Introduction
Primary liver cancer is the fifth most frequently
diagnosed cancer globally and the second leading cause of
cancer-related mortality. In developing countries, incidence rates
are 2- to 3-fold higher than in developed countries (1) and it currently results in 360,000
cases and 350,000 deaths a year in China. The clinical prognosis is
very poor with the medium survival time approaching 6 months
(2). Hepatocellular carcinoma (HCC)
is the most common type of liver cancer. Most cases of HCC are
induced by either a viral hepatitis infection (hepatitis B or C) or
cirrhosis. Despite recent discoveries in screening and early
detection, HCC exhibits a rapid clinical course with an average
survival of 6 months and an overall 5-year survival rate of 5%
(3). Therefore, there is an urgent
demand for biomarkers of early detection and targeted therapy.
Copy number variations (CNVs) are alterations of the
DNA and they are being identified with different genome analysis
platforms, such as array comparative genomic hybridization (aCGH),
single nucleotide polymorphism (SNP) genotyping platforms, and
next-generation sequencing. CNVs are involved in human health and
disease (4,5) and are currently being applied for the
diagnosis of various diseases (6,7).
CNVs also play important roles in the pathogenesis
of various types of cancer, such as CNVs of epidermal growth factor
receptor (EGFR), which have been associated with head and neck
squamous (8), non-small cell lung
(9), colorectal (10) and prostate cancer (11). Previous studies have indicated that
decrease in the copy number of mitochondrial DNA may be a critical
event during the early phase of liver carcinogenesis (12,13).
Guichard et al conducted an integrated analysis of somatic
mutations and focal copy-number changes and subsequently identified
several key genes and pathways in HCC (14).
In the present study, we carried out an integrated
analysis of liver cancer CNV data from The Cancer Genome Atlas
(TCGA) and liver cancer expression profile data from the EBI Array
Express database using bioinformatic tools, aiming to identify
CNV-driven genes. These CNV-related differentially expressed genes
(DEGs) may be potential biomarkers for early diagnosis or
treatment. In addition, they may aid in identifying underlying
mechanisms of liver cancer.
Materials and methods
Data sources
The CNV data set was obtained from TCGA database.
Genome-Wide SNP array 6.0 chip was used to detect CNV information
in 323 pairs of cases and controls with hg19 as the reference
genome. Level 3 data were adopted in the following analysis. CNV
sites and mean segment information were acquired in each sample.
Gene expression data set E-MTAB-950 in original CEL format were
downloaded from EBI Array Express. A total of 30 samples were
selected out, including 10 normal liver tissue samples and 20 liver
cancer samples.
Pretreatment of gene expression data
CEL format was converted into expression matrix
using the rma function from package affy of R. Probes were
then mapped into genes using Bioconductor with annotation files of
Affymetrix Human Genome U133 Plus 2.0 Array. Expression values were
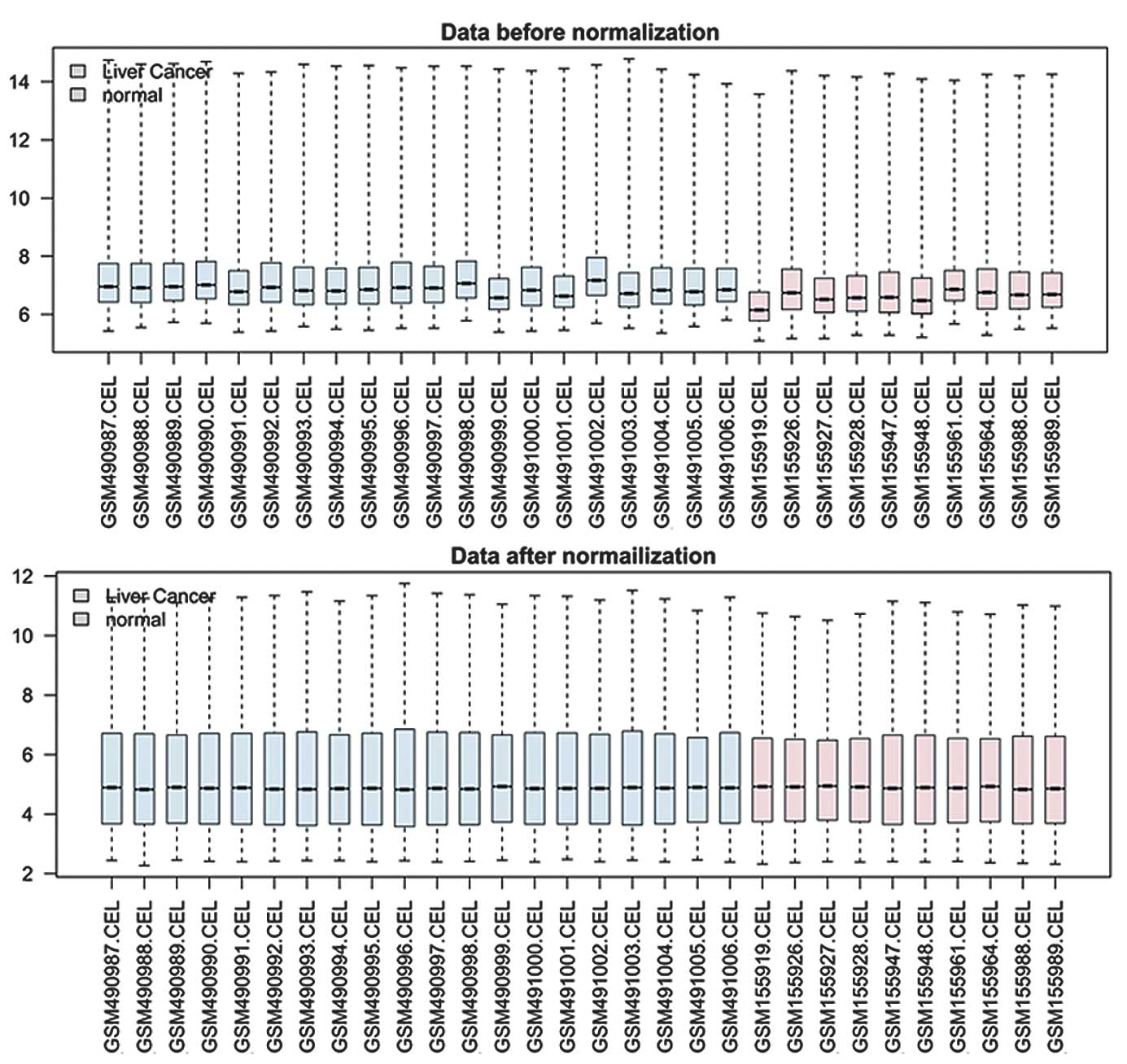
averaged when multiple probes were mapped into a single gene. Box
plots for gene expression data before and after normalization were
plotted using R.
Pretreatment of CNV data
The case and the control group were pretreated
separately. The distribution of CNVs on the 22 chromosomes was
analyzed in three intervals, 1–10, 10–50 and >50 kb,
respectively. P-values of difference in CNV distribution between
the case and the control group were calculated using permutation
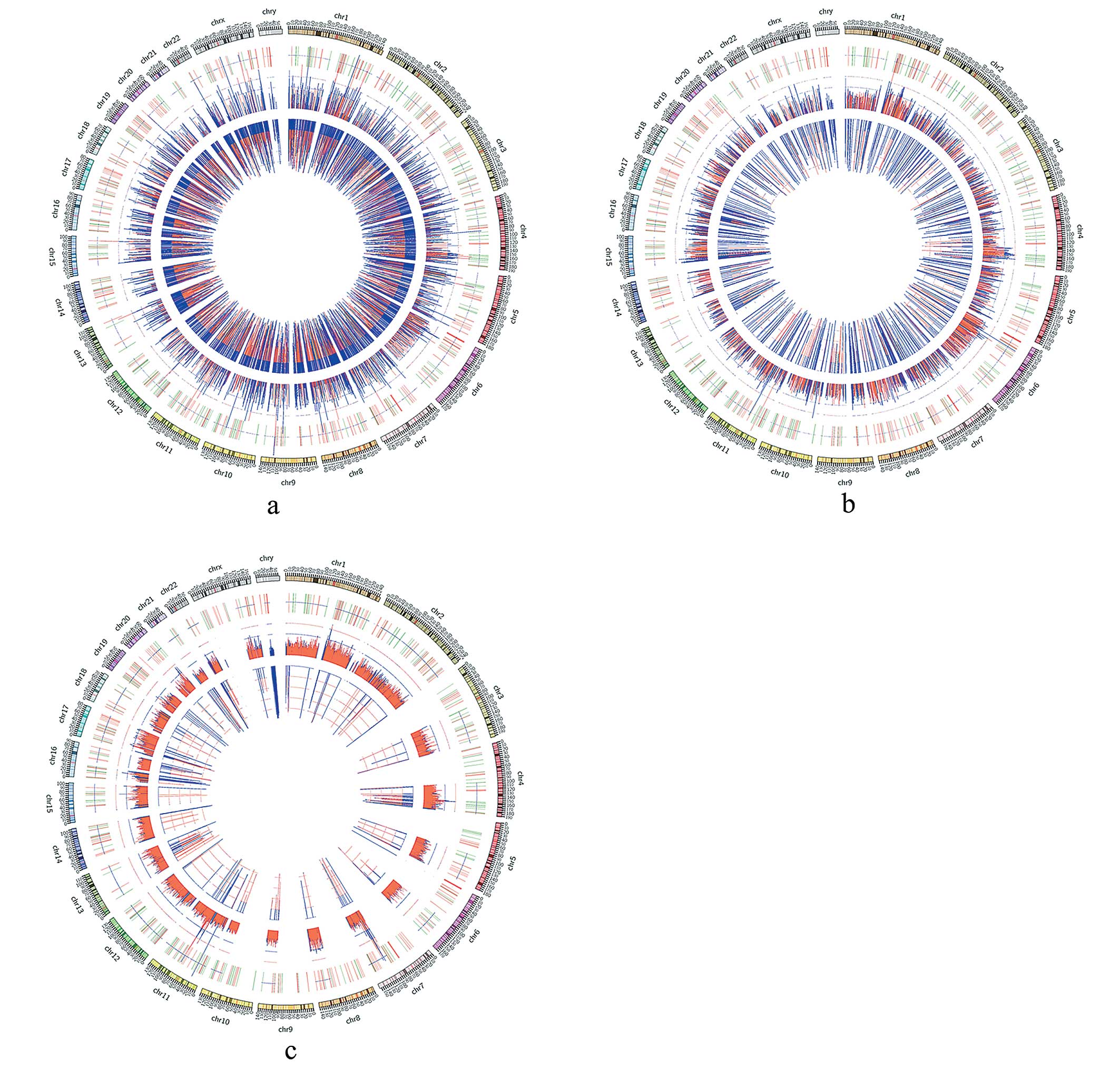
test. Circos circular diagram was plotted to display CNV
distribution. DEGs were also marked in the diagram.
Screening of DEGs
Differential analysis was performed with package
limma to screen out DEGs. |log2FC| >0.585 (i.e. absolute
fold-change >1.5) and adjusted p-value <0.05 were set as the
cut-offs.
Screening of potential liver
cancer-related CNVs
Using hg19 annotation information provided by UCSC
(15), genes in CNV regions and
values of CNVs were obtained. Liver cancer-related CNVs were then
screened out according to the criterion that it is not observed in
controls but is detected in >80% of cases. The gene-CNV matrix
was constructed and missing value was filled up with 0 (i.e. log2
(segment_mean) = 0, copy number 1).
Screening of CNV-driven genes
Matrix of CNVs and expression values were
constructed and correlation analysis was performed on genes with
both values. Genes showing same trends in significant differential
expression and CNV were termed as CNV-driven genes.
Functional enrichment analysis
Gene Ontology (GO) enrichment analysis and Kyoto
Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analysis were performed on DEGs and CNV-driven genes using Database
for Annotation, Visualization, and Integrated Discovery (DAVID)
(16) online tools. P-value
<0.05 was set as the threshold to filter out significant terms.
TF-target gene interactions were also predicted with information
from UCSC using DAVID online tools. The transcriptional regulatory
network was then visualized with Cytoscape (17).
Results
DEGs
A total of 19,944 gene expression values were
obtained in normal liver tissue samples and liver cancer samples.
Box plots for gene expression values before and after normalization
are shown in Fig. 1. A total of
1,675 DEGs were identified in liver cancer, of which 1,090 were
upregulated.
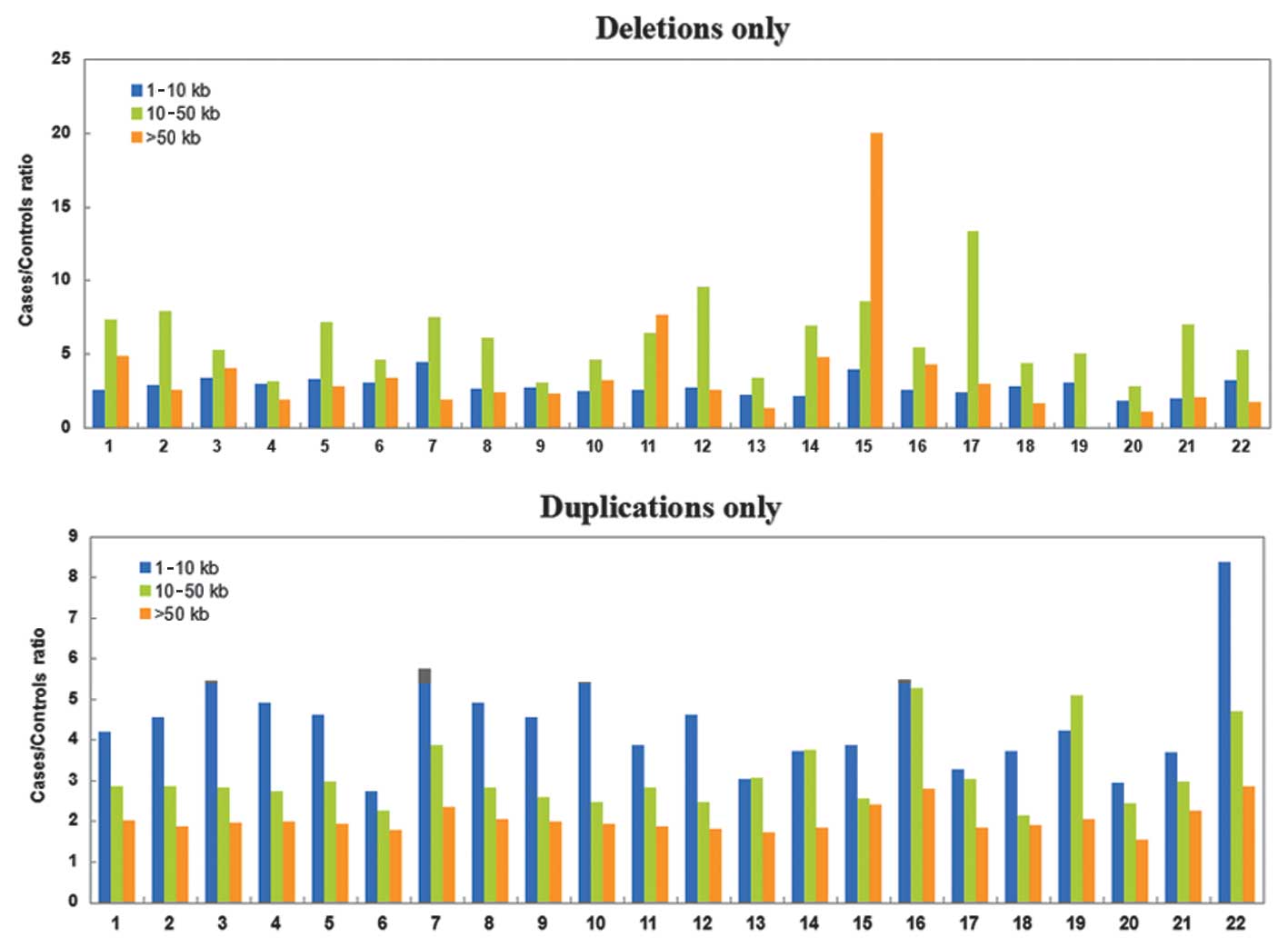
CNV data analysis results
CNV data were analyzed and distribution of CNVs in
chromosomes is shown in Tables I
and II, and Figs. 2 and 3.
 | Table IDistribution of depleted CNVs in
different chromosomes. |
Table I
Distribution of depleted CNVs in
different chromosomes.
| Deletions only |
|---|
|
|
|---|
| 1–10 kb | 10–50 kb | >50 kb |
|---|
|
|
|
|
|---|
| Chromosome | Observed CNV in cases
and controls | Ratio of
case/control | P-value | Observed CNV in cases
and controls | Ratio of
case/control | P-value | Observed CNV in cases
and controls | Ratio of
case/control | P-value |
|---|
| 1 | 487 | 2.479 | 0.055 | 100 | 7.333 | 0.007 | 41 | 4.857 | 0.3995 |
| 2 | 433 | 2.832 | 0.1975 | 89 | 7.900 | 0.0905 | 28 | 2.500 | 0.424 |
| 3 | 359 | 3.325 | 0.003 | 75 | 5.250 | 0.1075 | 20 | 4.000 | 0.0785 |
| 4 | 437 | 2.902 | 0.9775 | 95 | 3.130 | 0.6475 | 83 | 1.862 | 0.425 |
| 5 | 329 | 3.218 | 0.1825 | 73 | 7.111 | 0.4055 | 38 | 2.800 | 0.925 |
| 6 | 396 | 3.041 | 0.149 | 89 | 4.563 | 0.009 | 26 | 3.333 | 0.8285 |
| 7 | 358 | 4.424 | 0.057 | 76 | 7.444 | 0.0725 | 26 | 1.889 | 0.1775 |
| 8 | 432 | 2.600 | 0.99 | 99 | 6.071 | 0.0825 | 90 | 2.333 | 0.7685 |
| 9 | 299 | 2.646 | 0.553 | 77 | 3.053 | 0.889 | 65 | 2.250 | 0.8385 |
| 10 | 330 | 2.474 | 0.4795 | 72 | 4.538 | 0.103 | 71 | 3.176 | 0.6855 |
| 11 | 289 | 2.482 | 0.0125 | 59 | 6.375 | 0.1385 | 26 | 7.667 | 0.1415 |
| 12 | 274 | 2.653 | 0.0245 | 42 | 9.500 | 0.812 | 14 | 2.500 | 0.358 |
| 13 | 280 | 2.218 | 0.44 | 56 | 3.308 | 0.613 | 57 | 1.280 | 0.5635 |
| 14 | 177 | 2.105 | 0.044 | 71 | 6.889 | 0.211 | 23 | 4.750 | 0.816 |
| 15 | 192 | 3.923 | 0.104 | 67 | 8.571 | 0.0465 | 42 | 20.000 | 0.25 |
| 16 | 206 | 2.552 | 0.8335 | 70 | 5.364 | 0.9395 | 42 | 4.250 | 0.853 |
| 17 | 183 | 2.327 | 0.086 | 43 | 13.333 | 0.1795 | 39 | 2.900 | 0.0655 |
| 18 | 190 | 2.800 | 0.062 | 16 | 4.333 | 0.4735 | 13 | 1.600 | 0.8005 |
| 19 | 149 | 3.027 | 0.116 | 30 | 5.000 | 0.7145 | 10 | - | 0.0005 |
| 20 | 159 | 1.741 | 0.2635 | 15 | 2.750 | 0.083 | 2 | 1.000 | 0.5055 |
| 21 | 83 | 1.964 | 0.376 | 16 | 7.000 | 0.901 | 24 | 2.000 | 0.4615 |
| 22 | 121 | 3.172 | 0.228 | 25 | 5.250 | 0.208 | 27 | 1.700 | 0.87 |
 | Table IIDistribution of duplicated CNVs in
different chromosomes. |
Table II
Distribution of duplicated CNVs in
different chromosomes.
| Duplications
only |
|---|
|
|
|---|
| 1–10 kb | 10–50 kb | >50 kb |
|---|
|
|
|
|
|---|
| Chromosome | CNV in
cases/controls | Case/control
ratio | P-value | CNV in
cases/controls | Case/control
ratio | P-value | CNV in
cases/controls | Case/control
ratio | P-value |
|---|
| 1 | 697 | 4.201 | 0.0105 | 1152 | 2.853 | 0.0005 | 5202 | 2.007 | 0.0005 |
| 2 | 465 | 4.536 | 0.424 | 748 | 2.856 | 0.0715 | 3946 | 1.853 | 0.015 |
| 3 | 450 | 5.429 | 0.3125 | 689 | 2.828 | 0.0005 | 3272 | 1.958 | 0.0005 |
| 4 | 532 | 4.911 | 0.0015 | 758 | 2.734 | 0.0005 | 3824 | 1.990 | 0.1435 |
| 5 | 325 | 4.603 | 0.1385 | 493 | 2.976 | 0.0175 | 2934 | 1.922 | 0.0175 |
| 6 | 535 | 2.715 | 0.002 | 997 | 2.258 | 0.0005 | 3731 | 1.784 | 0.006 |
| 7 | 432 | 5.750 | 0.099 | 633 | 3.869 | 0.0005 | 3047 | 2.334 | 0.0075 |
| 8 | 472 | 4.900 | 0.0305 | 549 | 2.813 | 0.0005 | 3222 | 2.057 | 0.0005 |
| 9 | 272 | 4.551 | 0.2055 | 390 | 2.578 | 0.0015 | 2614 | 1.970 | 0.418 |
| 10 | 276 | 5.419 | 0.7085 | 438 | 2.449 | 0.129 | 2606 | 1.915 | 0.9845 |
| 11 | 345 | 3.859 | 0.1175 | 536 | 2.829 | 0.0435 | 2605 | 1.878 | 0.1035 |
| 12 | 297 | 4.604 | 0.0505 | 450 | 2.462 | 0.0075 | 2508 | 1.818 | 0.127 |
| 13 | 189 | 3.021 | 0.9835 | 235 | 3.052 | 0.792 | 1540 | 1.711 | 0.2585 |
| 14 | 141 | 3.700 | 0.992 | 322 | 3.735 | 0.4105 | 1788 | 1.825 | 0.1985 |
| 15 | 160 | 3.848 | 0.038 | 365 | 2.544 | 0.029 | 1791 | 2.411 | 0.007 |
| 16 | 149 | 5.478 | 0.714 | 300 | 5.250 | 0.371 | 2021 | 2.778 | 0.492 |
| 17 | 218 | 3.275 | 0.2245 | 297 | 3.014 | 0.0225 | 2070 | 1.836 | 0.0005 |
| 18 | 231 | 3.714 | 0.1515 | 314 | 2.140 | 0.002 | 1267 | 1.899 | 0.114 |
| 19 | 141 | 4.222 | 0.9115 | 243 | 5.075 | 0.266 | 1437 | 2.032 | 0.002 |
| 20 | 165 | 2.929 | 0.8945 | 264 | 2.429 | 0.055 | 1305 | 1.549 | 0.022 |
| 21 | 61 | 3.692 | 0.8655 | 107 | 2.963 | 0.423 | 608 | 2.251 | 0.2475 |
| 22 | 131 | 8.357 | 0.827 | 233 | 4.683 | 0.8505 | 1158 | 2.847 | 0.767 |
Functional enrichment analysis
results
Significant GO terms and KEGG pathways of
upregulated and downregulated genes are listed in Tables III and IV. Cell cycle and ECM-receptor
interaction were enriched in upregulated genes. Several metabolic
pathways were significant in downregulated genes, such as cellular
amino acid derivative metabolic process, metabolism of xenobiotics
by cytochrome P450 and glycolysis/gluconeogenesis.
 | Table IIITop 5 significant GO terms and KEGG
pathways in upregulated genes. |
Table III
Top 5 significant GO terms and KEGG
pathways in upregulated genes.
| Category | Term | Count | P-value | FDR |
|---|
| GOTERM_BP_FAT | GO:0000279 - M
phase | 24 | 7.32E-11 | 1.21E-07 |
| GOTERM_BP_FAT | GO:0022403 - Cell
cycle phase | 25 | 1.26E-09 | 2.08E-06 |
| GOTERM_BP_FAT | GO:0000278 -
Mitotic cell cycle | 23 | 4.11E-09 | 6.77E-06 |
| GOTERM_BP_FAT | GO:0000280 -
Nuclear division | 18 | 5.69E-09 | 9.38E-06 |
| GOTERM_BP_FAT | GO:0007067 -
Mitosis | 18 | 5.69E-09 | 9.38E-06 |
| GOTERM_CC_FAT | GO:0005819 -
Spindle | 15 | 2.52E-09 | 3.26E-06 |
| GOTERM_CC_FAT | GO:0015630 -
Microtubule cytoskeleton | 23 | 6.77E-07 | 8.76E-04 |
| GOTERM_CC_FAT | GO:0005581 -
Collagen | 7 | 3.37E-06 | 0.004363662 |
| GOTERM_CC_FAT | GO:0044430 -
Cytoskeletal part | 28 | 2.42E-05 | 0.031236022 |
| GOTERM_CC_FAT | GO:0000777 -
Condensed chromosome kinetochore | 7 | 6.70E-05 | 0.086584259 |
| GOTERM_MF_FAT | GO:0005201 -
Extracellular matrix structural constituent | 7 | 8.33E-04 | 1.135061593 |
| GOTERM_MF_FAT | GO:0005524 - ATP
binding | 31 | 0.007391687 | 9.66850349 |
| GOTERM_MF_FAT | GO:0048407 -
Platelet-derived growth factor binding | 3 | 0.008381921 | 10.89579452 |
| GOTERM_MF_FAT | GO:0032559 - Adenyl
ribonucleotide binding | 31 | 0.008892243 | 11.52223223 |
| GOTERM_MF_FAT | GO:0008022 -
Protein C-terminus binding | 7 | 0.009752326 | 12.56878133 |
| KEGG_PATHWAY | hsa04512:
ECM-receptor interaction | 9 | 1.46E-05 | 0.015673595 |
| KEGG_PATHWAY | hsa04510: Focal
adhesion | 11 | 3.29E-04 | 0.352863777 |
| KEGG_PATHWAY | hsa04110: Cell
cycle | 8 | 0.001395976 | 1.488280096 |
| KEGG_PATHWAY | hsa04612: Antigen
processing and presentation | 6 | 0.005091654 | 5.331859118 |
| KEGG_PATHWAY | hsa04062: Chemokine
signaling pathway | 8 | 0.012780092 | 12.89568049 |
 | Table IVSignificant GO terms and KEGG
pathways in downregulated genes. |
Table IV
Significant GO terms and KEGG
pathways in downregulated genes.
| Category | Term | Count | P-value | FDR |
|---|
| GOTERM_BP_FAT | GO:0009611 -
Response to wounding | 30 | 3.85E-11 | 6.54E-08 |
| GOTERM_BP_FAT | GO:0006575 -
Cellular amino acid derivative metabolic process | 17 | 4.32E-10 | 7.33E-07 |
| GOTERM_BP_FAT | GO:0006954 -
Inflammatory response | 20 | 4.19E-08 | 7.11E-05 |
| GOTERM_BP_FAT | GO:0051384 -
Response to glucocorticoid stimulus | 11 | 5.97E-08 | 1.01E-04 |
| GOTERM_BP_FAT | GO:0031960 -
Response to corticosteroid stimulus | 11 | 1.37E-07 | 2.33E-04 |
| GOTERM_CC_FAT | GO:0005615 -
Extracellular space | 34 | 2.20E-10 | 2.76E-07 |
| GOTERM_CC_FAT | GO:0005576 -
Extracellular region | 56 | 1.70E-07 | 2.13E-04 |
| GOTERM_CC_FAT | GO:0044421 -
Extracellular region part | 34 | 7.99E-07 | 0.001003709 |
| GOTERM_CC_FAT | GO:0005792 -
Microsome | 16 | 9.55E-07 | 0.001199343 |
| GOTERM_CC_FAT | GO:0042598 -
Vesicular fraction | 16 | 1.38E-06 | 0.001729266 |
| GOTERM_MF_FAT | GO:0048037 -
Cofactor binding | 15 | 6.92E-06 | 0.010006401 |
| GOTERM_MF_FAT | GO:0019842 -
Vitamin binding | 11 | 1.01E-05 | 0.014633569 |
| GOTERM_MF_FAT | GO:0009055 -
Electron carrier activity | 13 | 4.50E-05 | 0.065002326 |
| GOTERM_MF_FAT | GO:0008483 -
Transaminase activity | 5 | 1.60E-04 | 0.230550386 |
| GOTERM_MF_FAT | GO:0030246 -
Carbohydrate binding | 15 | 3.13E-04 | 0.45164934 |
| KEGG_PATHWAY | hsa00830: Retinol
metabolism | 10 | 1.91E-07 | 2.14E-04 |
| KEGG_PATHWAY | hsa00980:
Metabolism of xenobiotics by cytochrome P450 | 10 | 4.90E-07 | 5.47E-04 |
| KEGG_PATHWAY | hsa00982: Drug
metabolism | 9 | 7.09E-06 | 0.007928545 |
| KEGG_PATHWAY | hsa00010:
Glycolysis/gluconeogenesis | 7 | 4.44E-04 | 0.49547712 |
| KEGG_PATHWAY | hsa00380:
Tryptophan metabolism | 6 | 4.82E-04 | 0.537167906 |
Screening of potential liver
cancer-related CNVs
A total of 735 liver cancer-related CNVs were
obtained. Matrix of genes and CNVs was then constructed. A total of
251 genes with CNVs and gene expression values were selected out,
of which 46 genes showed significant differential expression
between liver cancer and normal liver tissue. Given CNVs in X and Y
chromosome from controls were not included, 11 genes located in X
and Y chromosome were excluded from subsequent analysis and 35
genes were retained for subsequent analysis (Fig. 4 and Table V).
 | Table VResult of correlation analysis
between copy number and differential expression for the 35
genes. |
Table V
Result of correlation analysis
between copy number and differential expression for the 35
genes.
| Chromosome | Gene | Log2 (copy
no.) | Log2 (FC) |
|---|
| chr11 | ATHL1 | 0.593809495 | 0.982449 |
| chr5 | CEP72 | 0.704121055 | 1.06131 |
| chr13 | CHAMP1 | 0.852111156 | 0.969549 |
| chr22 | CHKB | 0.753779885 | 0.96845 |
| chr4 | CPLX1 | 1.047010058 | 0.967371 |
| chr10 | CYP2E1 | −0.905659049 | 0.968898 |
| chr8 | DGAT1 | 0.597175013 | 1.11897 |
| chr4 | DGKQ | 0.747548576 | 0.964431 |
| chr17 | FAM101B | −0.916939916 | 0.880721 |
| chr2 | FAM110C | −1.322774314 | 1.0116 |
| chr1 | FAM132A | −0.712891007 | 0.920818 |
| chr2 | FAM150B | 0.969367389 | 1.01004 |
| chr1 | FAM213B | 0.700468993 | 0.91626 |
| chr8 | FBXO25 | −0.627491404 | 0.889618 |
| chr1 | HES4 | 0.942507623 | 0.91584 |
| chr18 | HSBP1L1 | −0.707252053 | 0.974772 |
| chr16 | ITFG3 | 0.611547777 | 0.953547 |
| chr8 | KIFC2 | 0.601630897 | 1.12167 |
| chr1 | LINC00115 | 0.656600032 | 0.939262 |
| chr7 | NCAPG2 | 0.689807598 | 1.0425 |
| chr2 | NEU4 | −0.943299743 | 0.967018 |
| chr5 | PP7080 | 0.792638133 | 1.05958 |
| chr12 | PXMP2 | −0.967032365 | 0.979198 |
| chr16 | RAB11FIP3 | 0.748865682 | 0.956749 |
| chr20 | RBCK1 | 0.725328672 | 1.04205 |
| chr22 | SHANK3 | 0.751687098 | 0.970314 |
| chr11 | SIGIRR | −1.372384603 | 0.987798 |
| chr19 | TRIM28 | 0.891861874 | 1.01467 |
| chr5 | TRIM41 | 0.978259149 | 1.03054 |
| chr5 | TRIM52 | 0.805628747 | 1.03054 |
| chr5 | TRIP13 | 0.89827027 | 1.05874 |
| chr22 | ZBED4 | 0.700677442 | 0.973172 |
| chr20 | ZGPAT | −1.477496713 | 1.05204 |
| chr20 | ZNF512B | 0.764282511 | 1.05494 |
| chr1 | ZNF692 | 0.605492215 | 1.16697 |
CNV-driven genes and transcriptional
regulatory network
A total of 25 CNV-driven genes were identified.
Functional enrichment analysis results of these genes are shown in
Table VI. In the transcriptional
regulatory network (Fig. 5), 16 TFs
regulated 21 CNV-driven genes. SP1, AP2, CREB, ELK1, PAX5, PPARA,
STAT3 and USF were recorded in TRED as known cancer-related TFs.
The other 8 TFs (AHRARNT, MAZR, NRF2, ROAZ, RORA1, SREBP1, TAXCREB
and ZIC1) may play roles in the development of liver cancer.
 | Table VIFunctional enrichment analysis
results for potential CNV-driven genes. |
Table VI
Functional enrichment analysis
results for potential CNV-driven genes.
| Category | Term | Count | P-value | Genes |
|---|
| GOTERM_MF_FAT | GO:0008270-Zinc ion
binding | 8 | 0.014354978 | ZNF512B
DGKQ
ZNF692
ZBED4
TRIM28
TRIM41
RBCK1
TRIM52 |
| GOTERM_MF_FAT |
GO:0046914-Transition metal ion
binding | 8 | 0.03796619 | ZNF512B
DGKQ
ZNF692
ZBED4
TRIM28
TRIM41
RBCK1
TRIM52 |
| GOTERM_MF_FAT |
GO:0008374-O-acyl-transferase
activity | 2 | 0.047021614 | DGAT1
CHKB |
| KEGG_PATHWAY | hsa00561:
Glycerolipid metabolism | 2 | 0.026319552 | DGKQ
DGAT1 |
| KEGG_PATHWAY | hsa00564:
Glycero-phospholipid metabolism | 2 | 0.039591583 | DGKQ
CHKB |
Discussion
In the present study, we carried out an integrated
analysis of copy number variation (CNV) data and gene expression
data for liver cancer. A total of 1,675 differentially expressed
genes (DEGs) were identified in liver cancer, of which 1,090 were
upregulated. According to the CNV distribution results, in liver
cancer, deletion and duplication of CNVs were common in all the 22
chromosomes. CNV repeats with length 1–10 kb were significantly
more than those with length >50 kb, suggesting CNVs in liver
cancer were likely to affect the expression of a single gene.
Thirty five genes with associated copy number and
differential expression were acquired, of which 25 genes showed the
same trends in the gene expression and CNV and they were regarded
as liver cancer-related CNV-driven genes. Zinc ion binding was
enriched in these genes, indicating zinc plays a role in liver
cancer, which was in accordance with previous studies (18,19).
Tripartite motif containing 28 (TRIM28) mediates transcriptional
control via interaction with the Kruppel-associated box repression
domain found in many transcription factors; it can suppress murine
HCC by forming regulatory complexes with TRIM24 and TRIM33
(20). RanBP-type and C3HC4-type
zinc finger containing 1 (RBCK1) can promote cancer cell
proliferation (21,22). Zinc finger protein 512B (ZNF512B) is
a transcription factor promoting the expression of a downstream
gene in the signal transduction pathway of the transforming growth
factor-β (TGF-β), which is essential for the protection and
survival of neurons, however the influence of the new SNP
(rs2275294) in actual ALS patients remained unknown (23). Diacylglycerol kinase theta (DGKQ)
has been reported to be associated with the risk of Parkinson’s
disease (PD) in Caucasian populations (24). Choline kinase β (CHKB) is both a
CNV-driven gene and a candidate for susceptibility to CNS
hypersomnias (EHS), as well as narcolepsy with cataplexy.
Therefore, the 25 CNV-driven genes may be potential markers for
liver cancer.
In the transcriptional regulatory network, 8 TFs
have been linked to cancers and the other 8 TFs (AHRARNT, MAZR,
NRF2, ROAZ, RORA1, SREBP1, TAXCREB and ZIC1) are implicated in
regulation of the 21 CNV-driven genes and may play roles in the
pathogenesis of liver cancer. The CD4 vs. CD8 lineage specification
of thymocytes is linked to co-receptor expression. The
transcription factor POZ (BTB) and AT hook containing zinc finger 1
(PATZ1, MAZR) has been identified as an important regulator of Cd8
expression (25). Transcription
factor nuclear factor erythroid-2-related factor 2 (NRF2) is
essential for the antioxidant responsive element (ARE)-mediated
induction of phase II detoxifying and oxidative stress enzyme genes
(26). Shibata et al
reported that mutations in NRF2 impair its recognition by
Keap1-Cul3 E3 ligase and promote malignancy (27). Zinc finger protein 423 (ZFP423,
ROAZ), a rat C2H2 zinc finger protein, plays a role in the
regulation of olfactory neuronal differentiation through its
interaction with the Olf-1/EBF transcription factor family
(28). Sterol regulatory
element-binding protein 1 (SREBP-1), a member of the
basic-helix-loop-helix-leucine zipper (bHLH-ZIP) family of
transcription factors, is synthesized as a 125 kd precursor that is
attached to the nuclear envelope and endoplasmic reticulum
(29). Human T-lymphotropic virus
type 1 Tax interacts specifically with the cellular transcription
factor CREB and the viral 21-bp repeat element to form a
Tax-CREB-DNA ternary complex which mediates activation of viral
mRNA transcription (30). These TFs
merit further study to delineate their roles in liver cancer.
Collectively, the present study identified DEGs in
liver cancer and disclosed a range of CNV-driven genes. Their
biological functions and regulatory network were also discussed.
These findings may improve our understanding of liver cancer and
advance therapy development.
Acknowledgements
This study was supported by the National High
Technology Research (863) Project of China (2012AA020204).
References
|
1
|
Bosch FX, Ribes J and Borràs J:
Epidemiology of primary liver cancer. Semin Liver Dis. 19:271–285.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ko YH, Pedersen PL and Geschwind JF:
Glucose catabolism in the rabbit VX2 tumor model for liver cancer:
characterization and targeting hexokinase. Cancer Lett. 173:83–91.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kiyosawa K, Umemura T, Ichijo T, et al:
Hepatocellular carcinoma: recent trends in Japan. Gastroenterology.
127(Suppl 1): S17–S26. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Redon R, Ishikawa S, Fitch KR, et al:
Global variation in copy number in the human genome. Nature.
444:444–454. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang F, Gu W, Hurles ME and Lupski JR:
Copy number variation in human health, disease, and evolution. Annu
Rev Genomics Hum Genet. 10:451–481. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Parajes S, Quinteiro C, Domínguez F and
Loidi L: High frequency of copy number variations and sequence
variants at CYP21A2 locus: implication for the genetic
diagnosis of 21-hydroxylase deficiency. PLoS One. 3:e21382008.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vissers LE, de Vries BB and Veltman JA:
Genomic microarrays in mental retardation: from copy number
variation to gene, from research to diagnosis. J Med Genet.
47:289–297. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Temam S, Kawaguchi H, El-Naggar AK, et al:
Epidermal growth factor receptor copy number alterations correlate
with poor clinical outcome in patients with head and neck squamous
cancer. J Clin Oncol. 25:2164–2170. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hirsch FR, Varella-Garcia M, Cappuzzo F,
et al: Combination of EGFR gene copy number and protein expression
predicts outcome for advanced non-small-cell lung cancer patients
treated with gefitinib. Ann Oncol. 18:752–760. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sartore-Bianchi A, Moroni M, Veronese S,
et al: Epidermal growth factor receptor gene copy number and
clinical outcome of metastatic colorectal cancer treated with
panitumumab. J Clin Oncol. 25:3238–3245. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Schlomm T, Kirstein P, Iwers L, et al:
Clinical significance of epidermal growth factor receptor protein
overexpression and gene copy number gains in prostate cancer. Clin
Cancer Res. 13:6579–6584. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lee HC, Li SH, Lin JC, Wu CC, Yeh DC and
Wei YH: Somatic mutations in the D-loop and decrease in the copy
number of mitochondrial DNA in human hepatocellular carcinoma.
Mutat Res. 547:71–78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yin PH, Lee HC, Chau GY, et al: Alteration
of the copy number and deletion of mitochondrial DNA in human
hepatocellular carcinoma. Br J Cancer. 90:2390–2396.
2004.PubMed/NCBI
|
|
14
|
Guichard C, Amaddeo G, Imbeaud S, et al:
Integrated analysis of somatic mutations and focal copy-number
changes identifies key genes and pathways in hepatocellular
carcinoma. Nat Genet. 44:694–698. 2012. View Article : Google Scholar
|
|
15
|
Fujita PA, Rhead B, Zweig AS, et al: The
UCSC Genome Browser database: update 2011. Nucleic Acids Res.
39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dennis G Jr, Sherman BT, Hosack DA, et al:
DAVID: Database for Annotation, Visualization, and Integrated
Discovery. Genome Biol. 4:P32003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: new features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Franklin RB, Levy BA, Zou J, et al: ZIP14
zinc transporter downregulation and zinc depletion in the
development and progression of hepatocellular cancer. JJ
Gastrointest Cancer. 43:249–257. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ebara M, Fukuda H, Hatano R, et al:
Relationship between copper, zinc and metallothionein in
hepatocellular carcinoma and its surrounding liver parenchyma. J
Hepatol. 33:415–422. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Herquel B, Ouararhni K, Khetchoumian K, et
al: Transcription cofactors TRIM24, TRIM28, and TRIM33 associate to
form regulatory complexes that suppress murine hepatocellular
carcinoma. Proc Natl Acad Sci USA. 108:8212–8217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gustafsson N, Zhao C, Gustafsson JA and
Dahlman-Wright K: RBCK1 drives breast cancer cell proliferation by
promoting transcription of estrogen receptor α and cyclin B1.
Cancer Res. 70:1265–1274. 2010.PubMed/NCBI
|
|
22
|
Donley C, McClelland K, McKeen HD, et al:
Identification of RBCK1 as a novel regulator of FKBPL: implications
for tumor growth and response to tamoxifen. Oncogene. Aug
5–2013.(Epub ahead of print). View Article : Google Scholar
|
|
23
|
Tetsuka S, Morita M, Iida A, Uehara R,
Ikegawa S and Nakano I: ZNF512B gene is a prognostic factor in
patients with amyotrophic lateral sclerosis. J Neurol Sci.
324:163–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen YP, Song W, Huang R, et al:
GAK rs1564282 and DGKQ rs11248060 increase the risk
for Parkinson’s disease in a Chinese population. J Clin Neurosci.
20:880–883. 2013. View Article : Google Scholar
|
|
25
|
Sakaguchi S, Hombauer M, Bilic I, et al:
The zinc-finger protein MAZR is part of the transcription factor
network that controls the CD4 versus CD8 lineage fate of
double-positive thymocytes. Nat Immunol. 11:442–448. 2010.
View Article : Google Scholar
|
|
26
|
Itoh K, Wakabayashi N, Katoh Y, et al:
Keap1 represses nuclear activation of antioxidant responsive
elements by Nrf2 through binding to the amino-terminal Neh2 domain.
Genes Dev. 13:76–86. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shibata T, Ohta T, Tong KI, et al: Cancer
related mutations in NRF2 impair its recognition by
Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci
USA. 105:13568–13573. 2008.
|
|
28
|
Tsai RY and Reed RR: Identification of DNA
recognition sequences and protein interaction domains of the
multiple-Zn-finger protein Roaz. Mol Cell Biol. 18:6447–6456.
1998.PubMed/NCBI
|
|
29
|
Wang X, Sato R, Brown MS, Hua X and
Goldstein JL: SREBP-1, a membrane-bound transcription factor
released by sterol-regulated proteolysis. Cell. 77:53–62. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tie F, Adya N, Greene WC and Giam CZ:
Interaction of the human T-lymphotropic virus type 1 Tax dimer with
CREB and the viral 21-base-pair repeat. J Virol. 70:8368–8374.
1996.PubMed/NCBI
|