Introduction
Despite the significant progress toward the
understanding of human lung cancer tumorigenesis in the past two
decades, lung cancer remains the leading cause of cancer-related
mortality worldwide (1,2). It is estimated that lung and bronchus
cancer account for only 12–14% of new cancer cases per year, but
its early metastatic spread results in a 5-year patient survival of
only 10–15% (3–5). Tumor invasion and metastasis predict
poor prognosis in lung cancer and are the main causes of treatment
failure and cancer mortality (6).
Therefore, it is critical to investigate the molecular mechanism of
lung cancer and to find an effective method by which to inhibit its
ability to invade and metastasize.
Increasing evidence demonstrates that sumoylation is
a multistep process analogous to the ubiquitin pathway and involves
maturation, activation, conjugation, ligation, and deconjugation
steps. Sumoylation, which includes small ubiquitin-like modifier
(SUMO) protein addition or removal from other proteins, is a
post-translational modification that is significantly involved in
diverse cellular processes, including transcriptional regulation,
nuclear transport, cell cycle control, and maintenance of genome
integrity through modulating protein-protein interactions of target
proteins (7–9). Ubiquitin-conjugating enzyme 9 (Ubc9),
the sole E2 conjugating enzyme for sumoylation, plays an important
role in tumorigenesis and progression (10), such as progression through M phase,
DNA damage repair, maintenance of nuclear integrity, and chromosome
segregation; in addition, several signaling molecules are targets
of Ubc9-mediated sumoylation (11–15).
Ubc9 has been reported to be expressed at high levels in advanced
melanomas and was found to shield melanoma cells from apoptosis.
Blocking Ubc9 expression sensitized melanoma cells to the cytotoxic
effects of chemotherapeutic drugs (16). Ubc9 was also reported to be
expressed at higher levels in head and neck tumor and lung tumor
specimens than in the matched normal tissues (17). However, little is known about the
role of Ubc9 in lung cancer and, in particular, about its role in
cell invasion and tumor metastasis.
In the present study, we report higher levels of
Ubc9 expression in primary lung cancer and metastatic nodules than
in premalignant and/or normal tissues. Furthermore, we observed
that inducing upregulation of Ubc9 expression in lung cancer cells
promotes migration and invasion. Based on these findings, we
concluded that Ubc9 may play an important role in cancer
progression and that Ubc9 promotes invasion and metastasis in lung
cancer.
Materials and methods
Cell lines and reagents
The lung cancer cell lines A549, NCI-H460, NCI-H446,
NCI-H292 were obtained from the American Type Culture Collection.
The MCF-7 cell line was a gift from the Department of Cell Biology,
China Medical University. Cell culture reagents were obtained from
Gibco BRL (Grand Island, NY, USA). Protease inhibitors (leupeptin,
aprotinin, Na3VO4, and
phenylmethylsulfonylfluoride) were purchased from Sigma Aldrich
(St. Louis, MO, USA). Goat anti-human Ubc9 antibody and GAPDH
antibody were purchased from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA). Horseradish peroxidase (HRP)-conjugated secondary
antibody was obtained from Santa Cruz Biotechnology, Inc. Reagents
for SDS-PAGE were obtained from Bio-Rad (Hercules, CA, USA).
Chemiluminescent developing reagents and a Micro BCA Protein Assay
Reagent kit were obtained from Pierce Chemical Co. (Rockford, IL,
USA). All other reagents were of analytical grades.
Patients and tissue samples
Tissue samples were obtained from a total of 143
patients with lung cancer who were treated at Liaoning Cancer
Hospital and Institute from 2006 to 2010. Institutional Review
Board approval was obtained to procure and analyze the tissues used
in this study. None of the patients had received preoperative
neo-adjuvant chemotherapy or radiation therapy. Histological types
included squamous carcinoma, adenocarcinoma and small cell lung
cancer (SCLC) (Table I). No other
previous or concomitant primary cancer was present. The surgical
specimens were fixed in formalin and embedded in paraffin before
they were archived. Ubc9 expression in the tumor specimens was
detected using immunohistochemical staining.
 | Table ICorrelation between Ubc9 expression
and clinical pathological factors. |
Table I
Correlation between Ubc9 expression
and clinical pathological factors.
| Variable | Case (n and %) | The level of Ubc9
expression | χ2 value
(P) |
|---|
| |
| |
|---|
| | > median values
(n) | < median values
(n) | |
|---|
| Gender |
| Male | 97 (67.83%) | 51 | 46 | 2.26 (>0.05) |
| Female | 46 (32.17%) | 18 | 28 | |
| Age (years) |
| >60 | 51 (35.66%) | 40 | 11 | 25.01 (<0.01) |
| ≤60 | 92 (64.34%) | 32 | 60 | |
| Histological
type |
| Squamous
carcinoma | 31 (21.68%) | 18 | 13 | 7.02 (>0.05) |
| Adenocarcinoma | 28 (19.58%) | 9 | 19 | |
| SCLC | 79 (55.25%) | 42 | 37 | |
| Adenosquamous
ca. | 1 (0.7%) | 1 | 0 | |
| Large cell
carcinoma | 1 (0.7%) | 0 | 1 | |
| Alveolar
carcinoma | 3 (1.09%) | 1 | 2 | |
| Grade of
differentiation |
| Poor | 95 (66.43%) | 50 | 45 | 0.45 (>0.05) |
| Moderate | 32 (22.38%) | 16 | 16 | |
| Well | 16 (11.19%) | 7 | 9 | |
| Nodal status |
| N (+) | 118 (82.52%) | 78 | 40 | 5.94
(<0.05) |
| N (−) | 25 (17.48%) | 10 | 15 | |
| TNM |
| I | 41 (28.67%) | 15 | 26 | 9.57
(<0.05) |
| II | 32 (20.98%) | 20 | 12 | |
| III | 50 (34.97%) | 28 | 22 | |
| IV | 20 (15.38%) | 15 | 5 | |
| Survival |
| Yes | 91 (63.64%) | 33 | 58 | 16.12
(<0.01) |
| No | 52 (36.36%) | 37 | 15 | |
Immunohistochemistry
All specimens for immunohistochemical staining were
fixed in 10% neutral formalin, embedded in paraffin and cut in 5-μm
serial sections. Immunohistochemical staining was performed using a
peroxidase-labeled streptavidin-biotin technique. Briefly, tissue
sections were deparaffinized and rehydrated. Then, sections were
heated in a microwave oven for 10 min to retrieve antigenicity and
were treated with 3% H2O2 in methanol for 10
min to quench endogenous peroxidase activity. After washing in 10
mM PBS (pH 7.6), sections were incubated with 10% normal goat serum
(Solarbio Science and Technology, Beijing, China) for 10 min to
block nonspecific antibody binding. Sections were then incubated
overnight at 4°C with goat anti-human Ubc9 polyclonal antibody
(1:100). After washing in PBS, sections were treated with a 1:100
dilution of biotinylated donkey anti-goat IgG for 30 min followed
by a streptavidin-peroxidase conjugate for 30 min (Dako). A
solution of 0.02% diaminobenzidine hydrochloride (DAB) containing
0.03% H2O2 was used as chromogen to visualize
peroxidase activity. The preparations were lightly counterstained
with hematoxylin, mounted with Permount (Thermo Fisher Scientific,
Waltham, MA, USA) and examined by light microscopy.
Plasmid construction and cell
transfection
Human Ubc9 cDNA (GenBank accession no. NM_003345)
was obtained by reverse transcription from NCI-H446 cells using the
following primers: 5′-CGGAATTCCCACCATGTCGGGGAT-3′ and
5′-CGGGATCCAATGAGGGCGCAAAC-3′. PCR conditions were: 95°C for 2 min
(94°C 10 sec, 58°C 1 min, 72°C 2.5 min) 30 cycles at 72°C for 2
min. The PCR product was cloned into the pGEM-T vector (Promega,
Madison, WI, USA). The resulting plasmid, pGEM-T-Ubc9, was
sequence-verified. The Ubc9 fragment was then sub-cloned into the
pEGFP-vector (Invitrogen, Carlsbad, CA, USA) using the XhoI
and BamHI restriction enzymes. The resulting pEGFP
recombinant plasmid containing Ubc9 was also sequence-verified.
Cell culture and transfection
The human lung cancer cells A549, NCI-H460,
NCI-H446, NCI-H292 and MCF-7 were maintained in RPMI-1640 medium
containing 10% FBS, 100 IU/ml penicillin and 100 μg/ml
streptomycin. All cell lines were incubated in 5% CO2 at
37°C. Stable transfections of NCI-H446 cells were carried out with
the following expression vectors: pEGFP-N1-vector and
pEGFP-N1-Ubc9. Single cell clones were selected using G418 and
confirmed by western blotting.
Western blotting
Western blotting was performed using standard
techniques as previously described. Briefly, cells were washed
twice with PBS buffer and lysed in RIPA lysis buffer (50 mM Tris-Cl
pH 7.4, 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 0.1% SDS,
1 mM EDTA, 100 mM NaF, 1 mM Na3VO4, 1 mM
PMSF, and 2 μg/ml aprotinin) on ice. Total proteins (50 μg) were
subjected to sodium dodecyl sulfate-polyacrylamide gel
electrophoresis (SDS-PAGE) and transferred to polyvinylidene
difluoride membranes. Membranes were blocked with 5% nonfat milk in
TBST (10 mM Tris, pH 7.4, 150 mM NaCl and 0.1% Tween-20) at room
temperature for 2 h and incubated with the indicated primary
antibodies at 4°C overnight with gentle rocking. After washing with
TBST, the membrane was reacted with the appropriate HRP-conjugated
secondary antibodies for 1 h at room temperature. After extensive
washing with TBST, proteins were visualized by the enhanced
chemiluminescence (ECL) detection kit in accordance with the
manufacturer’s recommendations.
Matrigel in vitro invasion assay
NCI-H446 cells (1.50×104) were placed in
the upper chamber of 8 μm Transwells (Costar Inc.) on 100 μl of
solid, growth factor reduced Matrigel (BD Biosciences, Bedford, MA,
USA). RPMI-1640 medium supplemented with 20% FBS was added to the
wells and then cells were incubated for 4 h at 37°C. At the end of
the migration assay, the filter side of the upper chamber was
cleansed with a cotton swab and the filter was stained for 1 h with
crystal violet (Sigma) in 2% ethanol and rinsed with water. The
filter was gently cut from the chamber and the cells that had
migrated through the filter pores from the underside of the filter
were counted. Four high-power fields per insert were counted and
the values were averaged. Both for NCI-H446+pEGFP-N1-vector and
NCI-H446+pEGFP-N1-Ubc9, three identical replicates were
performed.
Cell scratch migration assay
Cells were seeded in 6-well plates and cultured
until confluent. A pipette tip was used to make a straight scratch
across the diameter of the dish, simulating a wound. Cell migration
was documented using phase contrast microscopy at ×10 at 12, 24, 48
and 72 h.
In vivo metastasis assay
Nude mice were anesthetized by intraperitoneal
injection of Nembutal [45 mg/kg of pentobarbital (50
mg/ml)/saline/ethanol/propylene glycol 10/63/7/18]. The anterior
chest wall was scrubbed with 70% alcohol. A 30-gauge needle on a
tuberculin syringe was inserted into the second intercostal space 3
mm to the left of the sternum and aimed centrally. The spontaneous
and continuous entrance of pulsating blood into the transparent
needle hub indicated proper positioning of the needle into the left
ventricle of the heart. Tumor cells [106 cells in 0.1 ml
Hank’s balanced salt solution (HBSS)] were injected over a 20–40
sec period.
Statistical analysis
Statistical analysis was performed using the
Student’s t-test. Data are presented as the means ± SD and p-values
<0.05 were considered to indicate statistically significant
differences. All calculations were carried out using the SPSS13.0
statistical package.
Results
Expression of Ubc9 in lung cancer
patients and its clinical significance
We observed Ubc9 immunostaining in all human lung
tissue sections and normal tissue. The Ubc9 protein level was
markedly elevated in the lung tumor specimens compared with the
normal lung tissue, including both non-small cell lung cancer
(NSCLC) and SCLC tissue types. Moreover, the level of Ubc9 protein
in NSCLC was higher than in SCLC (Fig.
1A and B). The relationship between the level of Ubc9
expression and the clinicopathological characteristics of lung
cancer are summarized in Table I.
In this study, the staining intensity of Ubc9 in tumor tissues was
assessed as Ubc9 expression levels and was quantified by the IOD
values. The IOD values of Ubc9 for the tumor samples ranged from
1663.75 to 64533.7, with a median value of 16523.9. Patients were
divided into low and high expression groups, based on whether Ubc9
levels were above or below the median value. There were significant
differences correlation between the levels of Ubc9 expression and
age, nodal metastasis status, UICC-TNM classification and the
probabilities of cancer-specific survival. However, there were no
significant differences between the levels of Ubc9 expression and
gender, histological type and grade of differentiation. These data
suggested that patients with the high Ubc9 expression were more
likely to have advanced disease than those with the low Ubc9
expression, which correlated with survival of patients with lung
cancer.
The level of Ubc9 mRNA in lung cancer tissues was
quantitated using real-time PCR. Ubc9 mRNA expression of Ubc9 was
again detected in lung normal tissue and lung cancer tissue,
including NSCLC and SCLC. Similar to the levels of Ubc9 protein,
the level of Ubc9 mRNA in NSCLC samples was significantly higher
than in SCLC samples (Fig. 1C). We
also tested the level of Ubc9 mRNA and protein using RT-PCR and
western blotting in the cell lines, including A549, NCI-H460,
NCI-H446, NCI-H292 and MCF-7, and the level of Ubc9 mRNA in
NCI-H446 was significantly lower than in other cell lines, as the
levels of Ubc9 protein (Fig. 2A and
B).
The invasive ability of different lung cancer cells
was analyzed using a cell invasion assay (Fig. 2C). The results showed that the
ability of lung cancer cells to invade correlated with their levels
of Ubc9. These results suggest that Ubc9 promotes lung cancer
invasion and metastasis.
Overexpression of Ubc9 enhances NCI-H446
cell invasion and migration
To further investigate the role of Ubc9 in lung
cancer, we constructed the mammalian expression vector:
pEGFP-N1-Ubc9 and pEGFP-N1-vector, stably transfected NCI-H446
cells, a SCLC cell, generated a stable cell line expressing
pEGFP-N1-Ubc9 and pEGFP-N1-vector. Expression of the fusion protein
in the pEGFP-N1-Ubc9 clones was detected by western blotting
(Fig. 3A). The NCI-H446 cells
stably transfected with the pEGFP-N1-vector were used as the
control. Transfectants were selected using G418 and the resulting
cell lines were designated pEGFP-N1-Ubc9 and pEGFP-N1-vector,
respectively. Using a Matrigel cell invasion assay, we found that
NCI-H446 cells overexpressing Ubc9 had a significantly enhanced
ability to invade (Fig. 3B).
Furthermore, a cell scratch assay demonstrated that the ability of
NCI-H446 cells to migrate was also significantly enhanced (Fig. 3C). These data indicated that higher
levels of Ubc9 were sufficient to promote increased invasion and
migration in NCI-H446 cells.
Ubc9 promotes NCI-H446 cell lung
metastasis in vivo
To directly test the hypothesis that Ubc9 has a
causal role in the metastasis of lung cancer cells, we used stable
transfection of Ubc9 cDNA to increase the level of Ubc9 protein in
NCI-H446 cells. Then, we injected these cells into the left heart
ventricle of nude mice in order to evaluate their ability to
metastasize to the lung. Injection of either the
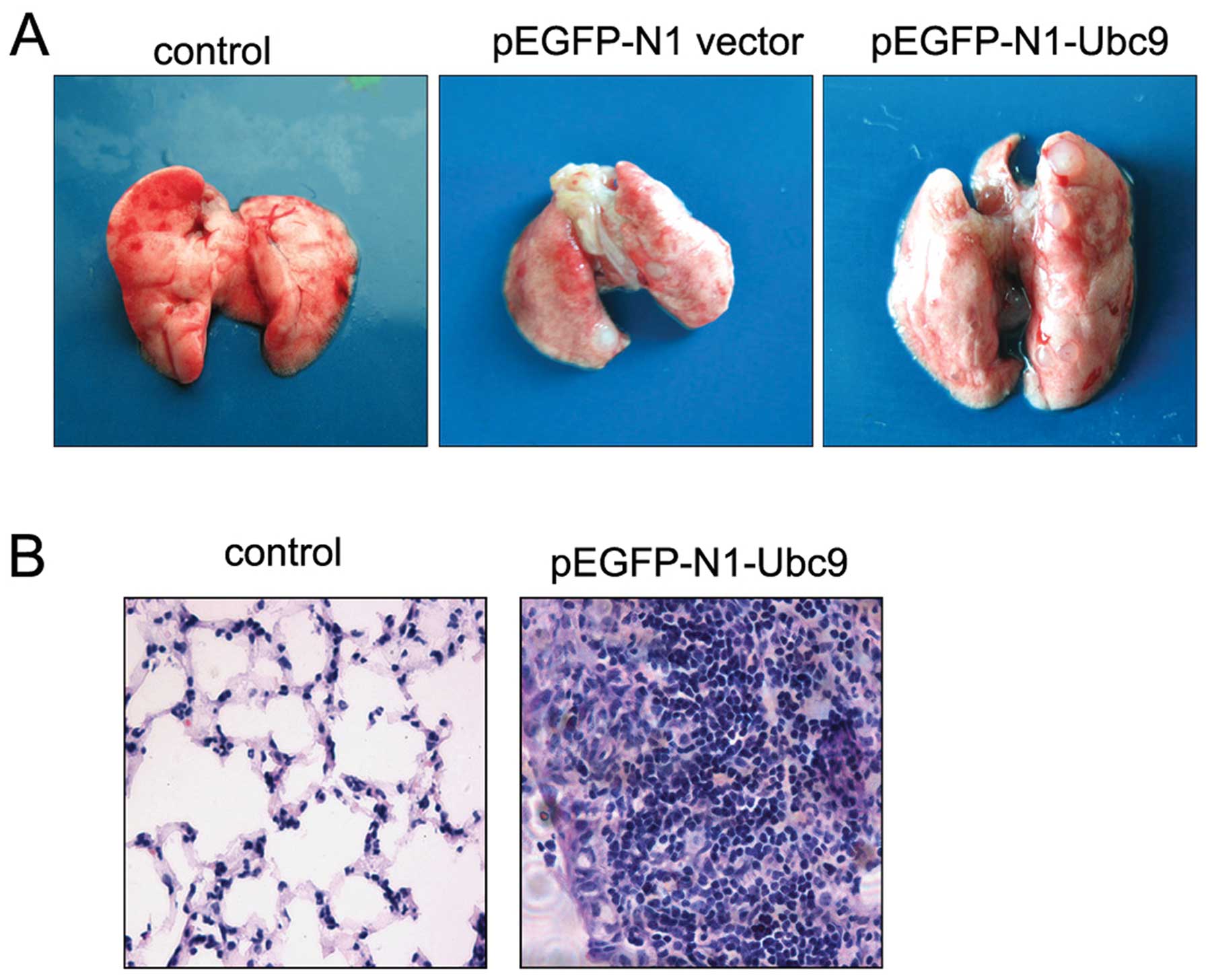
pEGFP-N1-vector/NCI-H446 cells or the pEGFP-N1-Ubc9/NCI-H446 cells
produced lung metastatic lesions in the injected mice. Some visible
metastatic nodules were observed even in nude mice injected with
either the pEGFP-N1 vector/NCI-H446 or the pEGFP-N1-Ubc9/NCI-H446
cells. However, the metastatic ability of pEGFP-N1-Ubc9/NCI-H446
cells was markedly increased compared with that of the control
cells (Table II). The pattern of
lung metastases in the pEGFP-Ubc9/NCI-H446 cells was characterized
by the formation of multiple foci; however, only one unique
metastatic nodule occurred in mice injected with the pEGFP-N1
vector/NCI-H446 cells (Fig. 4A).
The metastatic tumors formed in the lung had morphological
characteristics typical of SCLC tumors (Fig. 4B). By contrast, no metastatic nodule
formation was observed in the lungs of the mice injected with HBSS.
These results show that upregulation of Ubc9 expression enhances
lung metastasis of human SCLC cells.
 | Table IIEffects of upregulation of Ubc9
expression on lung metastasis in nude mice. |
Table II
Effects of upregulation of Ubc9
expression on lung metastasis in nude mice.
| | Lung
metastasis |
|---|
| |
|
|---|
| Cell line | Incidence | Survival |
|---|
| NCI-H446 cells | HBSS | 0/4 | All >90 |
|
pEGFP-N1-vector | 3/10 | 56 to >90 |
| pEGFP-N1-Ubc9 | 8/10 | 40 to >90 |
Discussion
As is well known, post-translational modifications
play an important role in protein function through the regulation
of their activity, turnover and localization and/or interactions.
One such modification involves the covalent attachment of the small
ubiquitin-related polypeptide SUMO (small ubiquitin-like modifier)
to different cellular protein substrates (8,18).
Sumoylation has been implicated in the regulation of protein
stability, protein-protein interactions, transcriptional activity
and subcellular localization (19,20).
As an essential E2 conjugating enzyme for sumoylation, Ubc9 plays a
key role in sumoylation-mediated cellular pathways. Evidence
suggests that Ubc9 is a tumor promoting factor. Ubc9 is well known
for its key function in protein sumoylation and
sumoylation-regulated cellular pathways, ultimately affecting tumor
initiation and progression (21).
However, little is known about the regulation of Ubc9 in tumor cell
invasion and metastasis. Our study provides evidence that Ubc9
promotes both lung cancer invasion and metastasis.
Ubc9 is the sole E2 conjugating enzyme required for
protein sumoylation (22). Ubc9
plays a role in a number of important biological processes, such as
progression through M phase, DNA damage repair, maintenance of
nuclear integrity and chromosome segregation. Several cell
signaling molecules such as p53, Rad51, MITF, Smad4, c-Jun, Daxx,
WT-1, NF-κB and sex hormone receptors are target proteins of Ubc9
(23–26). In contrast to the ubiquitin pathway
which utilizes several E2 conjugating enzymes, Ubc9 is the only
known SUMO E2 enzyme and therefore a key regulator of the
sumoylation pathway. As an essential E2 enzyme, Ubc9 is considered
to play a central role in sumoylation-regulated cellular pathways
which therefore suggests a putative role for sumo-regulation of
pathways linked to tumorigenesis and drug resistance.
Ubc9 has been specifically linked to tumorigenesis.
Ubc9 is overexpressed in several malignancies, such as lung
adenocarcinoma (27), ovarian
carcinoma (28), and melanoma
(16). Antagonizing Ubc9 function
in MCF-7 breast cancer cells transplanted in nude mice inhibited
cell growth and increased apoptosis via Bcl-2 dependent mechanisms
(29). These data suggest that Ubc9
may act by preventing activation of apoptotic pathways. In one
study, ectopic expression of Ubc9 was shown to enhance the invasive
ability of lung cancer cells. Additional studies have suggested
that the Ubc9 gene may play a pivotal role in lung tumorigenesis
and may potentially serve as a biomarker for both the diagnosis and
prognosis of human lung cancer.
In accordance with previous studies (16,30,31),
higher levels of Ubc9 expression were detected in primary lung
cancer as compared with normal lung tissue. Moreover, both protein
and mRNA level of Ubc9 in NSCLC were higher than in SCLC. In
patient tissue samples, the Ubc9 expression level was significantly
correlated with more invasive lung cancer. These data suggest that
Ubc9 is associated with the progression of lung cancer. Our study
also provides experimental evidence that Ubc9 plays a crucial role
in lung cancer cell invasion and migration. We transferred the
eukaryotic expression construct pEGFP-Ubc9 to NCI-H446 cells and
selected NCI-H446 single-clone cell lines stably overexpressing the
Ubc9 gene. Using a Matrigel invasion assay, we found that the
NCI-H446 cells overexpressing Ubc9 protein showed an enhanced
invasion ability as compared to control cells. We also analyzed the
effect of Ubc9 on cell migration and found similar results.
To further determine whether Ubc9 has a function in
tumor metastasis, we performed in vivo metastasis assays by
injecting transfected NCI-H446 cells into nude mice through the
cardiac ventricle and measured metastasis to the lung. Similar to
the results of our in vitro invasion assay, NCI-H446 cells
overexpressing Ubc9 protein induced more metastatic nodules than
the control cells. Moreover, the lung tumor nodules induced by the
Ubc9 overexpressing cells contained more foci than those induced by
the control cells. The H&E stain revealed that the tumor
nodules induced by NCI-H446 cells overexpressing Ubc9 displayed the
histomorphology of SCLC. These data indicate that NCI-H446 cells
overexpressing Ubc9 protein have increased tumorigenicity which
suggests that Ubc9 promotes lung cancer invasion and metastasis
in vivo. Our study suggests that Ubc9 plays a causal role in
cancer invasion and metastasis. This is likely due to the fact that
Ubc9 is an essential enzyme for sumoylation and numerous important
proteins, such as tumor suppressors or oncoproteins, are substrates
for sumoylation. Similarly, Ubc9-mediated sumoylation has been
shown to be involved in diverse cellular pathways (19). Therefore, cancer cells may have
evolved mechanisms to target the basic functions of these protein
modification pathways, involving microRNA regulation of Ubc9 at the
post-transcriptional level, leading to its upregulation in
tumors.
Metastasis is detected in 90% of lung cancer-related
deaths (25). As metastasis
involves different cellular processes than those that are involved
in the onset and early stages of tumorigenesis, metastatic cancer
can be more effectively treated if the metastasis process can be
specifically targeted by therapeutics. Then, identifying key
molecules involved in metastasis may aid substantially in the early
diagnosis of metastatic lung cancer, thus reducing patient
mortality. Our results indicate that Ubc9 may function as a
potential molecular marker in metastatic lung cancer, and may be a
new target for lung cancer therapy.
In summary, we provided clinical and experimental
evidence that Ubc9 is differentially expressed in lung cancer cells
and contributes to lung cancer cell migration and metastasis. Ubc9
may serve as a potential biomarker as a therapeutic target for
cancer intervention.
Acknowledgements
The study was supported by Outstanding Scientific
Fund of Shengjing Hospital.
References
|
1
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun M: Cancer statistics, 2008. CA Cancer J Clin. 58:71–96.
2008. View Article : Google Scholar
|
|
3
|
Hirsch FR, Franklin WA, Gazdar AF and Bunn
PA Jr: Early detection of lung cancer: clinical perspectives of
recent advances in biology and radiology. Clin Cancer Res. 7:5–22.
2001.PubMed/NCBI
|
|
4
|
Erridge S, Moller H, Price A and Brewster
D: International comparisons of survival from lung cancer: pitfalls
and warnings. Nat Clin Pract Oncol. 4:570–577. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kligerman S and White C: Epidemiology of
lung cancer in women: risk factors, survival, and screening. AJR Am
J Roentgenol. 196:287–295. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cagle PT, Allen TC, Dacic S, Beasley MB,
Borczuk AC, Chirieac LR, Laucirica R, Ro JY and Kerr KM: Revolution
in lung cancer: new challenges for the surgical pathologist. Arch
Pathol Lab Med. 135:110–116. 2011.PubMed/NCBI
|
|
7
|
Steeg PS: Metastasis suppressors alter the
signal transduction of cancer cells. Nat Rev Cancer. 3:55–63. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Muller S, Hoege C, Pyrowolakis G and
Jentsch S: SUMO, ubiquitin’s mysterious cousin. Nat Rev Mol Cell
Biol. 2:202–210. 2001.
|
|
9
|
Gill G: SUMO and ubiquitin in the nucleus:
different functions, similar mechanisms? Genes Dev. 18:2046–2059.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hay RT: SUMO: a history of modification.
Mol Cell. 18:1–12. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin D, Tatham MH, Yu B, Kim S, Hay RT and
Chen Y: Identification of a substrate recognition site on Ubc9. J
Biol Chem. 277:21740–21748. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Duan X, Trent JO and Ye H: Targeting the
SUMO E2 conjugating enzyme Ubc9 interaction for anti-cancer drug
design. Anticancer Agents Med Chem. 9:51–54. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Niedenthal R: Ubc9 fusion-directed
SUMOylation (UFDS). Biochem Soc Trans. 35:1430–1432. 2007.
View Article : Google Scholar
|
|
14
|
Galanty Y, Belotserkovskaya R, Coates J,
Polo S, Miller KM and Jackson SP: Mammalian SUMO E3-ligases PIAS1
and PIAS4 promote responses to DNA double-strand breaks. Nature.
462:935–939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vertegaal AC: SUMO chains: polymeric
signals. Biochem Soc Trans. 38:46–49. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ahn JH, Xu Y, Jang WJ, Matunis MJ and
Hayward GS: Evaluation of interactions of human cytomegalovirus
immediate-early IE2 regulatory protein with small ubiquitin-like
modifiers and their conjugation enzyme Ubc9. J Virol. 75:3859–3872.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moschos SJ, Smith AP, Mandic M, et al:
SAGE and antibody array analysis of melanoma-infiltrated lymph
nodes: identification of Ubc9 as an important molecule in
advanced-stage melanomas. Oncogene. 26:4216–4225. 2007. View Article : Google Scholar
|
|
18
|
Wu F, Zhu S, Ding Y, Beck WT and Mo YY:
MicroRNA-mediated regulation of Ubc9 expression in cancer cells.
Clin Cancer Res. 15:1550–1557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Johnson ES: Protein modification by SUMO.
Annu Rev Biochem. 73:355–382. 2004. View Article : Google Scholar
|
|
20
|
Geiss-Friedlander R and Melchior F:
Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol.
8:947–956. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Driscoll JJ, Pelluru D, Lefkimmiatis K, et
al: The sumoylation pathway is dysregulated in multiple myeloma and
is associated with adverse patient outcome. Blood. 115:2827–2834.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Moschos SJ, Jukic DM, Athanassiou C, et
al: Expression analysis of Ubc9, the single small ubiquitin-like
modifier (SUMO) E2 conjugating enzyme, in normal and malignant
tissues. Hum Pathol. 41:1286–1298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vertegaal AC: Small ubiquitin-related
modifiers in chains. Biochem Soc Trans. 35:1422–1423. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu SY and Chiang CM: p53 sumoylation:
mechanistic insights from reconstitution studies. Epigenetics.
4:445–451. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nowak M and Hammerschmidt M: Ubc9
regulates mitosis and cell survival during zebrafish development.
Mol Biol Cell. 17:5324–5336. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fu M, Wang C, Wang J, Zhang X, et al:
Androgen receptor acetylation governs transactivation and
MEKK1-induced apoptosis without affecting in vitro sumoylation and
trans-repression function. Mol Cell Biol. 22:3373–3388. 2002.
View Article : Google Scholar
|
|
27
|
Vatsyayan J, Qing G, Xiao G and Hu J:
SUMO1 modification of NF-kappaB2/p100 is essential for
stimuli-induced p100 phosphorylation and processing. EMBO Rep.
9:885–890. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Moschos SJ and Mo YY: Role of SUMO/Ubc9 in
DNA damage repair and tumorigenesis. J Mol Histol. 37:309–319.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Mo YY and Moschos SJ: Targeting Ubc9 for
cancer therapy. Expert Opin Ther Targets. 9:1203–1216. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lu Z, Wu H and Mo YY: Regulation of bcl-2
expression by Ubc9. Exp Cell Res. 312:1865–1875. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mo YY, Yu Y, Theodosiou E, Ee PL and Beck
WT: A role for Ubc9 in tumorigenesis. Oncogene. 24:2677–2683. 2005.
View Article : Google Scholar : PubMed/NCBI
|