Introduction
Malignant brain tumors, gliomas in particular, are
the most commonly diagnosed adult primary tumors of the central
nervous system. Gliomas can be classified into 3 main types:
astrocytomas, oligodendrogliomas and mixed oligoastrocytomas, which
are typically distinguished by their histological features. The
division of gliomas into World Health Organization (WHO) grades I
and II (low-grade), and WHO grades III and IV (high-grade or
malignant) is based on the presence of nuclear atypia, mitoses,
microvascular proliferation and necrosis (1). The treatment of high-grade gliomas has
posed a challenge to clinicians due to the lack of effective
therapeutic options. In fact, the 5-year survival rate for patients
with glioblastoma, a grade IV neoplasm, is approximately 3%
(2). Despite significant advances
in neurosurgical techniques, in radiation therapy, and in our
understanding of the molecular underpinnings of tumorigenesis, the
prognosis for patients with malignant glioma, and glioblastoma in
particular, remains poor (2,3).
Fibroblast growth factors (FGFs), FGF-1 to -23, are
heparin-binding ligands that exert their biological activities
through binding high-affinity tyrosine kinase FGF receptors (FGFRs)
on the surface of cells (4,5). FGFRs comprise four members, FGFR-1 to
-4. FGFRs 1–3 have two isoforms, IIIb and IIIc. In normal human
tissues, FGFR-2 IIIb, also known as keratinocyte growth factor
receptor, and FGFR-2 IIIc are primarily expressed in epithelial and
mesenchymal cells, respectively (6). FGFR-2 gene amplification, abnormal
activation, or single nucleotide polymorphisms (SNPs) reportedly
play important roles in the progression of cancers including those
of the breast, endometrium, colon and pancreas (7–12).
Studies have reported that FGFR-2 levels in human gliomas gradually
diminish or are lost, whereas FGFR-1 expression increases as the
tumor progresses from benign to malignant (13,14).
However, no studies have further evaluated the expression of FGFR-2
in gliomas and its clinicopathological significance.
In the present study, we evaluated the expression of
FGFR-2 and its major isoforms, FGFR-2 IIIb and FGFR-2 IIIc, in
gliomas of all histological grades. In addition, we assessed the
correlation between FGFR-2 expression levels, clinical features and
survival rates. We report here that FGFR-2 is expressed at lower
levels in high histological grade gliomas, and that decreased
FGFR-2 expression may be related to lower survival rates.
Materials and methods
Patients and samples
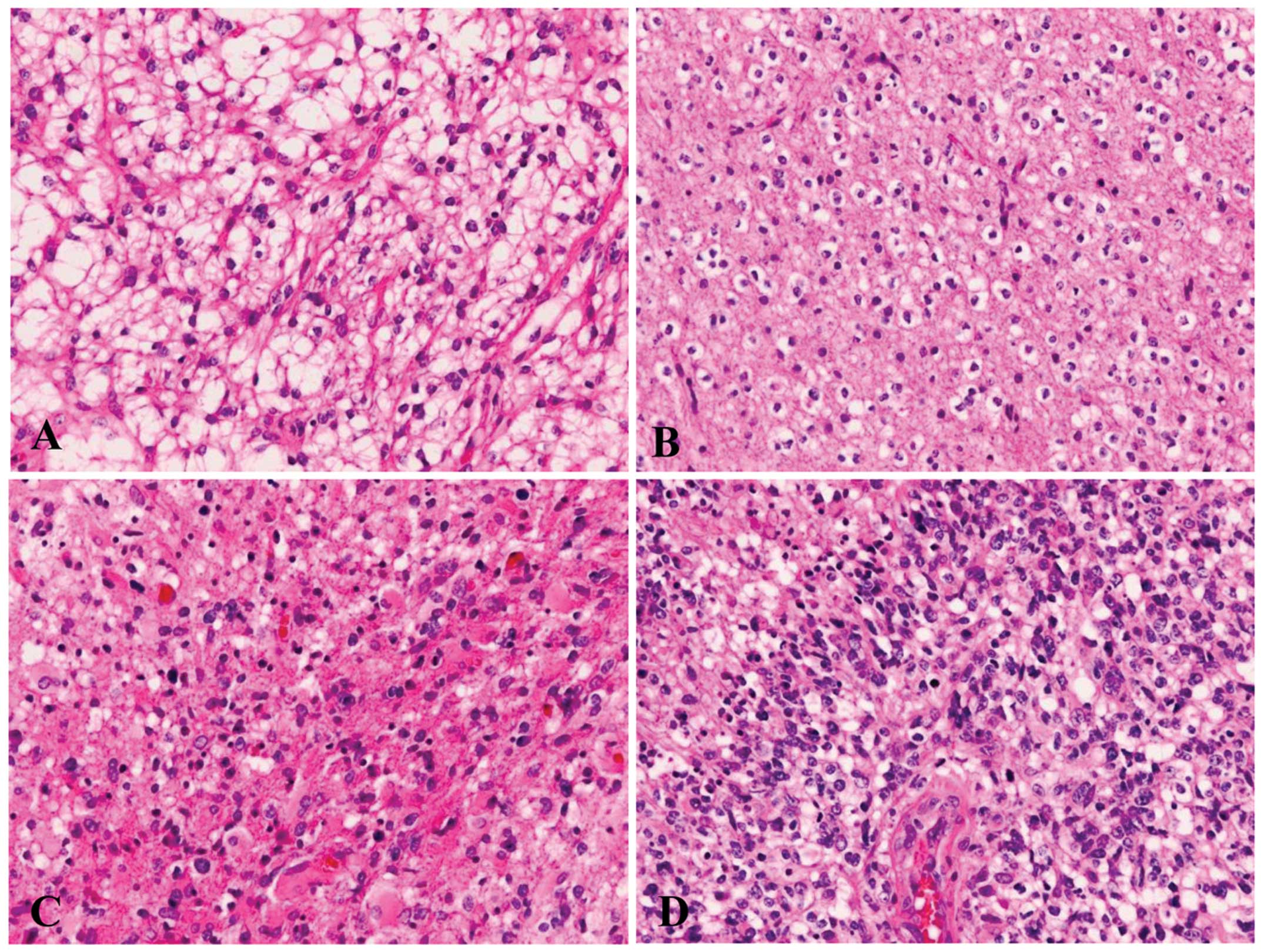
Tissues from 56 glioma patients who received
treatment at Nippon Medical School Hospital (Bunkyo-ku, Tokyo,
Japan) from 2007 to 2011 were utilized in this study: 7 patients
with grade I (pilocytic astrocytoma), 16 patients with grade II (14
diffuse astrocytomas and 2 oligodendrogliomas), 10 patients with
grade III (anaplastic astrocytoma), and 23 patients with grade IV
glioma (glioblastoma; Fig. 1).
Paraffin-embedded specimens were prepared for immunohistochemical
analysis as described previously (15). Normal brain tissues were obtained
from autopsy cases without brain disease (n=2). Non-neoplastic
brain tissues were obtained from the surgical margins of the
glioblastoma patients. This study was conducted in accordance with
the principles embodied in the Declaration of Helsinki (2008), and
informed consent was obtained from each patient for the use of
their tissues.
Immunohistochemistry
To investigate the protein levels of FGFR-2 IIIb and
FGFR-2 IIIc in gliomas, isoform-specific antibodies were prepared
as we described previously (12,16).
The expression of FGFR-2 was detected using goat polyclonal
anti-human FGFR2 antibody (N20) from Santa Cruz Biotechnology Inc.
(Santa Cruz, CA, USA), which recognizes both IIIb and IIIc isoforms
(11). Mouse monoclonal anti-Ki-67
antibody was purchased from Dako, Japan (Tokyo, Japan).
Paraffin-embedded sections (3-μm) were immunostained using the
Histofine Simple Stain Max Po (R) kit (Nichirei, Tokyo, Japan) for
FGFR-2 IIIb and FGFR-2 IIIc, the Max Po (G) Kit for FGFR-2, and the
Max Po (M) Kit for Ki-67. After deparaffinization, endogenous
peroxidase activity was blocked by incubating the sections in 0.3%
hydrogen peroxide in methanol for 30 min. The tissue sections were
then incubated with the anti-FGFR-2 (1:400), anti-FGFR-2 IIIb
(1:1,000), anti-FGFR-2 IIIc (1:200), or anti-Ki-67 antibody (1:100)
diluted in phosphate-buffered saline (PBS) containing 1% bovine
serum albumin (BSA) for 16 h at 4°C. Bound antibodies were detected
with the Histofine Simple Stain Max Po (R), (G), or (M) reagents,
using diaminobenzidine tetrahydrochloride as the substrate. The
sections were counterstained using Mayer’s hematoxylin. Negative
control tissue sections were prepared by omitting the primary
antibody from the staining procedure.
Evaluation of immunohistochemical
variables
To evaluate the expression of FGFR-2, FGFR-2 IIIb
and FGFR-2 IIIc in glioma tissues, we analyzed both the percentage
of positive cells and the intensity of staining for each antibody
in whole tumor lesions at ×200 magnification as described elsewhere
(11). The following scale was
employed to evaluate the intensity of staining: 0, no staining; 1+,
weak staining; 2+, moderate staining; and 3+, intense staining. The
percentage of positive cells in the entire field of the specimen
was determined at ×200 magnification. Blinded cell counts were
independently performed by three investigators (R.O., T.I. and
Y.M.). The mean values were used in subsequent analyses.
Glioma cell lines
KG-1-C and YKG-1 cell lines established from human
low-grade glioma and high-grade glioblastoma, respectively, were
obtained from Riken BioResource Center (Ibaraki, Japan). KG-1-C and
YKG-1 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)
containing 5% and 10% fetal bovine serum (FBS), respectively, at
37°C in a humidified 5% CO2 atmosphere.
Quantitative PCR
Glioma cells were grown in DMEM supplemented with 10
or 20% FBS for 48 h. Total-RNA was extracted using the FastPure RNA
kit (Takara Bio Inc., Shiga, Japan). Next, cDNA synthesis was
performed using a High-Capacity cDNA reverse transcription kit
according to the manufacturer’s protocol. Quantitative PCR (qPCR)
was performed using the ABI StepOnePlus System (Life Technologies)
as previously described (12).
The quantitative PCR primers nt 1693–1716 (5′-GGA
TAT CCT TTC ACT CTG CAT GGT-3′) and nt 1770–1794 (5′-TGG AGT AAA
TGG CTA TCT CCA GGTA-3′) were used to amplify a 102-bp fragment of
human FGFR-2 IIIc (accession no. NM_000141.4). The TaqMan probe
5′-CAG TTC TGC CAG CGC CTG GAA GA-3′ was used for FGFR-2 IIIc. PCR
primers nt 1587–1606 (5′-CAC TCG GGG ATA AAT AGT TC-3′) and nt
1719–1736 (5′-CGC TTG CTG TTT TGG CAG-3′) were used to amplify a
150-bp fragment of human FGFR-2 IIIb (accession no. NM_022970). The
TaqMan probe 5′-TGC CAA AAC AGC AAG CGC CTG G-3′ was used for
FGFR2IIIb. The PCR reaction mixture containing 2 μl of template
cDNA, 10 μl of TaqMan Fast Universal PCR Master Mix, and 1 μl of
each of TaqMan gene expression assay was placed in a 96-well
reaction plate. 18S rRNA, amplified using a TaqMan gene expression
assay, was used as an internal positive control. The optimized
program for FGFR-2 IIIb, FGFR-2 IIIc and 18S rRNA involved an
initial denaturation at 95°C for 20 sec, followed by 50 cycles of
amplification (95°C for 1 sec and 60°C for 20 sec). The internal
standard concentration ratio (target/18S rRNA) was calculated for
each gene. Gene expression measurements were performed in
triplicate.
Statistical analysis
All data are shown as mean values ± standard error
of the mean (SEM). The data between two groups were compared using
the Student’s t-test. The χ2 and Fisher’s exact tests
were used to analyze the correlation between FGFR-2 isotype
expression levels and clinicopathological features. The cumulative
survival rate was calculated using the Kaplan-Meier method and the
significance of the survival rate differences was analyzed by the
log-rank test. P<0.05 was considered to indicate a statistically
significant difference in all analyses.
Results
Clinicopathological characteristics of
the glioma patients
In total, 34 male and 22 female patients with a mean
age of 53±23 years (range 5–79 years) were enrolled in the study
(Table I). The tumors were
localized to the frontal (n=22), temporal (n=16), parietal (n=4)
and other brain regions (n=14). Follow-up data were available for
46 patients. The mean follow-up duration was 694.2±632 days (range
30–2,180 days). At the last follow-up, 30 patients were confirmed
to be alive and 16 patients were deceased.
 | Table IClinicopathological features of the
glioma cases with correlation to FGFR-2 isoform expression. |
Table I
Clinicopathological features of the
glioma cases with correlation to FGFR-2 isoform expression.
| FGFR2 | | FGFR2 IIIb | | FGFR2 IIIc | |
|---|
|
| |
| |
| |
|---|
| >30% | <29% | P-value | >40% | <39% | P-value | >40% | <39% | P-value |
|---|
| Sample size, n | 21 | 35 | | 23 | 33 | | 26 | 30 | |
| Gender |
| Male | 12 | 22 | 0.67 | 13 | 21 | 0.59 | 13 | 21 | 0.13 |
| Female | 9 | 13 | | 10 | 12 | | 13 | 9 | |
| Age (years) |
| ≥55 | 12 | 22 | 0.67 | 12 | 22 | 0.36 | 15 | 19 | 0.66 |
| <55 | 9 | 13 | | 11 | 11 | | 13 | 9 | |
| Tumor location |
| Frontal | 8 | 14 | 0.91 | 9 | 13 | 0.051 | 11 | 11 | 0.5 |
| Parietal | 1 | 3 | | 2 | 2 | | 1 | 3 | |
| Temporal | 7 | 9 | | 4 | 12 | | 7 | 9 | |
| Other site | 5 | 9 | | 10 | 4 | | 8 | 6 | |
| Tumor grade |
| High (III,
IV) | 5 | 28 | <0.01 | 5 | 28 | <0.01 | 5 | 28 | <0.01 |
| Low (I, II) | 16 | 7 | | 18 | 5 | | 21 | 2 | |
| Ki-67
expression |
| Positive
(>20%) | 3 | 17 | 0.011 | 4 | 16 | 0.0169 | 4 | 16 | <0.01 |
| Negative
(<19%) | 18 | 18 | | 19 | 17 | | 22 | 14 | |
Immunohistochemical analysis of FGFR-2,
FGFR-2 IIIb and FGFR-2 IIIc in non-neoplastic brain tissues and
gliomas
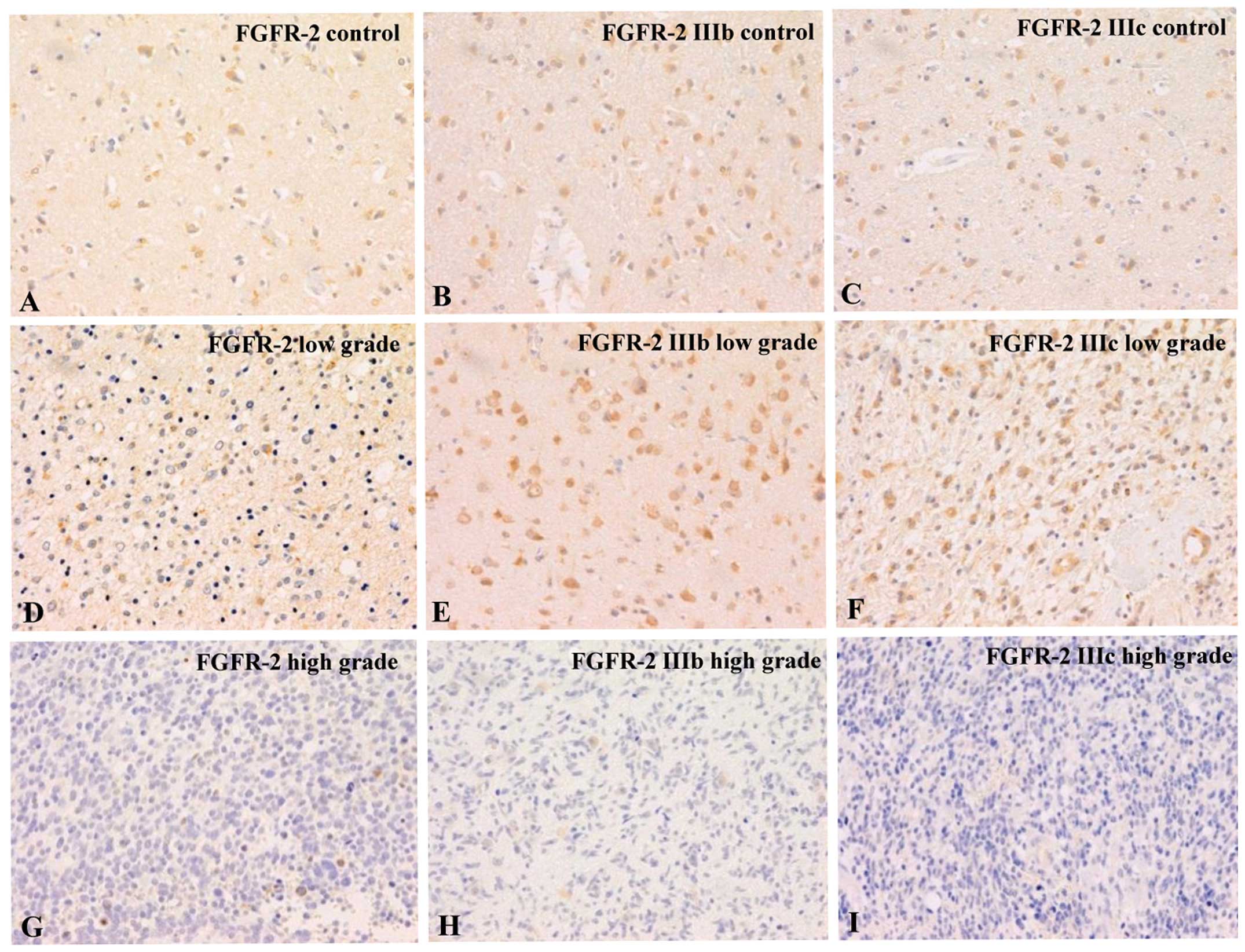
Immunohistochemical analyses showed that FGFR-2,
FGFR-2 IIIb and FGFR-2 IIIc were evenly distributed at low levels
in all the astrocytes present in the normal brain tissues collected
from autopsy cases (Fig. 2A–C). In
the low-grade astrocytomas (grade I or II), the expression of
FGFR-2, FGFR-2 IIIb and FGFR-2 IIIc did not differ from that in the
healthy brain tissues (Fig. 2D–F).
In contrast, high-grade astrocytomas (grade III or IV) showed a
significant decrease or loss of FGFR-2, FGFR-2 IIIb and FGFR-2 IIIc
expression. Notably, the expression of FGFR-2, FGFR-2 IIIb and
FGFR-2 IIIc was virtually absent in some of the glioblastoma cases
(Fig. 2G–I). To confirm the
histopathological grade of gliomas, we performed immunostaining for
Ki-67. High-grade astrocytomas demonstrated a marked increase in
Ki-67-positive cells relative to low-grade astrocytomas.
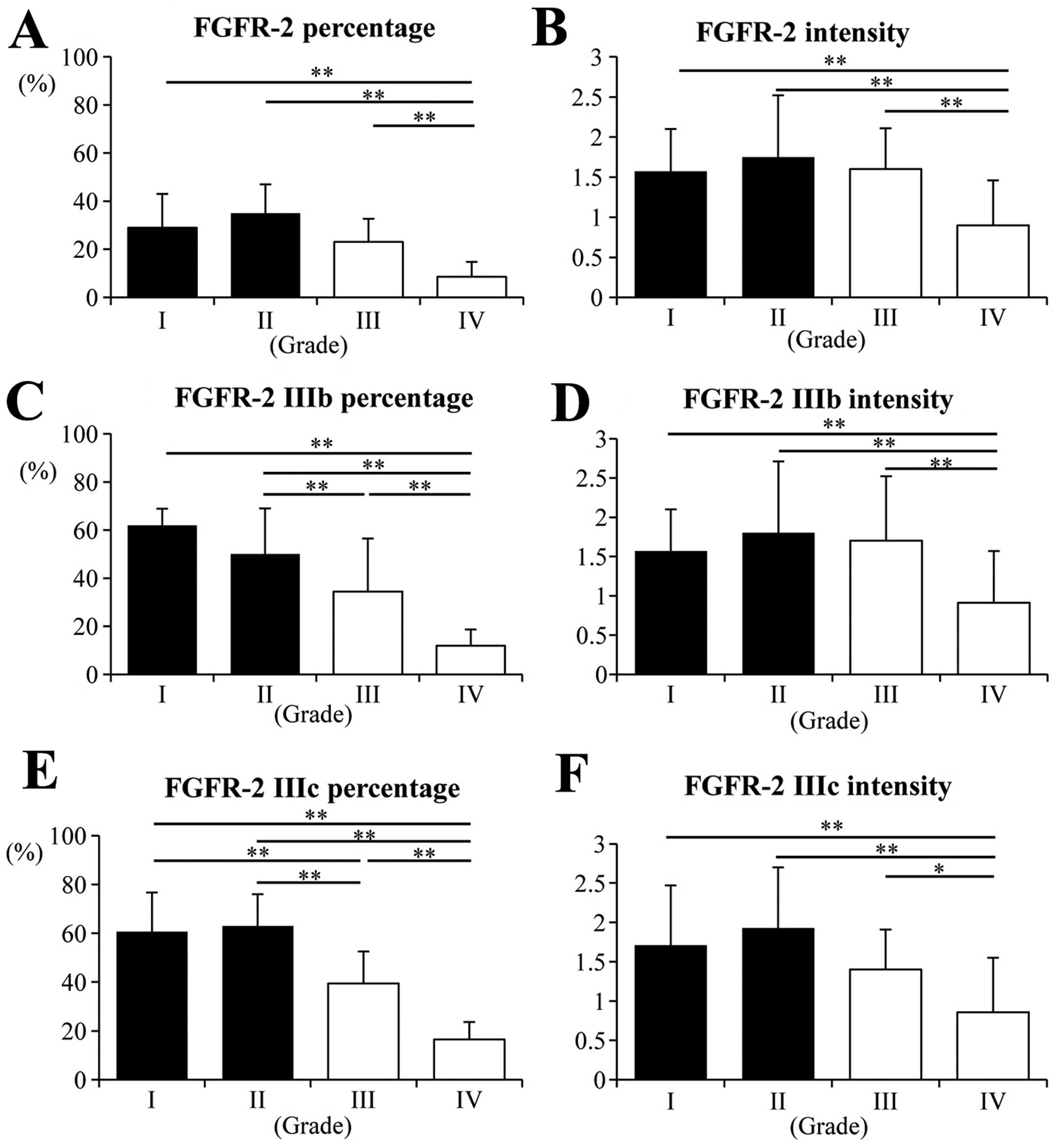
The percentages of cells positive for FGFR-2, FGFR-2
IIIb and FGFR-2 IIIc in grade IV astrocytomas were significantly
lower than these percentages in the grade I, II and III tumor
groups (Fig. 3A, C and E;
P<0.001). The percentages of FGFR-2 IIIb- and FGFR-2
IIIc-positive cells in the grade III gliomas were lower than these
percentages in the grade II tumors (Fig. 3C and E; P<0.001). Likewise, the
staining intensities for FGFR-2, FGFR-2 IIIb and FGFR-2 IIIc in the
grade IV astrocytomas were also lower than the intensities in the
other lower grade tumors (Fig. 3B, D
and F; P<0.001 or P<0.05). No significant differences
were observed in grade I and II with respect to any of the proteins
examined.
Correlation between FGFR-2 isoform
expression and the clinicopathological features of the gliomas
To evaluate the relationship between FGFR-2 and
FGFR-2 isoform expression in gliomas and the clinicopathological
features, we separated the patients into two groups: high
expression (>cutoff value) and low expression (<cutoff value)
(Table I). To select an appropriate
cutoff value, we conducted statistical analyses using different
percentages of positive cells (20, 30, 40 and 50%) as the cutoff.
For each protein, the cutoff value that yielded the most
statistically significant difference was used in the final
analysis. A cutoff value of 30% was used for FGFR-2, whereas 40%
was used for FGFR-2 IIIb and FGFR-2 IIIc. For FGFR-2 and both of
its isoforms, the degree of protein expression was negatively
correlated with the tumor grade (Table
I; P<0.01 for each protein). A high MIB-1 index, indicated
by Ki-67 expression in >20% of the cells, was also associated
with low expression of each protein (Table I; P=0.011 for FGFR-2, P=0.0169 for
FGFR-2 IIIb and P<0.01 for FGFR-2 IIIc). Gender, age and tumor
location did not show any significant difference between the groups
expressing low and high levels of FGFR-2 or its isoforms. The
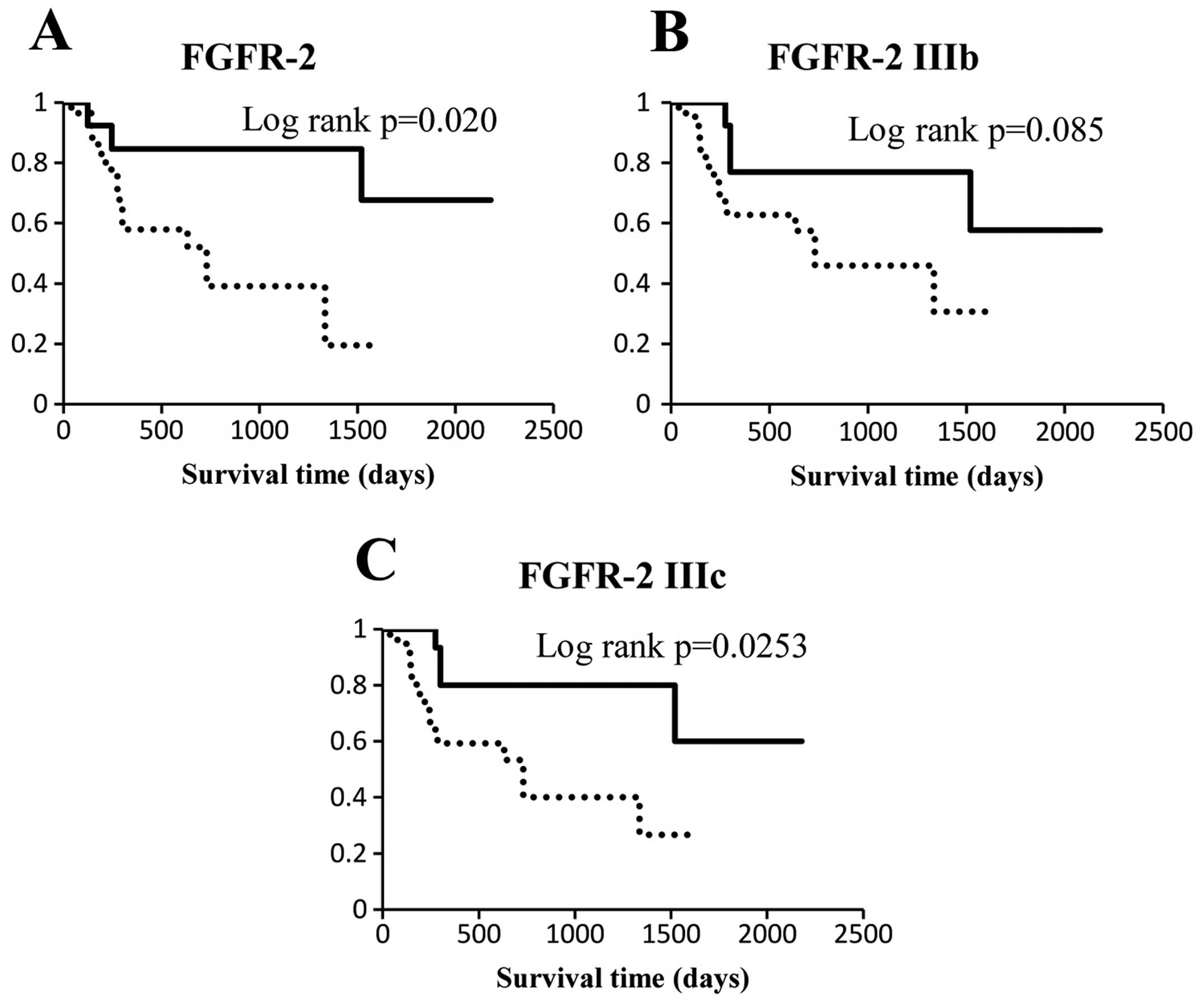
FGFR-2 and FGFR-2 IIIc low expression groups were associated with a
low survival rate (Fig. 4A and C;
P=0.020 and 0.0253, respectively). The FGFR-2 IIIb low expression
group showed a tendency towards a reduced survival rate (Fig. 4B; P=0.085).
Expression of FGFR-2 IIIb and FGFR-2 IIIc
in the glioma cell lines
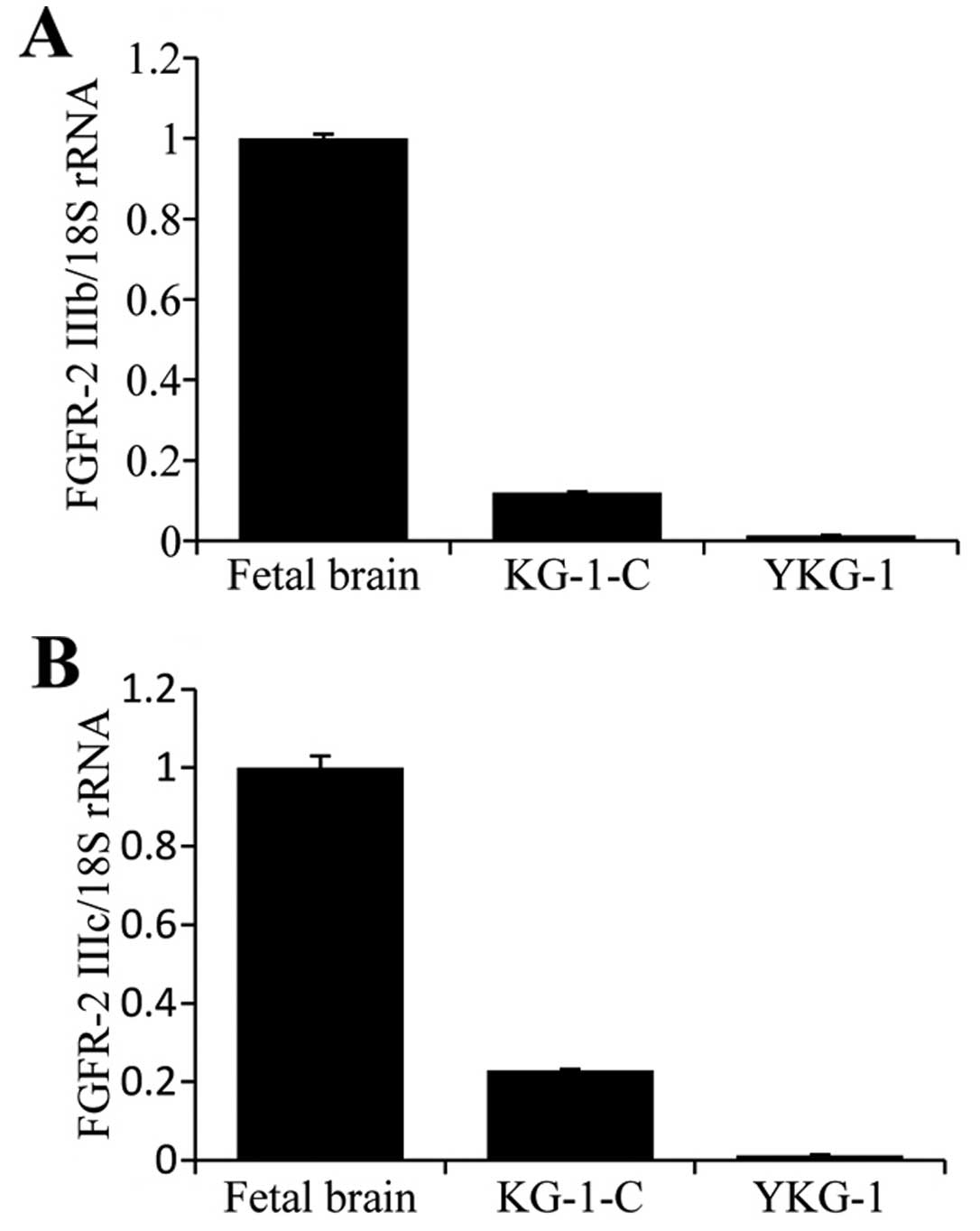
To examine the mRNA levels of FGFR-2 IIIb and FGFR-2
IIIc in human glioma cell lines, qPCR analysis was performed using
KG-1-C and YKG-1 cells that were derived from low-grade and
high-grade astrocytomas, respectively. FGFR-2 IIIb and FGFR-2 IIIc
mRNA was present in the KG-1-C cells. Notably, their expression
levels in the KG-1-C cells were lower than levels in the fetal
brain tissues. We attribute this to the high level of expression
and activity of FGF/FGFR signaling components in fetal brain
tissues (17) (Fig. 5). The levels of FGFR-2 IIIb and
FGFR-2 IIIc mRNA were considerably lower in the YKG-1 cells when
compared with these levels in the KG-1-C cells. The low expression
levels of FGFR-2 isoforms in the YKG-1 glioma cell line derived
from high-grade glioma were consistent with the results of our
immunohistochemical analysis.
Discussion
FGFRs are single transmembrane receptors that
contain extracellular, transmembrane and intracellular domains
(5). The extracellular domains of
FGFRs are typically composed of 3 immunoglobulin (Ig)-like domains
(I-III). Alternative splicing in the C-terminal half of the third
Ig-like domain of FGFRs 1–3 generates their IIIb and IIIc isoforms.
FGFR4 does not possess these alternatively spliced exons. The
FGF/FGFR pathway plays important roles in development and
differentiation in normal human tissues. Moreover, FGF/FGFR is
tightly linked to carcinogenesis and cancer progression in various
organs. We previously reported that FGFR-2 is overexpressed in the
invasive front of colorectal cancer (11). However, the expression of FGFRs and
their IIIb and IIIc isoforms have not been thoroughly examined in
the brain. In addition, their clinical significance in gliomas
remains unclear. In the present study, we examined the presence of
FGFR-2, FGFR-2 IIIb and FGFR-2 IIIc in normal human brain tissues
and gliomas using immunohistochemical staining. To our knowledge,
this is the first study reporting the correlation between the
expression levels of FGFR-2 and its two isoforms and
clinicopathological features in glioma. We demonstrated that the
decrease or loss of these molecules is highly related to the
progression of gliomas and a poor prognosis
The most notable finding in our study was that
FGFR-2, which is normally expressed in brain tissue, was
significantly diminished or nearly abolished as gliomas progressed
from low to high grade. To date, few studies have examined the
expression of FGFR-2 in the brain. Yamaguchi et al examined
the expression of FGFR-1 and FGFR-2 in normal human brain tissues
and astrocytomas (13). They showed
that FGFR-2 was abundantly expressed in normal brain tissues,
whereas FGFR-1 expression was minimal. Conversely, FGFR-2
expression was low or undetectable in high-grade astrocytomas, such
as glioblastomas, in which FGFR-1 was highly upregulated. They
speculated that the loss of FGFR-2 and gain of FGFR-1 are
associated with the malignant progression of astrocytomas from low-
to high-grade tumors. Although we did not assess FGFR-1 expression
in this study, the decreased FGFR-2 expression we observed in
high-grade astrocytomas is consistent with their findings. We
speculated that FGFR-1 and FGFR-2 play reciprocal roles in the
development of astrocytomas. FGFR-1 and FGFR-2 have different
ligand specificity. Therefore, examining the expression of the
FGFR-1 and FGFR-2 ligands in human brain specimens in future
studies may reveal a functional relationship between these two
molecules.
The mechanism underlying the loss of FGFR-2
expression in high-grade astrocytomas is unclear. The FGFR-2
locus is on the long arm of chromosome 10q26. Interestingly,
approximately 80% of all glioblastomas are associated with the loss
of an entire copy of chromosome 10 (18,19).
Therefore, the production of FGFR-2 is likely impaired as a result
of losing chromosome 10 during the astrocyte transformation
process. Alternatively, the FGFR-2 gene may still be present in
glioblastomas, but silent following translocation to another
chromosome. As it is lost in glioblastomas, chromosome 10 is the
proposed location of a tumor-suppressor gene. Since decreased
FGFR-2 expression was observed in higher-grade astrocytomas, we
postulate that FGFR-2 has a tumor-suppressing effect. On the other
hand, one may argue that the decrease in FGFR-2 expression is not
directly related to tumorigenesis. Instead, it may simply reflect
the loss of one copy of chromosome 10. We were unable to test
alterations of the chromosomes in our study, leaving the underlying
mechanism of FGFR-2 loss in gliomas unexplored. Additional studies
are required to elucidate the mechanism of FGFR-2 loss in the
future.
We present evidence that the levels of FGFR-2
isoforms, FGFR-2 IIIb and FGFR-2 IIIc, are altered in gliomas.
FGFR-2 IIIc is widely expressed in cancer cells including
adenocarcinomas, squamous cell carcinomas and urothelial cell
carcinomas (20–25). In the prostate, a switch from FGFR-2
IIIb to FGFR-2 IIIc expression is related to the progression of
cancer (26). FGFR-2 IIIc
expression in prostate cancer cells induces epithelial-mesenchymal
transition and alters splicing, which may contribute to cancer
metastasis (27). We recently
demonstrated that enhanced expression of FGFR-2 IIIc promotes
pancreatic cancer cell proliferation (12). In contrast to FGFR-2 IIIc, the role
of the FGFR-2 IIIb isoform is controversial. Overexpression of the
FGFR-2 IIIb isoform has been reported in breast, endometrial,
gastric, pancreatic and colorectal cancers; however, several of
these studies presented conflicting results regarding its clinical
roles (28–32). Higher expression of FGFR-2 IIIb in
pancreatic cancers was correlated with a poor prognosis (32). In contrast, expression of FGFR-2
IIIb in gastric cancer cells was associated with a better prognosis
(31). Here, we showed that the
expression of the FGFR-2 IIIb and FGFR-2 IIIc isoforms decreased
with the progression of gliomas from low to high grade. Lower
expression of the FGFR-2 isoforms was associated with a worse
prognosis. These findings indicate that these two isoforms are
synchronized in the development of glioma. Therefore, it is
unlikely that they have conflicting roles in gliomas. We anticipate
that the decrease or loss of FGFR-2 isoform expression can be used
as a marker to predict the malignant potential of gliomas.
Additional studies are warranted to validate this hypothesis.
Acknowledgements
The authors thank Ms. Taeko Suzuki and Ms. Yoko
Kawamoto (Departments of Pathology and Integrative Oncological
Pathology, Nippon Medical School) for the technical assistance.
This study was funded by Grants-in-Aid for Clinical Rebiopsy Bank
Project for Comprehensive Cancer Therapy Development to Z.N. from
the Ministry of Education, Culture, Sport, Science and Technology,
Japan (S1311022).
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, et al: The 2007 WHO classification of
tumours of the central nervous system. Acta Neuropathol.
114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bondy ML, Scheurer ME, Malmer B,
Barnholtz-Sloan JS, Davis FG, Il’yasova D, et al: Brain tumor
epidemiology: consensus from the Brain Tumor Epidemiology
Consortium. Cancer. 113:1953–1968. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gurney JG and Kadan-Lottick N: Brain and
other central nervous system tumors: rates, trends, and
epidemiology. Curr Opin Oncol. 13:160–166. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ornitz DM and Itoh N: Fibroblast growth
factors. Genome Biol. 2:reviews3005.1–reviews3005.12. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Itoh N and Ornitz DM: Evolution of the
Fgf and Fgfr gene families. Trends Genet. 20:563–569.
2004.
|
|
6
|
Miki T, Bottaro DP, Fleming TP, Smith CL,
Burgess WH, Chan AM and Aaronson SA: Determination of
ligand-binding specificity by alternative splicing: two distinct
growth factor receptors encoded by a single gene. Proc Natl Acad
Sci USA. 89:246–250. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hunter DJ, Kraft P, Jacobs KB, Cox DG,
Yeager M, Hankinson SE, et al: A genome-wide association study
identifies alleles in FGFR2 associated with risk of sporadic
postmenopausal breast cancer. Nat Genet. 39:870–874. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Byron SA, Gartside MG, Wellens CL, Mallon
MA, Keenan JB, Powell MA, et al: Inhibition of activated fibroblast
growth factor receptor 2 in endometrial cancer cells induces cell
death despite PTEN abrogation. Cancer Res. 68:6902–6907. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Byron SA and Pollock PM: FGFR2 as a
molecular target in endometrial cancer. Future Oncol. 5:27–32.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Byron SA, Gartside MG, Wellens CL,
Goodfellow PJ, Birrer MJ, Campbell IG and Pollock PM: FGFR2
mutations are rare across histologic subtypes of ovarian cancer.
Gynecol Oncol. 117:125–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Matsuda Y, Ishiwata T, Yamahatsu K,
Kawahara K, Hagio M, Peng WX, et al: Overexpressed fibroblast
growth factor receptor 2 in the invasive front of colorectal
cancer: a potential therapeutic target in colorectal cancer. Cancer
Lett. 309:209–219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ishiwata T, Matsuda Y, Yamamoto T, Uchida
E, Korc M and Naito Z: Enhanced expression of fibroblast growth
factor receptor 2 IIIc promotes human pancreatic cancer cell
proliferation. Am J Pathol. 180:1928–1941. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamaguchi F, Saya H, Bruner JM and
Morrison RS: Differential expression of two fibroblast growth
factor-receptor genes is associated with malignant progression in
human astrocytomas. Proc Natl Acad Sci USA. 91:484–488. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yamada SM, Yamaguchi F, Brown R, Berger MS
and Morrison RS: Suppression of glioblastoma cell growth following
antisense oligonucleotide-mediated inhibition of fibroblast growth
factor receptor expression. Glia. 28:66–76. 1999. View Article : Google Scholar
|
|
15
|
Matsuda Y, Fujii T, Suzuki T, Yamahatsu K,
Kawahara K, Teduka K, et al: Comparison of fixation methods for
preservation of morphology, RNAs, and proteins from
paraffin-embedded human cancer cell-implanted mouse models. J
Histochem Cytochem. 59:68–75. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishiwata T, Naito Z, Lu YP, Kawahara K,
Fujii T, Kawamoto Y, Teduka K and Sugisaki Y: Differential
distribution of fibroblast factor-7 and FGF-10 in
L-arginine-induced pancreatitis. Exp Mod Pathol. 73:181–190. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Eswarakumar VP, Lax I and Schlessinger J:
Cellular signaling by fibroblast growth factor receptors. Cytokine
Growth Factor Rev. 16:139–149. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mattei MG, Moreau A, Gesnel MC, Houssaint
E and Breathnach R: Assignment by in situ hybridization of a
fibroblast growth factor receptor gene to human chromosome band
10q26. Hum Genet. 87:84–86. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rasheed BK, Fuller GN, Friedman AH, Bigner
DD and Bigner SH: Loss of heterozygosity for 10q loci in human
gliomas. Genes Chromosomes Cancer. 5:75–82. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwabi-Addo B, Ropiquet F, Giri D and
Ittmann M: Alternative splicing of fibroblast growth factor
receptors in human prostate cancer. Prostate. 46:163–172. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Valve E, Martikainen P, Seppänen J,
Oksjoki S, Hinkka S, Anttila L, et al: Expression of fibroblast
growth factor (FGF)-8 isoforms and FGF receptors in human ovarian
tumors. Int J Cancer. 88:718–725. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Drugan CS, Paterson IC and Prime SS:
Fibroblast growth factor receptor expression reflects cellular
differentiation in human oral squamous carcinoma cell lines.
Carcinogenesis. 19:1153–1156. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cha JY, Lambert QT, Reuther GW and Der CJ:
Involvement of fibroblast growth factor receptor 2 isoform
switching in mammary oncogenesis. Mol Cancer Res. 6:435–445. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chaffer CL, Brennan JP, Slavin JL, Blick
T, Thompson EW and Williams ED: Mesenchymal-to-epithelial
transition facilitates bladder cancer metastasis: role of
fibroblast growth factor receptor-2. Cancer Res. 66:11271–11278.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marek L, Ware KE, Fritzsche A, Hercule P,
Helton WR, Smith JE, et al: Fibroblast growth factor (FGF) and FGF
receptor-mediated autocrine signaling in non-small-cell lung cancer
cells. Mol Pharmacol. 75:196–207. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan G, Fukabori Y, McBride G,
Nikolaropoulos S and McKeehan WL: Exon switching and activation of
stromal and embryonic fibroblast growth factor (FGF)-FGF receptor
genes in prostate epithelial cells accompany stromal independence
and malignancy. Mol Cell Biol. 13:4513–4522. 1993.PubMed/NCBI
|
|
27
|
Oltean S, Sorg BS, Albrecht T, Bonano VI,
Brazas RM, Dewhirst MW, et al: Alternative inclusion of fibroblast
growth factor receptor 2 exon IIIc in Dunning prostate tumors
reveals unexpected epithelial mesenchymal plasticity. Proc Natl
Acad Sci USA. 103:14116–14121. 2006. View Article : Google Scholar
|
|
28
|
Luqmani YA, Graham M and Coombes RC:
Expression of basic fibroblast growth factor, FGFR1 and FGFR2 in
normal and malignant human breast, and comparison with other normal
tissues. Br J Cancer. 6:273–280. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ishikawa A, Kudo M, Nakazawa N, Onda M,
Ishiwata T, Takeshita T and Naito Z: Expression of keratinocyte
growth factor and its receptor in human endometrial cancer in
cooperation with steroid hormones. Int J Oncol. 32:565–574.
2008.PubMed/NCBI
|
|
30
|
Kurban G, Ishiwata T, Kudo M, Yokoyama M,
Sugisaki Y and Naito Z: Expression of keratinocyte growth factor
receptor (KGFR/FGFR2 IIIb) in human uterine cervical cancer. Oncol
Rep. 11:987–991. 2004.PubMed/NCBI
|
|
31
|
Matsunobu T, Ishiwata T, Yoshino M,
Watanabe M, Kudo M, Matsumoto K, et al: Expression of keratinocyte
growth factor receptor correlates with expansive growth and early
stage of gastric cancer. Int J Oncol. 28:307–314. 2006.PubMed/NCBI
|
|
32
|
Cho K, Ishiwata T, Uchida E, Nakazawa N,
Korc M, Naito Z and Tajiri T: Enhanced expression of keratinocyte
growth factor and its receptor correlates with venous invasion in
pancreatic cancer. Am J Pathol. 170:1964–1974. 2007. View Article : Google Scholar : PubMed/NCBI
|