Introduction
Colon cancer is the third most common cancer
worldwide (1) and it is also the
fourth leading cause of mortality among all types of cancer in the
United States (National Cancer Institute, 2009). Distant metastasis
and recurrence of colon cancer are major causes of mortality
(2,3). With the advances in chemotherapy and
the development of multi-disciplinary collaboration of treatment,
the survival rate of colon cancer has increased significantly in
recent years.
With the potential ability of self-renewal and the
uncertain ability of differentiation, cancer stem cells are cell
populations with a stem cell nature presenting in tumor tissue and
are the root of tumor formation and metastasis (4–7). In
recent years, a growing number of researchers have considered that
cancer is a disease caused by the tumor stem cells (8). At present, cancer stem cells are
isolated with a variety of solid tumors and hematological neoplasms
(5). This shows that the cancer
stem cells are a common phenomenon in malignancies. Some studies
have confirmed the presence of ovarian cancer stem cells. These
cells are not sensitive to chemotherapy and radiotherapy (9–12).
Qiang et al identified stem-like cells in human glioblastoma
cell lines U251, U87MG, A172 (13).
microRNAs are a class of non-coding single-stranded
RNA molecules in eukaryote. Through mediating degradation of target
mRNA and inhibiting target mRNA translation, microRNA can regulate
its target gene expression with post-transcriptional regulation.
Through complementary combining with the target mRNA
3′-untranslated region (UTR) completely/incompletely (14–16),
microRNAs regulate a variety of pathophysiological processes, such
as stem cell differentiation, cell proliferation and tumor
formation (17–20). Some microRNAs are closely associated
with the ability of self-renewal and differentiation of cancer stem
cells (21–23). Qian et al(24) successfully separated tumor stem
cells from the lung tissue of mice, finding abnormal expression of
miR-142-3p, miR-451, miR-106a, miR-142-5p, miR-15b, miR-20a,
miR-106b, miR-25, miR-486 in the tumor stem cells. This suggests
that miRNA regulates differentiation, invasion, apoptosis and
several other characteristics. Silber et al(25) showed that miR-124 and miR-137 can
induce differentiation of adult mouse neural stem cells, mouse
oligodendrocyte glioma and human glioblastoma. microRNAs also
regulate the biological function of tumor stem cells. In pancreatic
cancer stem cells, miR-34 can inhibit the self-renewal ability of
tumor stem cells by regulating the Notch and the Bcl-2 gene
(26).
miR-449b can decrease SKOV32ip l adhesion ability,
arrest cell cycle, increase the number of G1 phase cells, decrease
the number of S phase cells and can downregulate cell cycle-related
proteins CDK6 and CDC25A (27). Bou
et al(28) found a
diminished expression of miR-449 in gastrin KO mice. Overexpressing
miR-449, they showed a significant increase in the sub-G1 fraction
indicative of apoptosis. Compared to normal human gastric tissues,
they also discovered a loss of miR-449 expression (16). Through targeting pocket proteins,
Chen et al(29) found that
miR-449a plays a role as a tumor suppressor in human bladder
cancer.
Cell cycle regulators and regulatory proteins play
an important role in the tumor genesis process. CCND1 and E2F3 are
key in cell cycle regulation. Cyclins, cyclin-dependent kinases and
cyclin-dependent kinase inhibitors all have the ability to regulate
cell cycle. The expression of CCND1 is closely linked to Ras, Raf,
MAPK pathway and pRb family (30–34).
The overexpression of CCND1 is the characteristic of a variety of
human primary tumors (35–37), with significance for the diagnosis
of cancer. E2F3 transcription factor is an important regulatory
factor of cell cycle G1 to S phase (38,39).
E2F3 includes two protein products (E2F3a and E2F3b) sharing DNA
binding. E2F3a is closely related to DNA synthesis and cell cycle
progression. E2F3b is expressed throughout the cell cycle (39) and it is also linked to tumor genesis
and proliferation (40). Studies
have shown that E2F3 plays a role as an oncogene in most
organizations (40–42). E2F3 overexpression can contribute to
the formation of skin tumors in p53-deficient mice (43).
Based on the above, we first identified
differentially expressing microRNAs in colon cancer stem cells and
we then explored the relationship between differential expressing
microRNA (miR-449b) in colon cancer stem cells and cell cycle
regulation.
Materials and methods
Cell line and cell culture
Human colon cancer cell line SW1116 was obtained
from the Shanghai Institute of Cell Biology, affiliated to the
Chinese Academy of Sciences, Shanghai, China. The cell culture
method was the same as the one we previously reported (44). We separated
CD133+CD44+ and
CD133−CD44− cells using the same method as we
previously reported (44).
Total cellular RNA extraction
We used TRIzol (Invitrogen, Carlsbad, CA, USA) to
extract total cellular RNA following the manufacturer’s
instructions. Extracted RNA samples were quantified with NanoDrop
1000 (NanoDrop Technologies, Wilmington, DE, USA). The extracted
RNA samples were treated by DNase (Ambion, Austin, TX, USA) to
remove genomic DNA contamination.
Analysis with microRNA chip
Human microRNA OneArray® v3 was used to
analyze samples. microRNA chip performance and analysis methods
were the same as we previously reported (44).
Real-time PCR
Total RNA was extracted as described above. We
extracted RNA reverse transcribed into cDNA using a
PrimeScript® RT Reagent kit (Takara, China). miRNA
samples were first polyadenylated and were then reverse transcribed
into cDNA with a One Step PrimeScript® miRNA cDNA Synthesis kit
(Takara). Quantitative real-time PCR reaction was performed with
7000 Sequence Detection System (ABI) and with the reaction system,
which included strand cDNA (0.5 μl), forward and reverse primers
(both 0.5 μl) and SYBR Green Supermix (12.5 μl) (Tianjin, China).
The formula 2−ΔΔCT values
(ΔCt=Ctgene-Ctcontrol) was used to calculate
relative expressions.
Dual-luciferase report system assay
The fragments of pMIR- CCND1, pMIR-E2F3 and mutant
variants were cloned downstream of the luciferase reporter into
pMIR-REPORT vector (Ambion). The sequences cloned were:
5′-UUUACAAUGUCAUAUACUGCCAU-3′ (CCND1),
5′-UUUACAAUGUCAUAUUGACGGUU-3′ (CCND1-mut),
5′-CAAUUAAUUUGUAAACACUGCCA-3′ (E2F3), 5′-CAAUUAAUUUGUAAAGUGACGGA-3′
(E2F3-mut). Cells were cultured in 24-well plates. Then, we added
siPORT NeoFX Transfection Agent (AM4510, 1 μl; ABI, USA),
pre-miR-449b (4464066, 40 nM; ABI)/miR-449b precursor molecule
negative control (4464058, 40 nM, ABI), pMIR-E2F3 (0.1 μg) + pRL-TK
vector (0.01 μg)/pMIR-CCND1 (0.1 μg) + pRL-TK vector (0.01
μg)/pMIR-E2F3-mut (0.1 μg )+ pRL-TK vector (0.01 μg)/pMIR-CCND1-mut
(0.1 μg) + pRL-TK vector (0.01 μg) into 25 μl of Opti-MEM medium.
Then, 450 μl of cell suspension containing 20,000 cells were added
into the above-mentioned reagents. The 24-well plates were placed
into the cell culture incubator and cell proliferation was observed
with an optical microscope at 24 and 48 h. Cell lysates were used
for Dual-Luciferase® Reporter Assay System analysis,
according to the manufacturer’s instructions (Promega, Madison, WI,
USA).
Transfection
We prepared pre-miR-449b precursor molecule,
pre-miR-449b precursor molecule negative control and their
inhibitors for transfection. SW116 cells were cultured in 6-well
plates at ~60% confluence. Cells were supplemented with siPORT
NeoFX Transfection Agent (ABI, AM4510, USA) after 24 h. We replaced
siPORT NeoFX Transfection Agent with complete medium containing FCS
6 h later. To complete real-time PCR and western blot analyses, we
lysated cells 48 h after transfection.
Cell growth and viability test
Tetrazolium blue method (MTT) was used to determine
cell growth and viability. Cells in each group were seeded in
96-well plates. Each group had three parallel holes and each hole
was supplemented with 5 mg/ml MTT 20 μl after culturing cells for
48 h. Then, dimethyl sulfoxide (DMSO) 150 μl was added after
incubating for 4 h. The microplate reader determined the absorbance
value (A) of each well at 640 nm wavelength. Proliferation ratio
represents the extent of cell proliferation. Proliferation ratio =
A value of experimental group/control group A value × 100%.
Cell cycle test
The treated cells were cultured for 72 h before
washing with PBS. The cells were fixed overnight using ice ethanol
(70%, 4°C). Cells were stained for 30 min with PI (Sigma, St.
Louis, MO, USA). Cell cycle analyses were performed with the
FACSCalibur (Becton-Dickinson). CellQuest (BD Bioscience, San Jose,
CA, USA) was used as data analyzing software.
Western blot analysis
We washed and lysed cells with lysis buffer. Lysates
were centrifuged to collect supernatants. Protein was denatured and
separated in 10% SDS-PAGE and was then transferred to
nitrocellulose membranes. Non-fat milk (5%) was used to block
nitrocellulose membranes. The membranes were incubated with
antibodies against CCND1, E2F3, pRb, Rb and p53 overnight and we
then incubated membranes with secondary antibodies (horseradish
peroxidase; Santa Cruz Biotechnology, Santa Cruz, CA, USA). Using
the ECL procedure (Amersham Biosciences, Piscataway, NJ, USA), we
detected secondary antibodies.
Results
CD133+CD44+ colon
stem cell isolation and stem characteristics
Several studies have proved that CD133 and CD44 were
powerful markers to detect colon cancer stem cells (45–51).
We isolated CD133+CD44+ and
CD133−CD44− cells from the SW1116 cell line
with FACS as we previously reported (44). Following amplification, flow
cytometry was used to detect cell surface molecules (CD133 and
CD44). Using real-time PCR, we examined several stem cell-specific
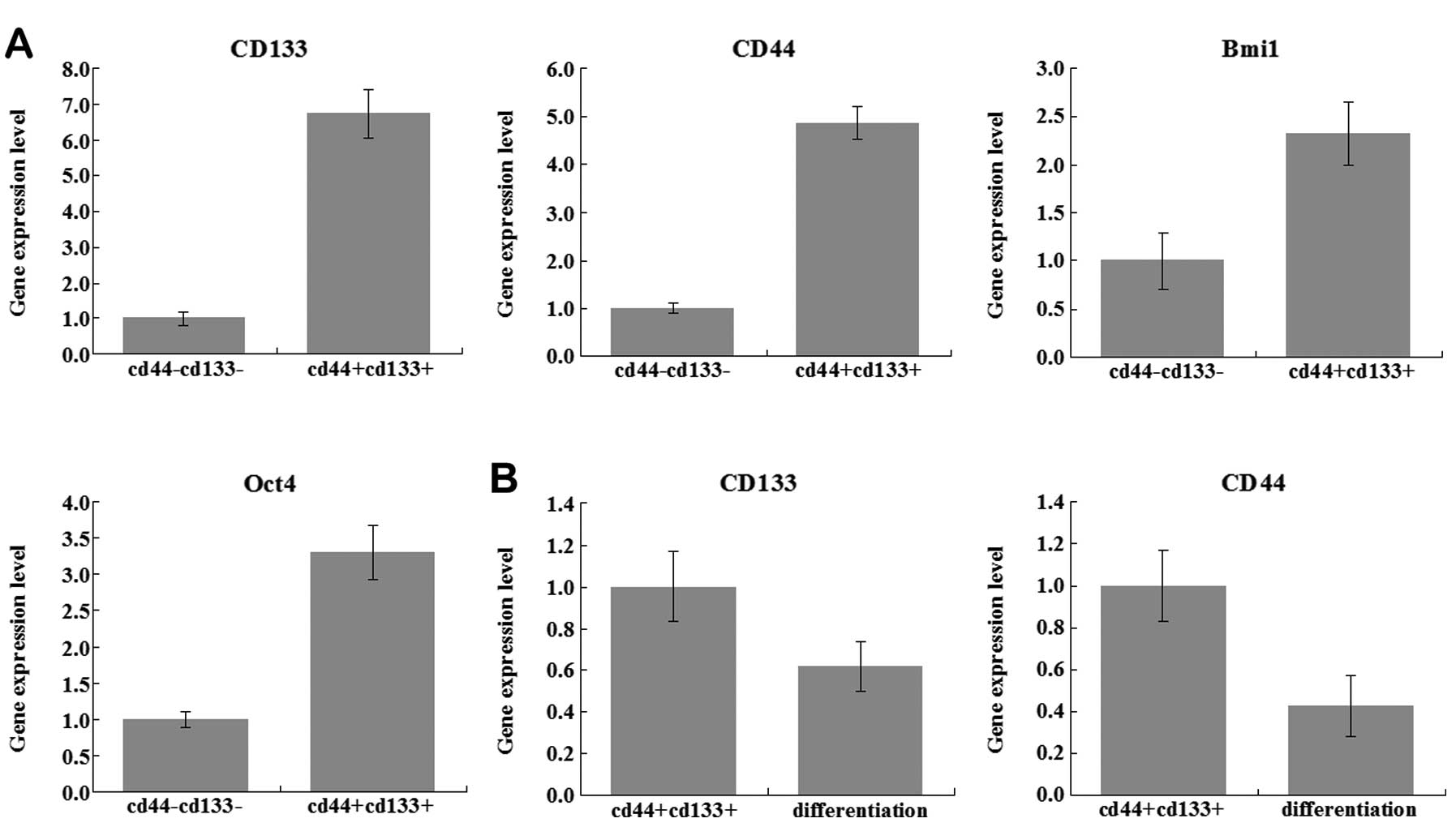
molecule expression levels; CD133, CD44, Bmi1 and Oct4. In the
CD133+CD44+ group, the expression levels of
these specific molecules were higher than in the
CD133−CD44− group (Fig. 1A). In the cell culture process, the
CD133+CD44+ cells can be gathered to become
balls of cells after ~10 days of culture, with EGF and β-EGF in
serum-free medium. Then, we moved the
CD133+CD44+ cells to normal medium culture.
Cells gradually restored the adherent nature. At the same time, we
found that the expression level of CD133, CD44 was also decreased
(Fig. 1B). Our study confirmed that
the CD133+CD44+ cells possess stem cell
phenotype, the specific molecular characteristics, the nature of
self-renewal and directed differentiation.
Differential microRNA expression profile
in colon cancer stem cells
With microRNA chip, we investigated the differential
microRNA expression profile in CD133+CD44+
colon cancer stem cells. We previously reported that there are 31
miRNAs upregulated and 31 miRNAs downregulated comparing the two
groups (44). miR-449b was included
in the most significantly downregulated list, whose average ratio
was 0.486804 (CD133+CD44+ cells vs.
CD133−CD44− cells).
Prediction of the target gene of
miR-449b
We thoroughly searched the microRNA database
(PicTar, TarBase, TargetScan and DIANA microT) bioinformatics
technical analysis to predict the candidate miR-449b target gene
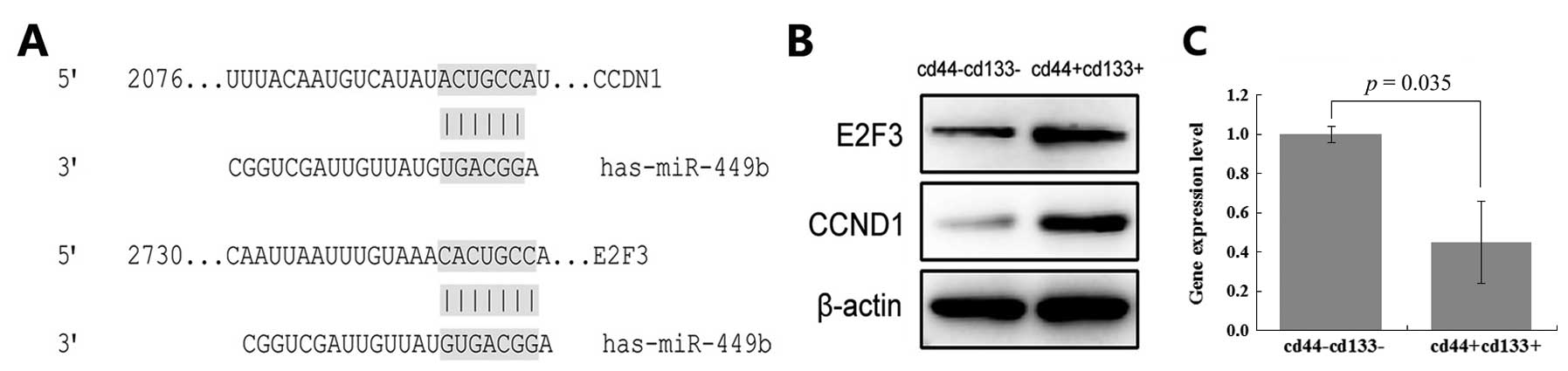
mRNA. The 3′UTRs of CCND1 and E2F3 contain miR-449 binding sites
(Fig. 2A). Thus, CCND1 and E2F3 may
target genes of miR-449b.
CCND1, E2F3 and miR-449b correlate
inversely in colon cancer stem cells
We verified the expression level of miR-449b in
CD133+CD44+ cells and
CD133−CD44− cells (Fig. 2C). miR-449b expression in the
CD133+CD44+ cells is only 0.455 times in the
CD133−CD44− cells. Then, we examined protein
levels of CCND1 and E2F3 (Fig. 2B).
Based on the above, CCND1, E2F3 and miR-449b correlate
inversely.
miR-449b regulates CCND1 and E2F3
expression in colon cancer stem cells
We tested CCND1 and E2F3 protein and mRNA expression
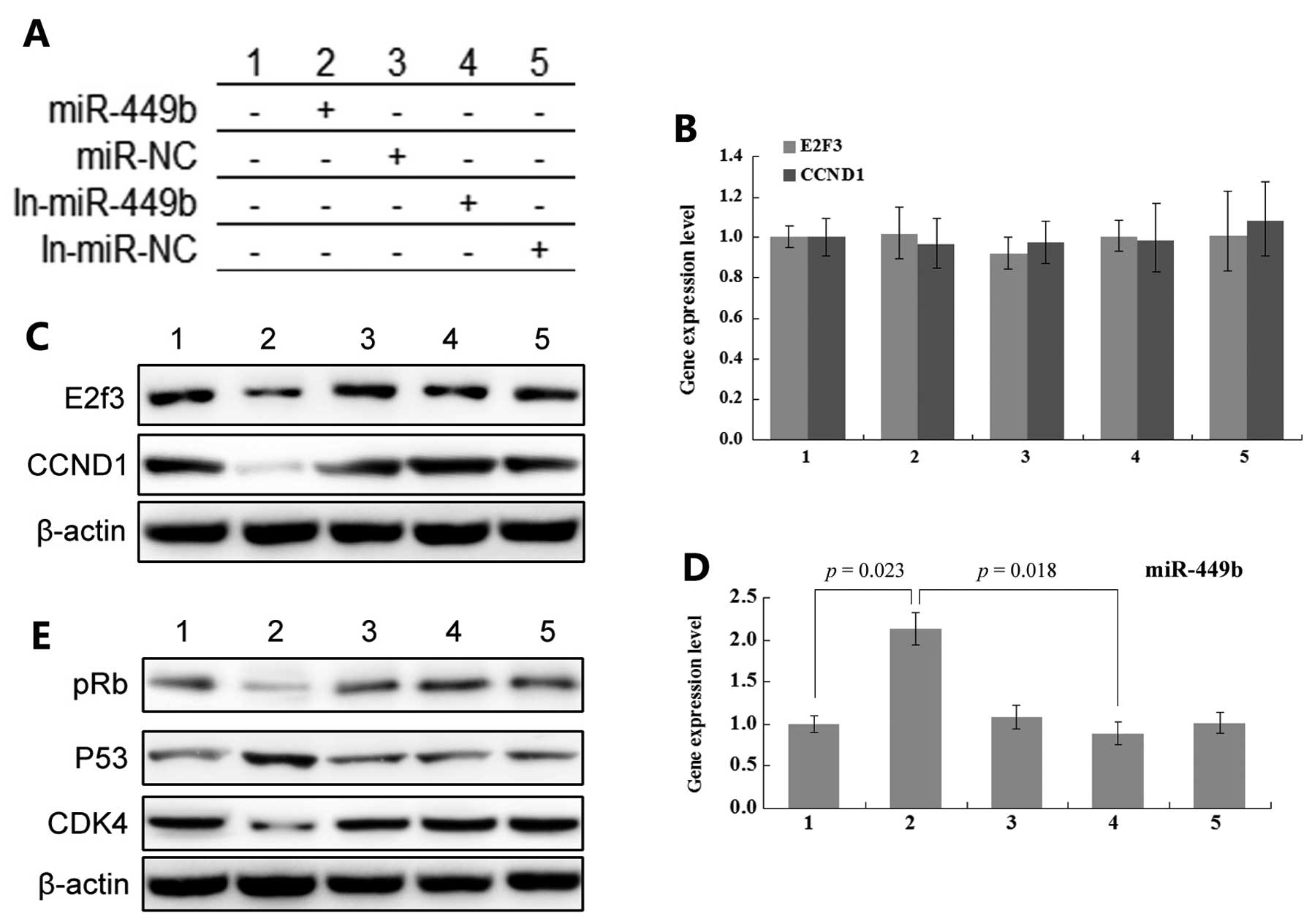
levels to evaluate the effect of miR-449b transfection. The
experiment set two groups of negative control; the blank control
group and the pre-miR-449b negative control group. The inhibitors
of pre-miR-449b and pre-miR-449b negative control were also
analyzed. In our experiment, transfection of pre-miR-449b reduced
the expression level of protein of CCND1 and E2F3. However, the
expression levels of mRNA of CCND1 and E2F3 presented only minor
changes. Moreover, the transfection of miR-449b inhibitor led to
the overexpression of protein of CCND1 and E2F3. The change of mRNA
level was not very sensitive (Fig.
3).
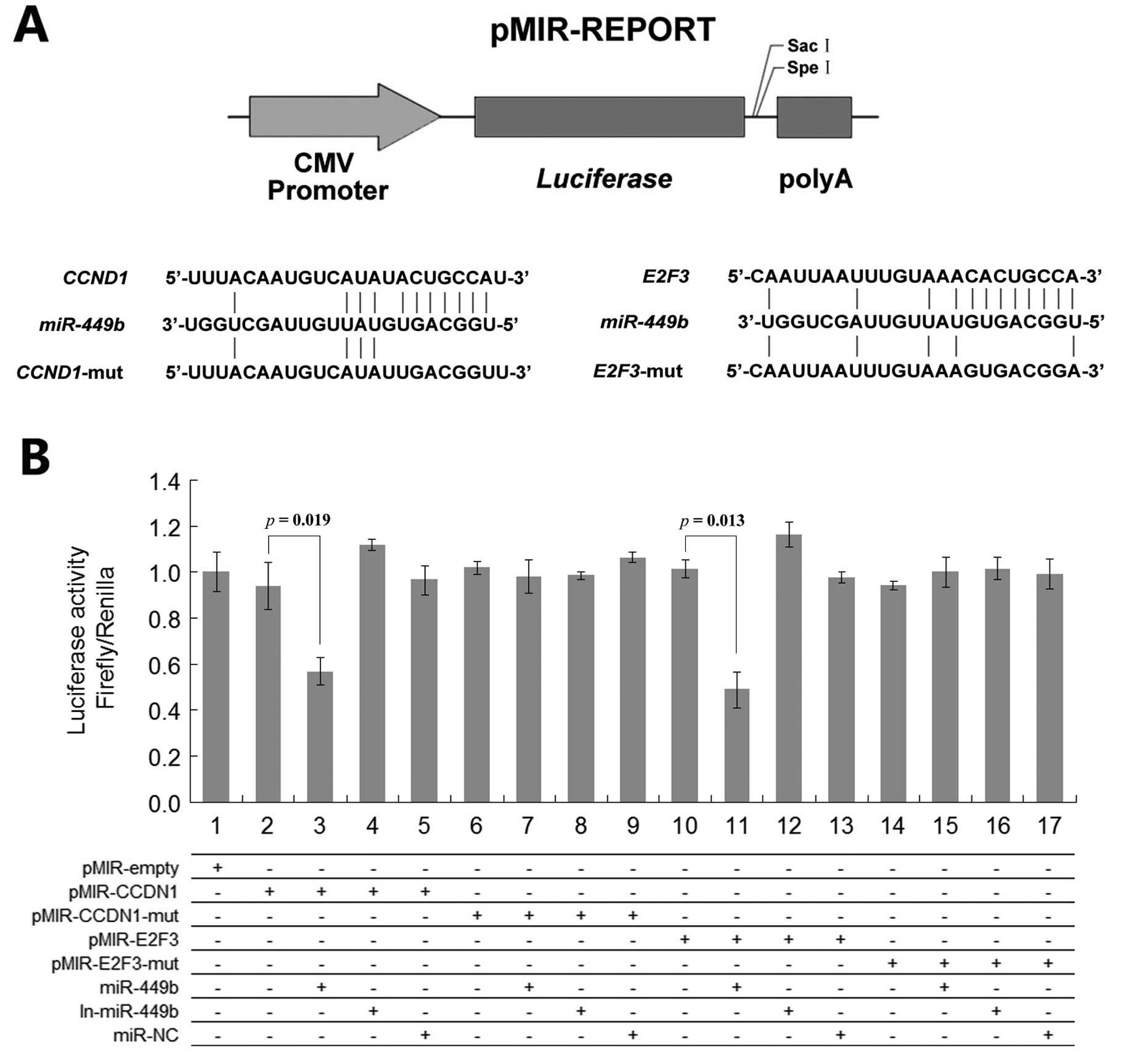
As the putative binding site of miR-449b in mRNA
3′-UTR region of CCND1 and E2F3, a Dual-Luciferase assay was
performed to determine the direct link between CCND1, E2F3 and
mir-449b. The mRNA region of CCND1 and E2F3 containing the miR-449b
putative binding site and mutant variants were cloned into
pMIR-REPORT vector. In Fig. 4,
fluorescence activity was significantly reduced in the pMIR-CCND1 +
pre-miR-449b group and in the pMIR-E2F3 + pre-miR-449b group.
Meanwhile, we found that transfecting miR-449b inhibitor can
increase the fluorescent expression while transfecting with
pre-miR-449b negative control had no effect (Fig. 4).
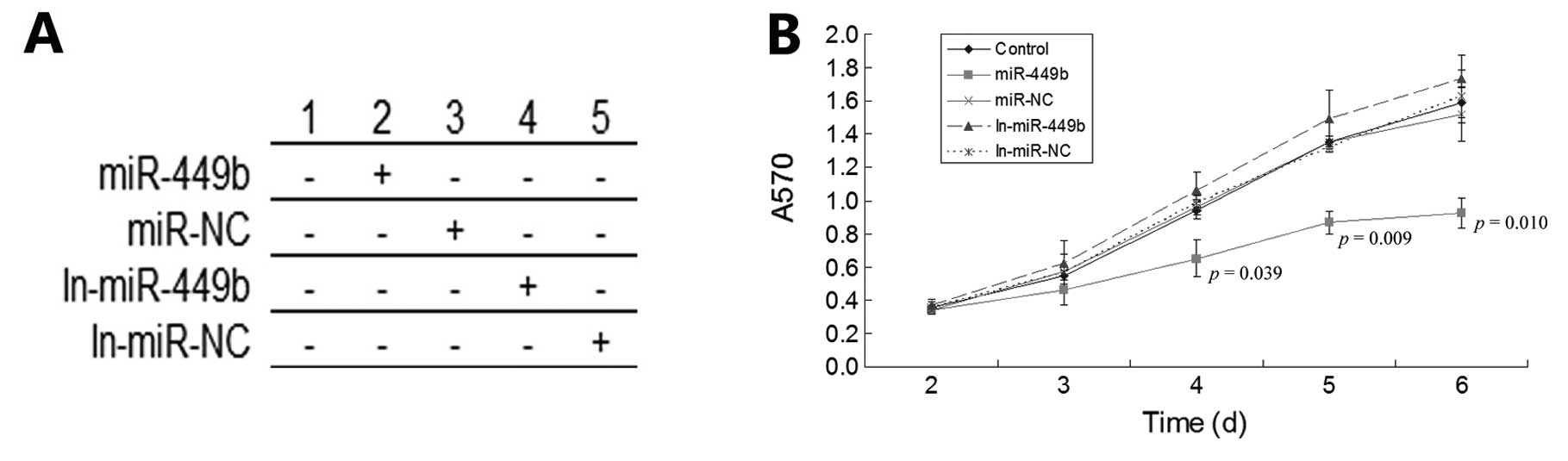
miR-449b influences cell growth and
viability
As mentioned above, the expression of CCND1 and E2F3
can promote the role of cell proliferation. Therefore, miR-449b can
be defined as a factor for the inhibition of cancer. In this
experiment, we detected cell proliferation in each group by MTT
assay. We observed cells in each group at different times after
transfection and we found that the pre-miR-449b group lost its
ability of proliferation significantly after 48 h. Conversely, in
the miR-449b inhibitor group, the proliferation ability of cells
increased slightly after 72 h (Fig.
5).
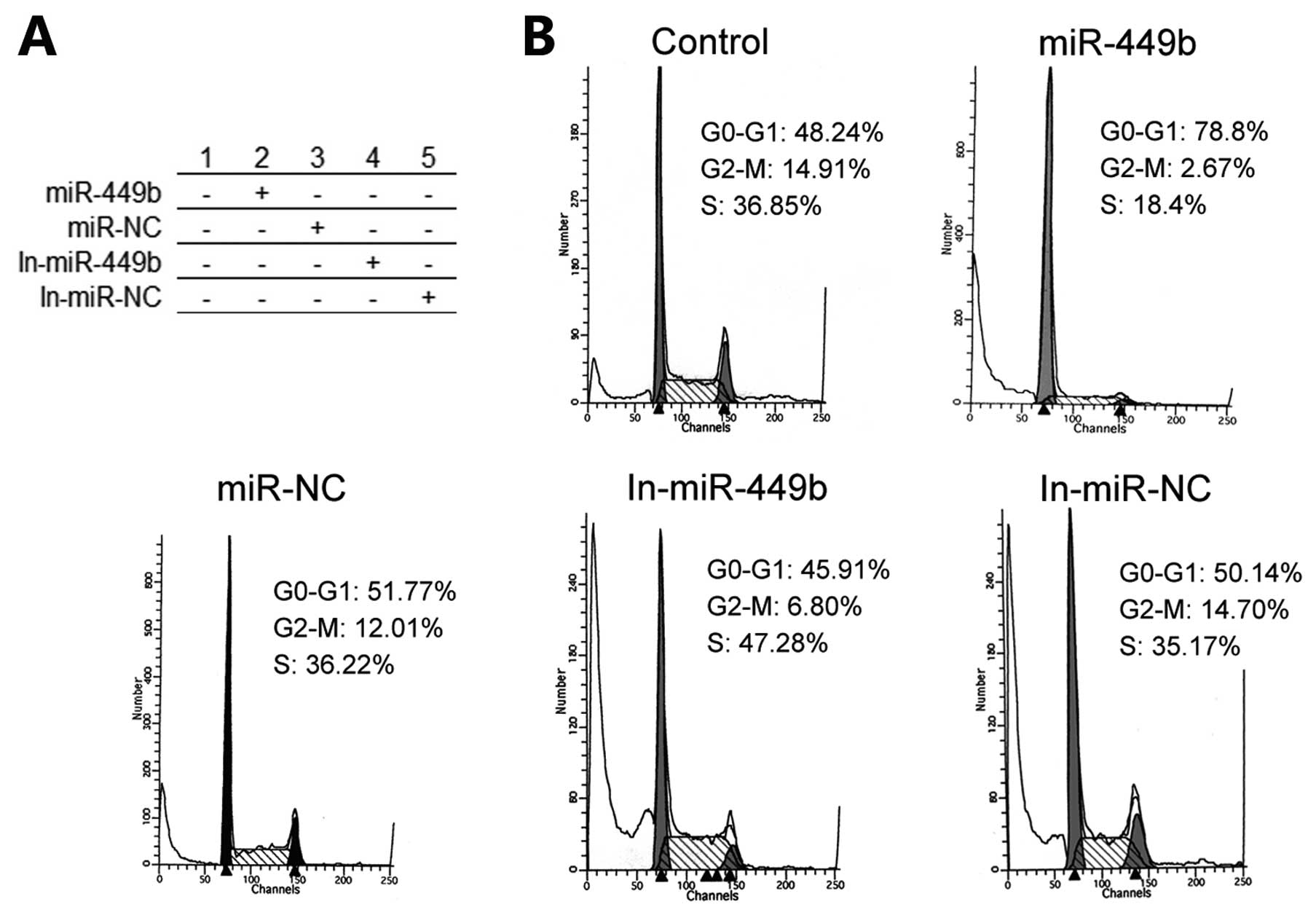
miR-449b affects the cell cycle
According to the above findings, miR-449b has the
ability to arrest cell cycle through controlling CCND1 and E2F3. A
flow cytometer was used to analyze the cell cycle of all treatment
groups. We found most of the pre-miR-449b group of cells in G
phases. Accordingly, the number of this group in the S phase was
significantly reduced. Other group processing by the miR-449b
inhibitor, cells in G phases were relatively reduced while cells in
S phase had a small increase (Fig.
6).
Expression level of pathway-related
molecules
By treating miR449b, we changed the expression level
of its target genes. This affects the expression levels of other
interrelated molecules in the pathway. Through the detection of
pRb, p53, and CDK4 molecular levels, we found an increased level of
expression of these molecules in the pre-miR-449b group (Fig. 3E).
Discussion
Previous studies suggested that differences of miRNA
expression levels play an important role in the physiological
function and phenotype of cells (52,53).
Cancer in various organs of the body have microRNA expression
profile changes. Iorio et al(54) reported that overall miRNA expression
can help separate normal vs. cancer tissues in human breast cancer.
Michael et al(55) found 28
different miRNAs identified in a colonic adenocarcinoma and normal
mucosa. They inhibited the expression of oncogenes while inhibiting
the expression of tumor suppressor genes, directly or
indirectly.
In cancer research, the hypothesis that cancer stem
cells are the root of tumor cells has gained attention. With
lineage tracing system, Schepers et al(56) confirmed a variety of different types
of tumor cells from the same stem cells expressing Lgr5+
in mice. Moreover, these stem cells are the driving force of tumor
development. Driessens et al(57) marked single tumor cells, while not
specifically labeling stem cells. They found that cells exhibit two
different modes of activities: before slowly depleting, they split
a few or several cells. This again confirmed a unique type of cell
subsets is the driving force of tumor growth. Therefore, the tumor
stem cell hypothesis is a scientific explanation of the occurrence
of cancer.
Based on these findings, it is evident that cancer
stem cells are special groups in the tumor cells. Tumor stem cells
may have a different microRNA expression profile vs. non-stem cell
groups. Liu et al(58)
showed miR-34a, a p53 target, was underexpressed in
CD44+ prostate cancer cells, and they confirmed that
miR-34a inhibits prostate cancer stem cells and metastasis by
directly suppressing CD44. Godlewski et al(59) showed that miR-128 caused a striking
decrease in the expression of the Bmi-1 oncogene, which is also a
stem cell renewal factor. Through access to the microRNA database,
several target genes of differentially expressing microRNAs are
closely related to the special biology phenotype of stem cells.
Possible target genes which have functions such as self-renewal,
differentiation, membrane receptor connection, metabolism,
tumor-related function are all regulated by differentially
expressing microRNAs.
Through further bioinformatics analysis, we found
that the expression levels of miR-449b and CCND1, E2F3 in colon
cancer stem cells and stem cells are negatively correlated. CCND1,
E2F3 have been proved to have the function of controlling cell
cycle. The same family members of miR-449b, miR-449a have also been
shown to be closely related to the miR-34 family. Target genes and
target pathways of the miR-34 family are closely related to cell
cycle regulation (29). Therefore,
miR-449b may also be related to the regulation of colon cancer stem
cell cycle. To prove this hypothesis, we transfected the precursor
of miR-449b and miR-449b inhibitor into the colon cancer stem
cells. As the colon cancer stem cell hypothesis demonstrated, the
expression level of CCND1, E2F3 and miR-449b correlated inversely
after transfection. However, this does not suffice to prove miR449b
directly regulated CCND1 and E2F3 through a silencing mechanism,
nor through other cellular factors or pathways. We performed a
Dual-Luciferase assay to prove this. We co-transfected miR-449b
precursor and reporter vectors containing E2F3 and CCND1 into
cells. A decrease in luciferase activity in groups containing
microRNA precursors and gene sequence reporter vectors was
successfully observed. This confirmed direct interaction between
CCND1, E2F3 and miR-449b. When we transfected mutants, these
phenomena were not observed. At the same time, we explored the
regulatory mechanism by which miR449b regulates E2F3 and CCND1. The
change of the target gene mRNA levels is less than the protein
level change. Thus, our evidence indicates that miR-449b uses two
ways to regulate target genes, degradation and reversible
combination.
We further studied the role of miR-449b in the colon
cancer stem cell cycle regulation. In the group transfected with
precursor of miR-449b, the number of cells in the G0–G1 phase
increased significantly. In the group transfected with inhibitor of
miR-449b, the opposite phenomenon was observed; the number of cells
in the G0–G1 phase decreased. This is also consistent with the
regulation of cell cycle function of E2F3 and CCND1. Similar to the
above, miR-449b also influences cell growth and viability. By MTT
assay, we found that the transfection of miR-449b precursors
significantly reduced the proliferation ability of colon cancer
stem cells; this may also be achieved through miR-449b direct
regulation of E2F3, CCND1. It was also consistent with the
phenomenon of E2F3 and CCND1 regulated by microRNAs confirmed by
previous reports (38,39,60,61).
These confirmed that miR-449b has a certain influence on cell
proliferation. Based on these, miR-449b, as its family member
miR-449a (29), plays a role of the
tumor suppressor gene.
In general, the regulation of cell signal is a
unified system. In the present study the colon cancer stem cell
expression levels of miR-449b changed. The level of its target
genes also changed. Some pathway-related molecule expression levels
have changed. In this study, in the pRb pathway, if active, the
complex of CCND-CDK4/6-Rb, Rb can be phosphorylated, promoting
cells from G1 to S phase. By inhibiting the expression of the Arf
gene, E2F3 can inhibit the expression of p53. This may result in
p53 tumor suppressor pathway dysfunction, promoting tumor
development. We have also seen these molecule expression levels
changed. This also shows miR-449b regulating cell cycle and
proliferation not only through E2F3 and CCND1, but also through
alteration of other molecules. miR-449b indirectly enhances p53
expression levels in colon cancer stem cells providing key evidence
to prove its tumor suppressor role.
In conclusion, our study shows that miR-449b may
have a direct relationship with E2F3 and CCND1 in colon cancer stem
cells in vitro. Notably, miR-449b can regulate the
expression level of E2F3 and CCND1 through the post-transcriptional
regulation. These in turn affect cell cycle regulation and
proliferation characteristics. The regulatory pathway related
molecules also change their expression levels. Regulating the
expression levels of miR-449b can affect a number of important stem
cell phenotypes in colon cancer stem cells. We can use it to
prevent the self-renewal and proliferation of colon cancer stem
cell, as the source of the colon cancer cells. miR-449b may become
an important target in the treatment of colon cancer in the
future.
References
|
1
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
2
|
Van Loon K and Venook AP: Adjuvant
treatment of colon cancer: what is next? Curr Opin Oncol.
23:403–409. 2011.PubMed/NCBI
|
|
3
|
Garcia-Foncillas J and Diaz-Rubio E:
Progress in metastatic colorectal cancer: growing role of cetuximab
to optimize clinical outcome. Clin Transl Oncol. 12:533–542. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clevers H: The cancer stem cell: premises,
promises and challenges. Nat Med. 17:313–319. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiao X, Katiyar S, Willmarth NE, et al:
c-Jun induces mammary epithelial cellular invasion and breast
cancer stem cell expansion. J Biol Chem. 285:8218–8226. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sullivan JP and Minna JD: Tumor
oncogenotypes and lung cancer stem cell identity. Cell Stem Cell.
7:2–4. 2010. View Article : Google Scholar
|
|
7
|
Li H and Tang DG: Prostate cancer stem
cells and their potential roles in metastasis. J Surg Oncol.
103:558–562. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zeki SS, Graham TA and Wright NA: Stem
cells and their implications for colorectal cancer. Nat Rev
Gastroenterol Hepatol. 8:90–100. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Silva IA, Bai S, McLean K, et al: Aldehyde
dehydrogenase in combination with CD133 defines angiogenic ovarian
cancer stem cells that portend poor patient survival. Cancer Res.
71:3991–4001. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Baba T, Convery PA, Matsumura N, et al:
Epigenetic regulation of CD133 and tumorigenicity of
CD133+ ovarian cancer cells. Oncogene. 28:209–218. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Landen CN Jr, Goodman B, Katre AA, et al:
Targeting aldehyde dehydrogenase cancer stem cells in ovarian
cancer. Mol Cancer Ther. 9:3186–3199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kurrey NK, Jalgaonkar SP, Joglekar AV, et
al: Snail and slug mediate radioresistance and chemoresistance by
antagonizing p53-mediated apoptosis and acquiring a stem-like
phenotype in ovarian cancer cells. Stem Cells. 27:2059–2068. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qiang L, Yang Y, Ma YJ, et al: Isolation
and characterization of cancer stem like cells in human
glioblastoma cell lines. Cancer Lett. 279:13–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim VN and Nam JW: Genomics of microRNA.
Trends Genet. 22:165–173. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shyu AB, Wilkinson MF and van Hoof A:
Messenger RNA regulation: to translate or to degrade. EMBO J.
27:471–481. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Garzon R, Fabbri M, Cimmino A, Calin GA
and Croce CM: MicroRNA expression and function in cancer. Trends
Mol Med. 12:580–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chan JA, Krichevsky AM and Kosik KS:
MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells.
Cancer Res. 65:6029–6033. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Li W, Nan F, et al: MicroRNA
expression profile of colon cancer stem-like cells in HT29
adenocarcinoma cell line. Biochem Biophys Res Commun. 404:273–278.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu F, Deng H, Yao H, Liu Q, Su F and Song
E: Mir-30 reduction maintains self-renewal and inhibits apoptosis
in breast tumor-initiating cells. Oncogene. 29:4194–4204. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Iliopoulos D, Lindahl-Allen M, Polytarchou
C, Hirsch HA, Tsichlis PN and Struhl K: Loss of miR-200 inhibition
of Suz12 leads to polycomb-mediated repression required for the
formation and maintenance of cancer stem cells. Mol Cell.
39:761–772. 2010. View Article : Google Scholar
|
|
24
|
Qian S, Ding JY, Xie R, et al: MicroRNA
expression profile of bronchioalveolar stem cells from mouse lung.
Biochem Biophys Res Commun. 377:668–673. 2008. View Article : Google Scholar
|
|
25
|
Silber J, Lim DA, Petritsch C, et al:
miR-124 and miR-137 inhibit proliferation of glioblastoma
multiforme cells and induce differentiation of brain tumor stem
cells. BMC Med. 6:142008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ji Q, Hao X, Zhang M, et al: MicroRNA
miR-34 inhibits human pancreatic cancer tumor-initiating cells.
PloS One. 4:e68162009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ma L, Li N, He X and Zhang Q: miR-449b and
miR-34c on inducing down-regulation of cell cycle-related proteins
and cycle arrests in SKOV3-ipl cell, an ovarian cancer cell line.
Beijing Da Xue Xue Bao. 43:129–133. 2011.(In Chinese).
|
|
28
|
Bou Kheir T, Futoma-Kazmierczak E,
Jacobsen A, et al: miR-449 inhibits cell proliferation and is
down-regulated in gastric cancer. Mol Cancer. 10:292011.PubMed/NCBI
|
|
29
|
Chen H, Lin YW, Mao YQ, et al:
MicroRNA-449a acts as a tumor suppressor in human bladder cancer
through the regulation of pocket proteins. Cancer Lett. 320:40–47.
2012. View Article : Google Scholar
|
|
30
|
Matsushime H, Roussel MF, Ashmun RA and
Sherr CJ: Colony-stimulating factor 1 regulates novel cyclins
during the G1 phase of the cell cycle. Cell. 65:701–713. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Buchkovich K, Duffy LA and Harlow E: The
retinoblastoma protein is phosphorylated during specific phases of
the cell cycle. Cell. 58:1097–1105. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen PL, Scully P, Shew JY, Wang JY and
Lee WH: Phosphorylation of the retinoblastoma gene product is
modulated during the cell cycle and cellular differentiation. Cell.
58:1193–1198. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Weinberg RA: The retinoblastoma protein
and cell cycle control. Cell. 81:323–330. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Harbour JW, Luo RX, Dei Santi A, Postigo
AA and Dean DC: Cdk phosphorylation triggers sequential
intramolecular interactions that progressively block Rb functions
as cells move through G1. Cell. 98:859–869. 1999. View Article : Google Scholar
|
|
35
|
Reis-Filho JS, Savage K, Lambros MB, et
al: Cyclin D1 protein overexpression and CCND1 amplification in
breast carcinomas: an immunohistochemical and chromogenic in situ
hybridisation analysis. Mod Pathol. 19:999–1009. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hosokawa Y and Arnold A: Mechanism of
cyclin D1 (CCND1, PRAD1) overexpression in human cancer cells:
analysis of allele-specific expression. Genes Chromosomes Cancer.
22:66–71. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Betticher DC, Heighway J, Hasleton PS, et
al: Prognostic significance of CCND1 (cyclin D1) overexpression in
primary resected non-small-cell lung cancer. Br J Cancer.
73:294–300. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Leone G, DeGregori J, Yan Z, et al: E2F3
activity is regulated during the cell cycle and is required for the
induction of S phase. Genes Dev. 12:2120–2130. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Humbert PO, Verona R, Trimarchi JM, Rogers
C, Dandapani S and Lees JA: E2f3 is critical for normal cellular
proliferation. Genes Dev. 14:690–703. 2000.PubMed/NCBI
|
|
40
|
Oeggerli M, Tomovska S, Schraml P, et al:
E2F3 amplification and overexpression is associated with invasive
tumor growth and rapid tumor cell proliferation in urinary bladder
cancer. Oncogene. 23:5616–5623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Olsson AY, Feber A, Edwards S, et al: Role
of E2F3 expression in modulating cellular proliferation rate in
human bladder and prostate cancer cells. Oncogene. 26:1028–1037.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cooper CS, Nicholson AG, Foster C, et al:
Nuclear overexpression of the E2F3 transcription factor in human
lung cancer. Lung Cancer. 54:155–162. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Pierce AM, Gimenez-Conti IB,
Schneider-Broussard R, Martinez LA, Conti CJ and Johnson DG:
Increased E2F1 activity induces skin tumors in mice heterozygous
and nullizygous for p53. Proc Natl Acad Sci USA. 95:8858–8863.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fang Y, Xiang J, Chen Z, et al: miRNA
expression profile of colon cancer stem cells compared to non-stem
cells using the SW1116 cell line. Oncol Rep. 28:2115–2124.
2012.PubMed/NCBI
|
|
45
|
Ieta K, Tanaka F, Haraguchi N, et al:
Biological and genetic characteristics of tumor-initiating cells in
colon cancer. Ann Surg Oncol. 15:638–648. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
O’Brien CA, Pollett A, Gallinger S and
Dick JE: A human colon cancer cell capable of initiating tumour
growth in immunodeficient mice. Nature. 445:106–110.
2007.PubMed/NCBI
|
|
47
|
Ricci-Vitiani L, Lombardi DG, Pilozzi E,
et al: Identification and expansion of human
colon-cancer-initiating cells. Nature. 445:111–115. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Vermeulen L, Todaro M, de Sousa Mello F,
et al: Single-cell cloning of colon cancer stem cells reveals a
multi-lineage differentiation capacity. Proc Natl Acad Sci USA.
105:13427–13432. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dalerba P, Dylla SJ, Park IK, et al:
Phenotypic characterization of human colorectal cancer stem cells.
Proc Natl Acad Sci USA. 104:10158–10163. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li C, Heidt DG, Dalerba P, et al:
Identification of pancreatic cancer stem cells. Cancer Res.
67:1030–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wang HJ, Ruan HJ, He XJ, et al:
MicroRNA-101 is down-regulated in gastric cancer and involved in
cell migration and invasion. Eur J Cancer. 46:2295–2303. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang X, Lam EK, Zhang J, Jin H and Sung
JJ: MicroRNA-122a functions as a novel tumor suppressor downstream
of adenomatous polyposis coli in gastrointestinal cancers. Biochem
Biophys Res Commun. 387:376–380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Michael MZ, SM OC, van Holst Pellekaan NG,
Young GP and James RJ: Reduced accumulation of specific microRNAs
in colorectal neoplasia. Mol Cancer Res. 1:882–891. 2003.PubMed/NCBI
|
|
56
|
Schepers AG, Snippert HJ, Stange DE, et
al: Lineage tracing reveals Lgr5+ stem cell activity in
mouse intestinal adenomas. Science. 337:730–735. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Driessens G, Beck B, Caauwe A, Simons BD
and Blanpain C: Defining the mode of tumour growth by clonal
analysis. Nature. 488:527–530. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Liu C, Kelnar K, Liu B, et al: The
microRNA miR-34a inhibits prostate cancer stem cells and metastasis
by directly repressing CD44. Nat Med. 17:211–215. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Godlewski J, Nowicki MO, Bronisz A, et al:
Targeting of the Bmi-1 oncogene/stem cell renewal factor by
microRNA-128 inhibits glioma proliferation and self-renewal. Cancer
Res. 68:9125–9130. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sun F, Fu H, Liu Q, et al: Downregulation
of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett.
582:1564–1568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Aslanian A, Iaquinta PJ, Verona R and Lees
JA: Repression of the Arf tumor suppressor by E2F3 is required for
normal cell cycle kinetics. Genes Dev. 18:1413–1422. 2004.
View Article : Google Scholar : PubMed/NCBI
|