’, ‘G2M checkpoint’, ‘MYC targets’ and
‘TGFB’. These findings revealed that high expression of HSPA5 may
promote tumor progression through multiple mechanisms.
As there are no proven targeted drugs for LSCC,
cisplatin-containing chemotherapy regimens (in combination with
gemcitabine, docetaxel, paclitaxel and vinorelbine) are the
first-line treatment option for patients with metastatic LSCC
(58). However, not all patients
are sensitive to chemotherapy. The IC50 is a method for
assessing the effectiveness of a drug. The present study found that
patients with LSCC with a higher expression of HSPA5 had lower
IC50 scores for gemcitabine, docetaxel, paclitaxel and
vinorelbine. However, there were no significant differences in
IC50 values for cisplatin for high and low expression of
HSPA5. In combination, the results highlight that the expression of
HSPA5 may be regarded as a sufficient biomarker for predicting
clinical responses to gemcitabine, docetaxel, paclitaxel and
vinorelbine in the context of LSCC. Although a high HSPA5
expression predicts that LSCC patients should be more sensitive to
chemotherapy, patients with high HSPA5 expression have worse DFS
and PFS, which suggests rapid recurrence/metastasis after treatment
in this group of patients. As mentioned above, the expression of
HSPA5 was positively correlated with the tumor proliferation
signature. Therefore, it is unsurprising that the worse prognosis
of patients with high HSPA5 expression who are sensitive to
chemotherapy may be related to the high proliferative
characteristics of this group of patients. Therefore, it is
important to assess the IC50 scores of other drugs for
LSCC in the future, and it is also important to explore the
potential mechanism of HSPA5 expression and chemosensitivity.
The present study has several limitations. It
remains unclear how ferroptosis may contribute to the outcome of
LSCC and how HSPA5 may initiate ferroptosis in LSCC. In addition,
the expression of HSPA5 was only confirmed in metastatic LSCC. More
studies should be conducted to confirm the present results and
samples from patients with early- or late-stage LSCC should also be
included. However, the expression of HSPA5 may still be an
effective predictor of LSCC prognosis. Therefore, further studies
are required to verify the role and function of HSPA5 in LSCC.
Supplementary Material
Supporting Data
Supporting Data
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
DG designed the study, performed the research,
analyzed the data and drafted the manuscript. YF, PL, SY and WZ
collected samples and performed analyses. HL designed the study and
edited the manuscript. All authors have read and approved the final
manuscript. DG and HL confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
Institutional review board approval and data sharing
agreements were obtained from Zhengzhou University (Zhengzhou,
China; approval no. ZZU-LAC-20220415). All procedures performed
were in accordance with the ethical standards of the Institutional
and/or National Research Committee and with the 1964 Helsinki
Declaration and its later amendments or comparable ethical
standards. Written informed consent was obtained from all patients
for the retention of diagnostic samples for future experimental use
at the time of collection.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NSCLC
|
non-small cell lung cancer
|
|
LSCC
|
lung squamous cell carcinoma
|
|
TCGA
|
The Cancer Genome Atlas
|
|
ICB
|
immune checkpoint blockade
|
|
RNA-Seq
|
RNA-sequencing
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor
1A
|
|
HSP
|
heat shock protein
|
|
TTC35/EMC2
|
endoplasmic reticulum membrane protein
complex subunit 2
|
|
SLC
|
solute carrier
|
|
NFE2L2
|
nuclear factor erythroid 2 like 2
|
|
MT1G
|
metallothionein-1G
|
|
GPX4
|
glutathione peroxidase 4
|
|
FANCD2
|
Fanconi anemia complementation group
D2
|
|
CISD1
|
CDGSH iron sulfur domain 1
|
|
FDFT1
|
farnesyl-diphosphate
farnesyltransferase 1
|
|
SAT1
|
spermidine/spermine
N1-acetyltransferase 1
|
|
TFRC
|
transferrin receptor
|
|
RPL8
|
ribosomal protein L8
|
|
NCOA4
|
nuclear receptor coactivator 4
|
|
LPCAT3
|
lysophosphatidylcholine
acyltransferase 3
|
|
GLS2
|
glutaminase 2
|
|
DPP4
|
dipeptidyl peptidase-4
|
|
CS
|
citrate synthase
|
|
ALOX15
|
arachidonate 15-lipoxygenase
|
|
ACSL4
|
acyl-CoA synthetase long-chain family
member 4
|
References
|
1
|
Wang M, Herbst RS and Boshoff C: Toward
personalized treatment approaches for non-small-cell lung cancer.
Nat Med. 27:1345–1356. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Global Burden of Disease Cancer
Collaboration, . Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chaft JE, Rimner A, Weder W, Azzoli CG,
Kris MG and Cascone T: Evolution of systemic therapy for stages
I–III non-metastatic non-small-cell lung cancer. Nat Rev Clin
Oncol. 18:547–557. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li BT, Smit EF, Goto Y, Nakagawa K,
Udagawa H, Mazières J, Nagasaka M, Bazhenova L, Saltos AN, Felip E,
et al: Trastuzumab deruxtecan in HER2-mutant non-small-cell lung
cancer. N Engl J Med. 386:241–251. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sezer A, Kilickap S, Gümüş M, Bondarenko
I, Özgüroğlu M, Gogishvili M, Turk HM, Cicin I, Bentsion D, Gladkov
O, et al: Cemiplimab monotherapy for first-line treatment of
advanced non-small-cell lung cancer with PD-L1 of at least 50%: A
multicentre, open-label, global, phase 3, randomised, controlled
trial. Lancet. 397:592–604. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Han J, Liu Y, Yang S, Wu X, Li H and Wang
Q: MEK inhibitors for the treatment of non-small cell lung cancer.
J Hematol Oncol. 14:12021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paz-Ares L, Ciuleanu TE, Cobo M, Schenker
M, Zurawski B, Menezes J, Richardet E, Bennouna J, Felip E,
Juan-Vidal O, et al: First-line nivolumab plus ipilimumab combined
with two cycles of chemotherapy in patients with non-small-cell
lung cancer (CheckMate 9LA): An international, randomised,
open-label, phase 3 trial. Lancet Oncol. 22:198–211. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hanna NH, Robinson AG, Temin S, Baker S
Jr, Brahmer JR, Ellis PM, Gaspar LE, Haddad RY, Hesketh PJ, Jain D,
et al: Therapy for stage IV non-small-cell lung cancer with driver
alterations: ASCO and OH (CCO) joint guideline update. J Clin
Oncol. 39:1040–1091. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Niu Z, Jin R, Zhang Y and Li H: Signaling
pathways and targeted therapies in lung squamous cell carcinoma:
Mechanisms and clinical trials. Signal Transduct Target Ther.
7:3532022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li L, Li WJ, Zheng XR, Liu QL, Du Q, Lai
YJ and Liu SQ: Eriodictyol ameliorates cognitive dysfunction in
APP/PS1 mice by inhibiting ferroptosis via vitamin D
receptor-mediated Nrf2 activation. Mol Med. 28:112022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen X, Kang R, Kroemer G and Tang D:
Broadening horizons: The role of ferroptosis in cancer. Nat Rev
Clin Oncol. 18:280–296. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Feng L, Zhao K, Sun L, Yin X, Zhang J, Liu
C and Li B: SLC7A11 regulated by NRF2 modulates esophageal squamous
cell carcinoma radiosensitivity by inhibiting ferroptosis. J Transl
Med. 19:3672021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang C, Yu M, Ma J and Zhu Y: Metabolic
classification of bladder cancer based on multi-omics integrated
analysis to predict patient prognosis and treatment response. J
Transl Med. 19:2052021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen X, Kang R, Kroemer G and Tang D:
Targeting ferroptosis in pancreatic cancer: A double-edged sword.
Trends Cancer. 7:891–901. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tang R, Xu J, Zhang B, Liu J, Liang C, Hua
J, Meng Q, Yu X and Shi S: Ferroptosis, necroptosis, and pyroptosis
in anticancer immunity. J Hematol Oncol. 13:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Z, Zhao Q, Zuo ZX, Yuan SQ, Yu K,
Zhang Q, Zhang X, Sheng H, Ju HQ, Cheng H, et al: Systematic
analysis of the aberrances and functional implications of
ferroptosis in cancer. iScience. 23:1013022020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang H, Yan M, Li W and Xu L: SIRPα and
PD1 expression on tumor-associated macrophage predict prognosis of
intrahepatic cholangiocarcinoma. J Transl Med. 20:1402022.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cao W, Fan W, Wang F, Zhang Y, Wu G, Shi
X, Shi JX, Gao F, Yan M, Guo R, et al: GM-CSF impairs
erythropoiesis by disrupting erythroblastic island formation via
macrophages. J Transl Med. 20:112022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li W, Wang Y, Zhao H, Zhang H, Xu Y, Wang
S, Guo X, Huang Y, Zhang S, Han Y, et al: Identification and
transcriptome analysis of erythroblastic island macrophages. Blood.
134:480–491. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang Y, Li W, Schulz VP, Zhao H, Qu X, Qi
Q, Cheng Y, Guo X, Zhang S, Wei X, et al: Impairment of human
terminal erythroid differentiation by histone deacetylase 5
deficiency. Blood. 138:1615–1627. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu L, Yang H, Yan M and Li W: Matrix
metalloproteinase 1 is a poor prognostic biomarker for patients
with hepatocellular carcinoma. Clin Exp Med. 23:2065–2083. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li W, Li T, Sun C, Du Y, Chen L, Du C, Shi
J and Wang W: Identification and prognostic analysis of biomarkers
to predict the progression of pancreatic cancer patients. Mol Med.
28:432022. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Malta TM, Sokolov A, Gentles AJ,
Burzykowski T, Poisson L, Weinstein JN, Kamińska B, Huelsken J,
Omberg L, Gevaert O, et al: Machine learning identifies stemness
features associated with oncogenic dedifferentiation. Cell.
173:338–354.e15. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ralser DJ, Klümper N, Gevensleben H, Zarbl
R, Kaiser C, Landsberg J, Hölzel M, Strieth S, Faridi A, Abramian A
and Dietrich D: Molecular and immune correlates of PDCD1 (PD-1),
PD-L1 (CD274), and PD-L2 (PDCD1LG2) DNA methylation in triple
negative breast cancer. J Immunother. 44:319–324. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang M, Du Q, Jin J, Wei Y, Lu Y and Li Q:
LAG3 and its emerging role in cancer immunotherapy. Clin Transl
Med. 11:e3652021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ascierto PA, Puzanov I, Agarwala SS, Blank
C, Carvajal RD, Demaria S, Dummer R, Ernstoff M, Ferrone S, Fox BA,
et al: Perspectives in melanoma: Meeting report from the ‘melanoma
bridge’ (December 5th-7th, 2019, Naples, Italy). J Transl Med.
18:3462020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Z, Zhang Y, Shi C, Zhou X, Xu K, Jiao
D, Sun Z and Han X: A novel immune classification reveals distinct
immune escape mechanism and genomic alterations: Implications for
immunotherapy in hepatocellular carcinoma. J Transl Med. 19:52021.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okła K, Rajtak A, Czerwonka A, Bobiński M,
Wawruszak A, Tarkowski R, Bednarek W, Szumiło J and Kotarski J:
Accumulation of blood-circulating PD-L1-expressing M-MDSCs and
monocytes/macrophages in pretreatment ovarian cancer patients is
associated with soluble PD-L1. J Transl Med. 18:2202020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhao B, Xia Y, Yang F, Wang Y, Wang Y,
Wang Y, Dai C, Wang Y and Ma W: Molecular landscape of IDH-mutant
astrocytoma and oligodendroglioma grade 2 indicate tumor purity as
an underlying genomic factor. Mol Med. 28:342022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ravi R, Noonan KA, Pham V, Bedi R,
Zhavoronkov A, Ozerov IV, Makarev E, V Artemov A, Wysocki PT, Mehra
R, et al: Bifunctional immune checkpoint-targeted antibody-ligand
traps that simultaneously disable TGFβ enhance the efficacy of
cancer immunotherapy. Nat Commun. 9:7412018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zeng D, Li M, Zhou R, Zhang J, Sun H, Shi
M, Bin J, Liao Y, Rao J and Liao W: Tumor microenvironment
characterization in gastric cancer identifies prognostic and
immunotherapeutically relevant gene signatures. Cancer Immunol Res.
7:737–750. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang J, Sun J, Liu LN, Flies DB, Nie X,
Toki M, Zhang J, Song C, Zarr M, Zhou X, et al: Siglec-15 as an
immune suppressor and potential target for normalization cancer
immunotherapy. Nat Med. 25:656–666. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X,
Li Z, Traugh N, Bu X, Li B, et al: Signatures of T cell dysfunction
and exclusion predict cancer immunotherapy response. Nat Med.
24:1550–1558. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wei J, Huang K, Chen Z, Hu M, Bai Y, Lin S
and Du H: Characterization of glycolysis-associated molecules in
the tumor microenvironment revealed by pan-cancer tissues and lung
cancer single cell data. Cancers (Basel). 12:17882020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wood DE: National comprehensive cancer
network (NCCN) Clinical practice guidelines for lung cancer
screening. Thorac Surg Clin. 25:185–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Magaki S, Hojat SA, Wei B, So A and Yong
WH: An introduction to the performance of immunohistochemistry.
Methods Mol Biol. 1897:289–298. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Friedmann Angeli JP, Krysko DV and Conrad
M: Ferroptosis at the crossroads of cancer-acquired drug resistance
and immune evasion. Nat Rev Cancer. 19:405–414. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu Q, Qian W, Sun X and Jiang S:
Small-molecule inhibitors, immune checkpoint inhibitors, and more:
FDA-approved novel therapeutic drugs for solid tumors from 1991 to
2021. J Hematol Oncol. 15:1432022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Passaro A, Brahmer J, Antonia S, Mok T and
Peters S: Managing resistance to immune checkpoint inhibitors in
lung cancer: Treatment and novel strategies. J Clin Oncol.
40:598–610. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang D, Tang DG and Rycaj K: Cancer stem
cells: Regulation programs, immunological properties and
immunotherapy. Semin Cancer Biol. 52:94–106. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Clara JA, Monge C, Yang Y and Takebe N:
Targeting signalling pathways and the immune microenvironment of
cancer stem cells-a clinical update. Nat Rev Clin Oncol.
17:204–232. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hänzelmann S, Castelo R and Guinney J:
GSVA: Gene set variation analysis for microarray and RNA-seq data.
BMC Bioinformatics. 14:72013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cescon DW, She D, Sakashita S, Zhu CQ,
Pintilie M, Shepherd FA and Tsao MS: NRF2 pathway activation and
adjuvant chemotherapy benefit in lung squamous cell carcinoma. Clin
Cancer Res. 21:2499–2505. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ruiz EJ, Diefenbacher ME, Nelson JK,
Sancho R, Pucci F, Chakraborty A, Moreno P, Annibaldi A, Liccardi
G, Encheva V, et al: LUBAC determines chemotherapy resistance in
squamous cell lung cancer. J Exp Med. 216:450–465. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Geeleher P, Cox NJ and Huang RS: Clinical
drug response can be predicted using baseline gene expression
levels and in vitro drug sensitivity in cell lines. Genome Biol.
15:R472014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Goldstraw P, Chansky K, Crowley J,
Rami-Porta R, Asamura H, Eberhardt WE, Nicholson AG, Groome P,
Mitchell A, Bolejack V, et al: The IASLC lung cancer staging
project: Proposals for revision of the TNM stage groupings in the
forthcoming (eighth) edition of the TNM classification for lung
cancer. J Thorac Oncol. 11:39–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo L, Ma Y, Ward R, Castranova V, Shi X
and Qian Y: Constructing molecular classifiers for the accurate
prognosis of lung adenocarcinoma. Clin Cancer Res. 12:3344–3354.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Selvaraj G, Kaliamurthi S, Kaushik AC,
Khan A, Wei YK, Cho WC, Gu K and Wei DQ: Identification of target
gene and prognostic evaluation for lung adenocarcinoma using gene
expression meta-analysis, network analysis and neural network
algorithms. J Biomed Inform. 86:120–134. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shi X, Li R, Dong X, Chen AM, Liu X, Lu D,
Feng S, Wang H and Cai K: IRGS: An immune-related gene classifier
for lung adenocarcinoma prognosis. J Transl Med. 18:552020.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cui J, Wen Q, Tan X, Chen Z and Liu G: A
genomic-clinicopathologic nomogram predicts survival for patients
with laryngeal squamous cell carcinoma. Dis Markers.
2019:59805672019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Cui J, Wang L, Tan G, Chen W, He G, Huang
H, Chen Z, Yang H, Chen J and Liu G: Development and validation of
nomograms to accurately predict risk of recurrence for patients
with laryngeal squamous cell carcinoma: Cohort study. Int J Surg.
76:163–170. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen P, Wu Q, Feng J, Yan L, Sun Y, Liu S,
Xiang Y, Zhang M, Pan T, Chen X, et al: Erianin, a novel dibenzyl
compound in Dendrobium extract, inhibits lung cancer cell growth
and migration via calcium/calmodulin-dependent ferroptosis. Signal
Transduct Target Ther. 5:512020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Fu X, Liu J, Liu D, Zhou Y, Guo Y, Wang Z,
Yang S, He W, Chen P, Wang X, et al: Glucose-regulated protein 78
modulates cell growth, epithelial-mesenchymal transition, and
oxidative stress in the hyperplastic prostate. Cell Death Dis.
13:782022. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xia S, Duan W, Liu W, Zhang X and Wang Q:
GRP78 in lung cancer. J Transl Med. 19:1182021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Xu H, Ye D, Ren M, Zhang H and Bi F:
Ferroptosis in the tumor microenvironment: Perspectives for
immunotherapy. Trends Mol Med. 27:856–867. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Torres-Ayuso P, An E, Nyswaner KM, Bensen
RC, Ritt DA, Specht SI, Das S, Andresson T, Cachau RE, Liang RJ, et
al: TNIK Is a therapeutic target in lung squamous cell carcinoma
and regulates FAK activation through merlin. Cancer Discov.
11:1411–1423. 2021. View Article : Google Scholar : PubMed/NCBI
|