Introduction
Lung cancer, with an estimated patient survival rate
of ~5 years, has become a leading cause of cancer-related death
(1). It has been proposed that
tumors are sustained by a cell subpopulation with tumor-initiating
properties. These cells, known as tumor-initiating cells (TICs) or
cancer stem cells (CSCs), have self-renewal capacity and the
ability to create the characteristic cellular heterogeneity found
in solid tumors. TICs have been previously identified and isolated
using specific cell membrane markers, such as OV-6, CD13 and CD44
in liver cancer (2) and EpCAM,
CD44, CD24 in breast cancer (3).
In 2008, Eramo et al identified for the first
time tumor-initiating cells from lung tumors using CD133 as a
biomarker (4–6). However, this result has been disputed
by several authors. For example, in 2009, Meng et al found
that both CD133− and CD133+ cell populations
from lung cancer possess TIC properties (7), and more recently Qiu et al
found no statistical difference between the ability of
CD133− and CD133+ cell populations to form
pneumospheres (8,9). The predictive value to detect this
subpopulation in lung cancer cell lines of other TIC biomarkers,
including ALDH1 and CD24, remains controversial (10–12).
To date, there are no reliable biomarkers for the detection of
tumor-initiating cells in lung cancer.
Neuropilin 1 (NRP1) is a transmembrane glycoprotein
involved in various cellular processes that include angiogenesis,
cell migration, T cell activation, survival and axon growth
(13,14). Existing data suggest an association
between NRP1 expression and a tumor-initiating cell phenotype. For
example, endothelial progenitors can be identified by NRP1
expression (15). In addition, it
has been shown that NRP1 is essential for proliferation and cell
migration of adult mesenchymal stem cells (16). NRP1 promotes TIC-related cellular
processes, such as angiogenesis, cell migration, invasion and
metastasis in cancer tissue (17,18).
Moreover, NRP1 overexpression induces a poorly differentiated
phenotype in renal carcinoma cells (19). Furthermore, NRP1 also maintains a
tumor-initiating phenotype in glioma and skin cancer cells
(20). In addition, Barr et
al reported that NRP1 is a critical co-receptor in
VEGF-mediated survival and tumor growth of NSCLC cells (21).
In the present study, we analyzed whether NRP1
expression was able to identified a TIC subpopulation in lung
cancer cell lines and is involved in the maintenance of these
cells. We found that NRP1-expressing cells exhibited TIC-like
properties, i.e. stemness and high clonogenic capability.
Concordant with this, NRP1 downregulation inhibited the expression
of stemness markers and prevented cell migration and pneumosphere
formation. Finally, a genome-wide expression analysis in
NRP1-knockdown cells revealed differentially expressed genes that
could be involved in the maintenance of the TIC phenotype.
Materials and methods
Cell culture
Lung cancer cell lines A549 and Calu-1 were obtained
from the American Type Culture Collection (CCL-185 and HTB-54;
ATCC; Manassas, VA, USA). Cell lines were maintained in Dulbecco's
modified Eagles medium (DMEM) (Corning Life Sciences, Corning, NY,
USA) supplemented with 5% fetal bovine serum (FBS) (#30-2020;
ATCC), and kept at 37°C, with 5% CO2 and 95%
humidity.
Flow cytometry
Cells were detached from the plates using StemPro
Accutase (Thermo Scientific, Waltham, MA, USA), washed with 1X
phosphate-buffered saline (PBS), and suspended in 1% FBS.
Subsequently, 1×107 cells were incubated in ice with the
antibodies APC-NRP1 (130-090-900) at a 1:10 dilution for 40 min.
The isotype control antibodies IgG1-APC (130-092-214) were used.
All antibodies were obtained from Miltenyi Biotech (Bergisch
Gladbach, Germany). The cells were sorted with a FACSAria flow
cytometer (Becton-Dickinson, Franklin Lakes, NJ, USA), according to
their phenotype into NRP1-negative (NRP1−) and
NRP1-positive (NRP1+) subpopulations. All of the sorted
populations were maintained under standard growth conditions.
Semi-quantitative PCR analysis
Total RNA was extracted from the cells using TRIzol
reagent (Thermo Fisher Scientific, Waltham, MA USA) following the
manufacturer's instructions. RNA concentration and purity were
determined using a NanoDrop (Thermo Scientific, Wilmington, DE,
USA). Subsequently, 1 µg of total RNA was reverse transcribed to
cDNA using the High Capacity cDNA reverse transcription kit
(Applied Biosystems, Foster City, CA, USA) in a total volume of 20
µl. The cDNA was amplified by semi-quantitative PCR using specific
primers for each tested gene. Saturation curves for each amplified
fragment was carried out at different cycles. TATA-box binding
protein (TBP) or 18S gene expression were used as an internal
reference control. PCR products were resolved by electrophoresis on
a 1.5% agarose gel. The DNA bands were visualized using Gel-Red
staining (Biotium, Hayward, CA, USA).
Colony forming assays
To evaluate the clonogenic capacity of each isolated
population, 4×103 cells were plated in DMEM with 0.3%
low melting point agarose and 10% FBS (as the upper layer). As a
basecoat, a mixture of 1% low-melting agarose with 2X DMEM medium
in a 1:1 ratio was used. The cells were incubated for 4 weeks at
37°C with 5% CO2 and 95% humidity. To prevent the agar
plate from dehydrating, 200 µl of DMEM with 10% FBS was added every
3 days. At the end of the incubation period, the colonies were
stained with 0.01% crystal violet and counted using an inverted
microscope (Olympus, Inc., New Orleans, LA, USA).
Pneumosphere formation assay
To assess the ability of each population to form
pneumospheres, 5×104 cells were seeded in 12.5
cm2 flasks in Leibovitz's medium (both from Corning
Inc., Corning, NY, USA), supplemented with 5% FBS. Cells were
maintained at 37°C with a constant 50 rpm orbital agitation for
several days. Medium was changed every day. After 5 days, the
pneumospheres were counted and dissociated into a single-cell
suspension using 2.5% trypsin (Thermo Fisher Scientific). The cells
were counted and seeded into new flasks for the following serial
passage assays.
NRP1 knockdown
Two shRNAs targeting NRP1 (shNRP1) were designed
using the online tool from Clontech Laboratories, Inc. (Mountain
View, CA, USA) (http://www.clontech.com). The shRNA oligonucleotides
were annealed and ligated into the vector pSIREN-RetroQ (631526;
Clontech Laboratories, Inc.) using the Quick Ligation kit from
Fermentas (K142; Waltham, MA, USA). An shRNA targeting the
luciferase gene (shLuc) was used as control. Lipofectamine 2000
(Thermo Fisher Scientific) was used in the transfection assays as
instructed by the manufacturer. Positive selection of A549 cells
was performed with 7.5 µg/ml puromycin (Sigma-Aldrich, St. Louis,
MO, USA), whereas Calu-1 cells were selected with 5 µg/ml puromycin
for 2 weeks. The gene expression level of NRP1 in the transfected
cells was analyzed by semi-quantitative PCR.
Western blot analysis
A549 cells were washed with 1X PBS, harvested and
centrifuged at 1,000 rpm for 5 min; the cell pellets were
resuspended in 300 µl of buffer A (1X protease inhibitor, 10 mM
HEPES pH 7.5, 15 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA and 2 mM
MgCl2), incubated on ice for 15 min, after which 0.01%
NP40 was added for 1 min. The lysates were centrifuged at 6,000
rpm/5 min. The cytosolic fraction (supernatant) was recovered and
stored at −20°C for later analysis. The remaining pellet was
resuspended in 200 µl of buffer C (400 mM NaCl, 1 mM EDTA, 25 mM
HEPES pH 7.5 and 20% glycerol) and incubated for 30 min on ice.
After incubation, the lysates were centrifuged at 8,000 rpm/5 min
and the nuclear fraction (supernatant) was recovered and stored at
−20°C. Fractions were resolved by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred
to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford,
MA, USA) and blocked with 5% albumin in Tris-buffered saline (TBS)
with 0.1% Tween for 1 h. The membranes were then incubated with the
primary antibody overnight at 4°C. Antibodies were diluted in 5%
bovine serum albumin, 1X TBS and 0.1% Tween. For the NRP1 antibody
(36–1400; Invitrogen, Waltham, MA, USA) a concentration of 2 µg/ml
was used. For p63 and phospho-p63 antibodies (#4892 and #4981; Cell
Signaling Technology Inc., Danvers, MA USA) a dilution of 1:1,000
was used. As loading controls, lamin B (sc-374015) and tubulin
(sc-53646) (Santa Cruz Biotechnology, Dallas, TX, USA) were used.
The membranes were washed with TBS-Tween 0.2% and incubated with
the secondary antibodies, anti-mouse IgG HRP-conjugated W402B or
anti-rabbit IgG HRP-conjugated W401B (Promega, Madison, WI, USA) at
room temperature for 1 h. Proteins were detected using the Western
Kit Immobilon (Millipore) and visualized with VersaDoc Imaging
System (Bio-Rad, Hercules, CA, USA).
Wound healing assay
Cell migration capacity was determined by seeding
1×106 cells into 6-well plates (Corning) with DMEM and
5% FBS. After 12 h, a wound was made to the confluent cell layer
using a 200-µl pipette tip. Images of the wounded area were
captured at 0, 24 and 48 h. Cell migration capacity was calculated
determining the wound opening area using ImageJ software (22).
Microarray analysis
Total RNA from shNRP1- and shLuc-transfected cells
was extracted from 3 independent experiments. RNA integrity was
assessed on a Bioanalyzer 2100 (Agilent Technologies, Inc., Santa
Clara, CA, USA), and samples with a RNA integrity number (RIN)
>9 were used. Microarray experiments were performed using the
Affymetrix GeneChip platform (HuGene 2.0 ST) in the National
Institute of Genomic Medicine's Microarray High Technology Core
Center, Mexico. Raw data were normalized using Robust Multi-array
Average (RMA) (23). Significance
of the differential expression was calculated with one-way analysis
of variance (ANOVA). Genes with P<0.05 and a fold change higher
or lower than1.5 were considered for further analysis.
Immunofluorescence
A549 and Calu-1 cell lines were culture over glass
coverslips in 6-well plates for 24 h after cell transfection. Cells
were rinsed with PBS and fixed with 4% paraformaldehyde (PFA)
(Sigma-Aldrich) for 30 min. Subsequently, cells were blocked in PBS
and 5% bovine serum albumin (Sigma-Aldrich) during 2 h and were
then incubated overnight with the NRP1 primary antibody
(Thermo-Fisher, Waltham, MA USA). Cells were washed and incubated
with a Cy3 linked secondary antibody (AP124C; Millipore) for 1 h.
Finally, cells were washed with PBS and slides were mounting in
EverBrite mounting medium with 4,6-diamidino-2-phenylindole (DAPI)
(Biotium) and stored at 4°C. Fluorescence analysis was performed on
a confocal microscope (Zeiss LSM 510; Carl Zeiss, Jena,
Germany).
Cell migration and invasion
assays
The cell migration ability of A549 and Calu-1
NRP1-deficient cells was examined using Transwell polycarbonate
permeable supports (BD Biosciences, Franklin Lakes, NJ, USA). A
total of 3.8×104 cells in 300 µl of culture medium
without FBS were placed in the upper chamber and 700 µl of medium
with 10% FBS were added to the lower chamber. Cells were culture
for 24 h, and then the inserts were removed from the wells. Cells
on the upper surface of the membrane were removed and migrating
cells located on the lower surface were washed with PBS and fixed
with 4% PFA and finally stained with 0.1% crystal violet. Images
were captured in a stereomicroscope and the number of migrating
cells were counted with ImageJ software (24).
Cell proliferation assays
Cell proliferation of A549 and Calu-1 NRP1-deficient
cells was analyzed using an MTS method. Briefly, 5×103
transiently transfected cells were seeded in triplicates onto
96-well plates and incubated at 37°C under a 5% CO2
atmosphere. Forty-eight hours after transfection, cells were
cultured for 24, 48 and 72 h. On the indicated days, cell numbers
were measured in triplicate using the CellTiter-96 Non-Radioactive
Cell Proliferation Assay kit (Promega). A549 and Calu-1 cells were
incubated with the MTS reagent during 1 and 2 h, respectively.
Then, absorbance was measured at an optical density of 590 nm using
a multi-detector reader (Beckman Coulter, Inc., Brea, CA, USA).
Proliferation curves were constructed by calculating the mean value
of optical density measurements in each condition.
Statistical analysis
FACS data were analyzed using the FlowJo 8.7
software (TreeStar, Inc., Ashland, OR, USA). One-way ANOVA and a
t-test with Bonferroni correction were applied. Analysis of the
microarray data was performed using the Bioconductor software.
Results
Isolation and expression of
pluripotency markers in NRP1+ lung cancer cells
The percentage of NRP1+ cells in A549 and
Calu-1 lung cancer cell lines was determined using flow cytometry.
As shown in Fig. 1A and C, NRP1 was
present in 76.8±13% of A549 and 86.6±4% of Calu-1 cells. We then
analyzed key stemness markers in the sorted NRP1+ and
NRP1− subpopulations. NRP1+ subpopulations of
both cell lines presented higher OCT-4 and NANOG expression when
compared with NRP1− cells, as assessed by
semi-quantitative PCR (Fig. 1B and
D).
NRP1+ cells exhibit high
clonogenic capacity and self-renewal ability
In order to assess whether NRP1+ cells
exhibit TIC abilities, we compared the clonogenic and self-renewal
capacities of enriched NRP1+ and NRP1−
subpopulations using a combination of soft agar and serial-passage
pneumosphere formation assays. As shown in Fig. 2A and C, NRP1+ cells
presented higher clonogenic capacity (represented by colony number
and size) compared with NRP1− cells in both A549 and
Calu-1 cell lines. Similarly, NRP1+ cells derived from
both A549 and Calu-1 cell lines exhibited a higher self-renewal
capacity, as assessed by serial-passage pneumosphere assays
(Fig. 2B and D).
NRP1+ cells express
additional stemness markers
To further support our hypothesis that
NRP1+ cells represent an enriched lung TIC
subpopulation, we analyzed an expression panel of stemness markers
(24,25). As shown in Fig. 3A, the NRP1+ subpopulation
had higher expression of Bmi-1, ALDH-1, KRT-5, CD133 and p63 than
the NRP1− subpopulation derived from both cell lines
(Fig. 3A).
It has been reported that epithelial-mesenchymal
transition (EMT) is associated with the generation and maintenance
of TICs (26). Concordant with
this, both A549 and Calu-1 NRP1+ cell subpopulations
presented higher Snail and vimentin expression when compared to the
NRP1− subpopulations (Fig.
3B).
NRP1 expression is associated with a
TIC phenotype in lung cancer cells
Seeking to further establish the association between
NRP1 expression and a stem cell phenotype, we performed
loss-of-function experiments by means of NRP1 gene knockdown. The
decrease in NRP1 expression was confirmed by RT-PCR (Fig. 3C). Downregulated NRP1 expression was
accompanied by a concomitant decrease in OCT-4 and Bmi-1
expression, when compared to the shLuc control cells (Fig. 3C). Likewise, NRP1 knockdown also
decreased the expression of p63 and its ΔNp63 mRNA isoform
(Fig. 3D, left panel).
Downregulation of p63 was corroborated at the protein level
(Fig. 3D, right panel). We found
that total p63 and phosphorylated p63 (P-p63) proteins were
predominantly expressed in the nuclear fraction. Furthermore,
nuclear P-p63 was detected at lower levels in the A459 cells with
silenced NRP1 expression when compared to the control cells.
Next, we tested the effect of NRP1 knockdown on the
clonogenic ability of lung cancer cells. Fig. 4A and B shows that the inhibition of
NRP1 resulted in a significantly reduced number of colonies for
both the A549 and Calu-1 cell lines, as measured by soft agar
colony formation assays and limiting dilution clonogenic
analyses.
We then analyzed whether lung cancer cells with low
NRP-1 expression exhibited a compromised self-renewal ability. As
shown Fig. 4C and D, middle and
right panel, a lower pneumosphere forming efficiency (PFE),
calculated by counting the number of spheres by serial passages,
was observed in both cell lines. In A549 cells, but not in Calu-1
cells, we were able to find smaller pneumospheres after NRP1
depletion (Fig. 4C and D).
Since TICs are known to possess higher cell
migration capability (27), we also
analyzed whether NRP1 knockdown had an impact on cell migration.
Using wound healing and Transwell cell migration assays, we
detected a reduced cell migration capacity of sh-NRP1 A549
(Fig. 5A and B) and shNRP1 Calu-1
cells (Fig. 5C and D), when
compared with shLuc control cells. No effect was observed for cell
proliferation for any of the NRP1-knockdown cell lines (data not
shown).
NRP1 downregulation modifies different
transcriptional networks
To identify the potential signaling pathways
modulated by NRP1, we performed a genome-wide analysis in
NRP1-depleted cells (using an Affymetrix microarray platform). Our
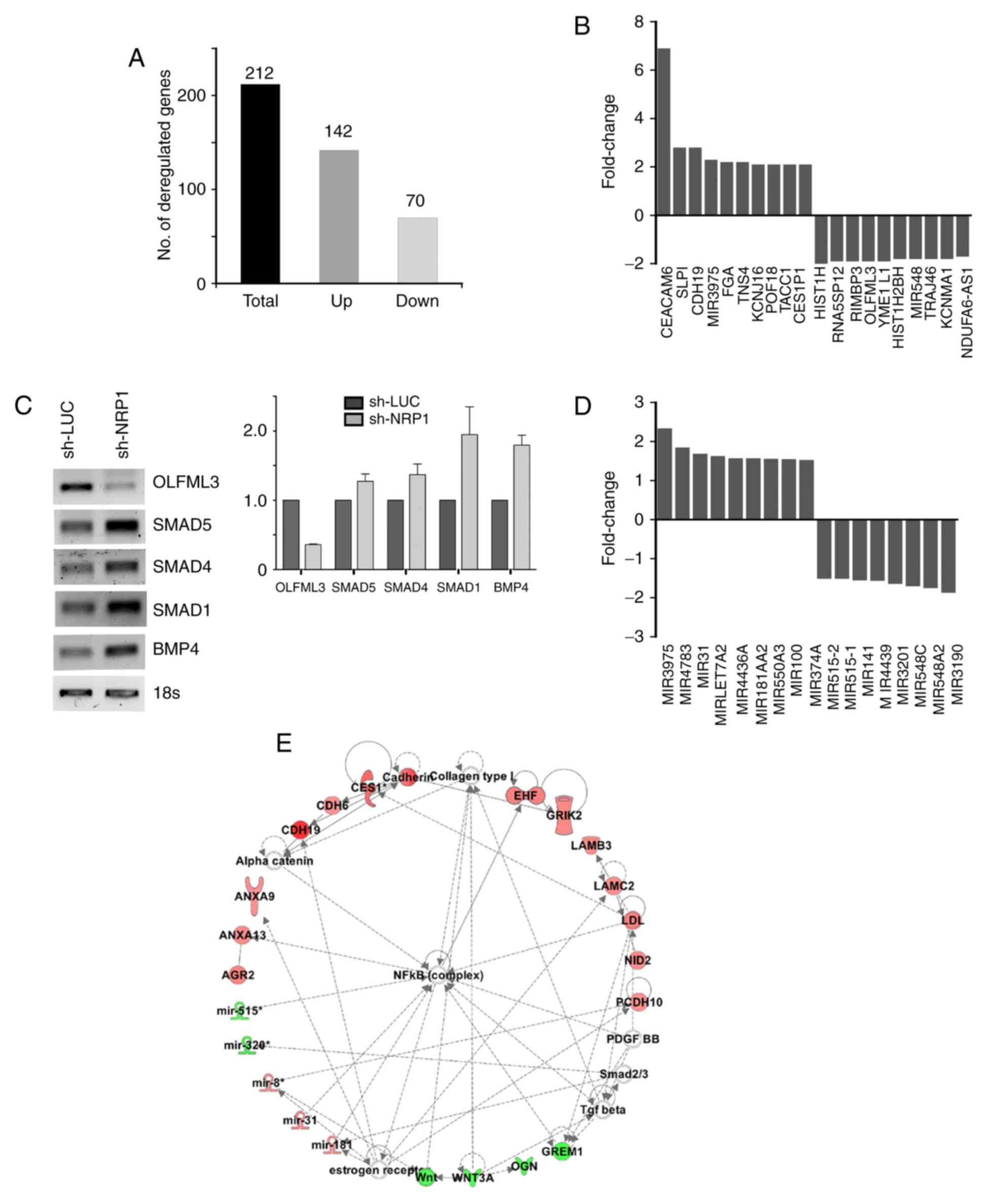
analysis showed 212 deregulated transcripts (142 upregulated and 70
downregulated) (Fig. 6A), including
17 deregulated miRNAs (Fig. 6D). A
network analysis using the Ingenuity Pathway Analysis (IPA)
platform showed that NRP1 modulates several molecular and cellular
functions, such as cell migration, cell-to-cell signaling, and drug
metabolism. Fig. 6B shows the top
upregulated/downregulated genes observed in these analyses, 13 of
which are associated with lung cancer. Notably, CDH19 and TACC1
have been associated with a TIC phenotype. Since it has been
reported that OLFML3 regulates angiogenesis via BMP4 and SMADs
(28) we analyzed the expression
level of these genes in the NRP1-depleted cells. Fig. 6C shows that NRP1 inhibition is
clearly associated with OLMF3 downregulation. Unexpectedly, this
was also accompanied by an increase in SMAD and BMP4 expression,
suggesting that the OLMF3/BMP4/SMAD signaling pathway is
deregulated in lung cancer cells (Fig.
6C). It has been reported that miR-31, miR-181 and miR-515
expression (which we found deregulated) is controlled by the NF-κB
signaling pathway, a master controller of the TIC phenotype
(29). Supporting this, the IPA
algorithm showed that the NF-κB pathway is probably deregulated in
A549-shNRP1 cells (Fig. 6E).
Discussion
NRP1 is a membrane protein expressed in embryonic
(15,30) and mesenchymal stem cells that
regulates cell migration and proliferation (16). It has been recently demonstrated
that NRP1 knockdown is sufficient to abrogate the tumor formation
potential of human fibrosarcoma HT1080 cells (31). These and other reports strongly
suggest that NRP-1 has a role in the regulation of a TIC phenotype
(15,16,20).
In the present study, we showed that
NRP1+ lung cancer cells have a TIC phenotype. This
conclusion is supported by the higher expression level of TIC
markers such as OCT-4, ALDH1, p63, NANOG and EMT-associated genes
(e.g. Snail). In accordance, the A549 and Calu-1 NRP1+
cell subpopulations presented higher clonogenic, self-renewal and
cell migration abilities when compared with the NRP1−
cell subpopulations. More important, we found that NRP1 is not only
a marker indicating TIC properties, but is also a key regulator of
this phenotype, since RNAi loss-of-function analyses resulted in
the reduced expression of molecular and phenotypic stemness
markers.
Another striking result of the present study is that
the p53 family member, p63 and its ΔNp63 isoform, were dysregulated
in NRP1+ A549 cells. These proteins are known key
regulators of the TIC phenotype and EMT processes in epithelial
tissue (32). Our results are
further supported by Rock et al, who identified a stem cell
population from human lung airway characterized by the expression
of p63 (33). Furthermore, it has
been reported that ΔNp63 is involved in the regulation of CD44
expression, a known cancer stem cell surface marker (34–36).
The microarray analyses performed on the shNRP1-A459
cells revealed significant changes in the transcriptome profile of
these cells, and among them, clear OLFML3 downregulation. Notably,
we were not able to find an association of OLFML3 with BMP4 and
SMAD expression in lung cancer cells, as is the case for other cell
types and stimuli (28). Therefore,
an alternative OLFML3 signaling pathway induced by NRP1 could be
acting in these cells. Finally, it is important to note that three
of the observed downregulated genes (YME1L1, KCNMA1 and ΔNp63)
(37–39) are associated with mitochondrial
function. Future research aimed at the association between NRP1 and
mitochondrial function in tumor cells is warranted. It would be of
great interest to conduct a pilot study to determine whether
NRP-1/OLFML3 levels determined by qRT-PCR or immunodetection in
tumor tissue obtained from biopsies, broncoalveolar fluid or serum
or alternatively, miRNAs regulated by NRP-1 (miR548, miR3201 and
miR3190) serve as stem cell biomarkers in lung cancer.
In conclusion, the present study demonstrated that
NRP1+ cells possess TIC properties, including the
preferential expression of stemness markers, higher clonogenic and
self-renewal potential, and increased cell migration capacity.
These findings support an important role for NRP1 in the biology of
lung cancer TICs.
Acknowledgements
We thank Raul Mojica and his staff for the
microarray processing and Nelly Patiño for the flow cytometric
assays (High Technology Units, National Institute of Genomic
Medicine, Mexico). Luis Enrique Jiménez Hernández is a doctoral
student from Programa de Doctorado en Ciencias Biomédicas,
Universidad Nacional Autónoma de México (UNAM) and received
fellowship 336320 from CONACyT.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun JH, Luo Q, Liu LL and Song GB: Liver
cancer stem cell markers: Progression and therapeutic implications.
World J Gastroenterol. 22:3547–3557. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao J, Fan W, Ma B and Wu Y: Breast
cancer stem cells expressing different stem cell markers exhibit
distinct biological characteristics. Mol Med Rep. 14:4991–4998.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Eramo A, Lotti F, Sette G, Pilozzi E,
Biffoni M, Di Virgilio A, Conticello C, Ruco L, Peschle C and De
Maria R: Identification and expansion of the tumorigenic lung
cancer stem cell population. Cell Death Differ. 15:504–514. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen YC, Hsu HS, Chen YW, Tsai TH, How CK,
Wang CY, Hung SC, Chang YL, Tsai ML, Lee YY, et al: Oct-4
expression maintained cancer stem-like properties in lung
cancer-derived CD133-positive cells. PLoS One. 3:e26372008.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tirino V, Camerlingo R, Franco R, Malanga
D, La Rocca A, Viglietto G, Rocco G and Pirozzi G: The role of
CD133 in the identification and characterisation of
tumour-initiating cells in non-small-cell lung cancer. Eur J
Cardiothorac Surg. 36:446–453. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Meng X, Li M, Wang X, Wang Y and Ma D:
Both CD133+ and CD133- subpopulations of A549 and H446 cells
contain cancer-initiating cells. Cancer Sci. 100:1040–1046. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qiu X, Wang Z, Li Y, Miao Y, Ren Y and
Luan Y: Characterization of sphere-forming cells with stem-like
properties from the small cell lung cancer cell line H446. Cancer
Lett. 323:161–170. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hongo K, Tanaka J, Tsuno NH, Kawai K,
Nishikawa T, Shuno Y, Sasaki K, Kaneko M, Hiyoshi M, Sunami E, et
al: CD133(−) cells, derived from a single human colon cancer cell
line, are more resistant to 5-fluorouracil (FU) than CD133(+)
cells, dependent on the β1-integrin signaling. J Surg Res.
175:278–288. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Roudi R, Madjd Z, Ebrahimi M, Samani FS
and Samadikuchaksaraei A: CD44 and CD24 cannot act as cancer stem
cell markers in human lung adenocarcinoma cell line A549. Cell Mol
Biol Lett. 19:23–36. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Xu H, Mu J, Xiao J, Wu X, Li M, Liu T and
Liu X: CD24 negative lung cancer cells, possessing partial cancer
stem cell properties, cannot be considered as cancer stem cells. Am
J Cancer Res. 6:51–60. 2015.PubMed/NCBI
|
|
12
|
Huo W, Du M, Pan X, Zhu X and Li Z:
Prognostic value of ALDH1 expression in lung cancer: A
meta-analysis. Int J Clin Exp Med. 8:2045–2051. 2015.PubMed/NCBI
|
|
13
|
Sulpice E, Plouët J, Bergé M, Allanic D,
Tobelem G and Merkulova-Rainon T: Neuropilin-1 and neuropilin-2 act
as coreceptors, potentiating proangiogenic activity. Blood.
111:2036–2045. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ango F: NRP1 and synapse formation.
Oncotarget. 7:81975–81976. 2016.PubMed/NCBI
|
|
15
|
Cimato T, Beers J, Ding S, Ma M, McCoy JP,
Boehm M and Nabel EG: Neuropilin-1 identifies endothelial
precursors in human and murine embryonic stem cells before CD34
expression. Circulation. 119:2170–2178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ball SG, Bayley C, Shuttleworth CA and
Kielty CM: Neuropilin-1 regulates platelet-derived growth factor
receptor signalling in mesenchymal stem cells. Biochem J.
427:29–40. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hong TM, Chen YL, Wu YY, Yuan A, Chao YC,
Chung YC, Wu MH, Yang SC, Pan SH, Shih JY, et al: Targeting
neuropilin 1 as an antitumor strategy in lung cancer. Clin Cancer
Res. 13:4759–4768. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng Y, Liu YM, Li LC, Wang LL and Wu XL:
MicroRNA-338 inhibits growth, invasion and metastasis of gastric
cancer by targeting NRP1 expression. PLoS One. 9:e944222014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cao Y, Wang L, Nandy D, Zhang Y, Basu A,
Radisky D and Mukhopadhyay D: Neuropilin-1 upholds
dedifferentiation and propagation phenotypes of renal cell
carcinoma cells by activating Akt and sonic hedgehog axes. Cancer
Res. 68:8667–8672. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hamerlik P, Lathia JD, Rasmussen R, Wu Q,
Bartkova J, Lee M, Moudry P, Bartek J Jr, Fischer W, Lukas J, et
al: Autocrine VEGF-VEGFR2-Neuropilin-1 signaling promotes glioma
stem-like cell viability and tumor growth. J Exp Med. 209:507–520.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Barr MP, Gray SG, Gately K, Hams E, Fallon
PG, Davies AM, Richard DJ, Pidgeon GP and O'Byrne KJ: Vascular
endothelial growth factor is an autocrine growth factor, signaling
through neuropilin-1 in non-small cell lung cancer. Mol Cancer.
14:452015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schneider CA, Rasband WS and Eliceiri KW:
NIH Image to ImageJ: 25 years of image analysis. Nat Methods.
9:671–675. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Irizarry RA, Bolstad BM, Collin F, Cope
LM, Hobbs B and Speed TP: Summaries of Affymetrix GeneChip probe
level data. Nucleic Acids Res. 31:e152003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Skirecki T, Hoser G, Kawiak J, Dziedzic D
and Domagała-Kulawik J: Flow cytometric analysis of CD133- and
EpCAM-positive cells in the peripheral blood of patients with lung
cancer. Arch Immunol Ther Exp. 62:67–75. 2014. View Article : Google Scholar
|
|
25
|
Kajstura J, Rota M, Hall SR, Hosoda T,
D'Amario D, Sanada F, Zheng H, Ogórek B, Rondon-Clavo C,
Ferreira-Martins J, et al: Evidence for human lung stem cells. N
Engl J Med. 364:1795–1806. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamb R, Lisanti MP, Clarke RB and Landberg
G: Co-ordination of cell cycle, migration and stem cell-like
activity in breast cancer. Oncotarget. 5:7833–7842. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Miljkovic-Licina M, Hammel P,
Garrido-Urbani S, Lee BP, Meguenani M, Chaabane C, Bochaton-Piallat
ML and Imhof BA: Targeting olfactomedin-like 3 inhibits tumor
growth by impairing angiogenesis and pericyte coverage. Mol Cancer
Ther. 11:2588–2599. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kastrati I, Canestrari E and Frasor J:
PHLDA1 expression is controlled by an estrogen
receptor-NFκB-miR-181 regulatory loop and is essential for
formation of ER+ mammospheres. Oncogene. 34:2309–2316. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Brusselmans K, Bono F, Collen D, Herbert
JM, Carmeliet P and Dewerchin M: A novel role for vascular
endothelial growth factor as an autocrine survival factor for
embryonic stem cells during hypoxia. J Biol Chem. 280:3493–3499.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Misra RM, Bajaj MS and Kale VP:
Vasculogenic mimicry of HT1080 tumour cells in vivo: Critical role
of HIF-1α-neuropilin-1 axis. PLoS One. 7:e501532012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Oh JE, Kim RH, Shin KH, Park NH and Kang
MK: DeltaNp63α protein triggers epithelial-mesenchymal transition
and confers stem cell properties in normal human keratinocytes. J
Biol Chem. 286:38757–38767. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rock JR, Onaitis MW, Rawlins EL, Lu Y,
Clark CP, Xue Y, Randell SH and Hogan BL: Basal cells as stem cells
of the mouse trachea and human airway epithelium. Proc Natl Acad
Sci USA. 106:pp. 12771–12775. 2009; View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen Y, Zhao J, Luo Y, Wang Y, Wei N and
Jiang Y: Isolation and identification of cancer stem-like cells
from side population of human prostate cancer cells. J Huazhong
Univ Sci Technolog Med Sci. 32:697–703. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Leung EL, Fiscus RR, Tung JW, Tin VP,
Cheng LC, Sihoe AD, Fink LM, Ma Y and Wong MP: Non-small cell lung
cancer cells expressing CD44 are enriched for stem cell-like
properties. PLoS One. 5:e140622010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Boldrup L, Coates PJ, Gu X and Nylander K:
DeltaNp63 isoforms regulate CD44 and keratins 4, 6, 14 and 19 in
squamous cell carcinoma of head and neck. J Pathol. 213:384–391.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ruan Y, Li H, Zhang K, Jian F, Tang J and
Song Z: Loss of Yme1L perturbates mitochondrial dynamics. Cell
Death Dis. 4:e8962013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Singh H, Lu R, Bopassa JC, Meredith AL,
Stefani E and Toro L: MitoBKCa is encoded by the Kcnma1 gene, and a
splicing sequence defines its mitochondrial location. Proc Natl
Acad Sci USA. 110:pp. 10836–10841. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Matin RN, Chikh A, Chong SL, Mesher D,
Graf M, Sanza' P, Senatore V, Scatolini M, Moretti F, Leigh IM, et
al: p63 is an alternative p53 repressor in melanoma that confers
chemoresistance and a poor prognosis. J Exp Med. 210:581–603. 2013.
View Article : Google Scholar : PubMed/NCBI
|