Introduction
Oral squamous cell carcinoma (OSCC) is the sixth
most commonly occurring cancer in the world and a lethal disease,
with a 5-year survival rate of ~50% (1). Despite continuing improvements in
therapy, the poor prognosis of OSCC patients remains unsolved
around the world. Carcinogenesis is a multistep process including
initiation, promotion and progression, and oral cancer usually
develops from premalignant lesions of oral mucosa into OSCC
(2). It has been revealed that risk
factors for human oral carcinogenesis include alcohol consumption,
tobacco and human papillomavirus infection (3). However, the underlying molecular
mechanism of dynamic oral carcinogenesis has not been completely
elucidated.
Macroautophagy (autophagy) is an evolutionarily
conserved and self-consumption process involved in preserving
organelle function, removing cellular waste products and providing
metabolic substrates (4,5). It has been reported that autophagy is
an evolving and multifaceted process during cancer initiation and
progression (6). In normal cells,
autophagy prevents excess reactive oxygen species, DNA damage and
genome instability, known causes of cancer initiation and
progression (7). In these contexts,
autophagy likely serves as a tumor suppressor in the tumor
initiation stages (8). However, in
the late stages of tumorigenesis, autophagic responses maintain
tumor metabolism and promote tumor cell survival. In this sense,
autophagy exhibits a pro-oncogenic role (6,8).
In addition to a cellular mechanistic role,
autophagy has been demonstrated to orchestrate the tumor
microenvironment by regulating the inflammation response (9–11).
Autophagy enhances the processing and presentation of tumor
antigens and thereby activates antitumor immunity (9). Martinez-Outschoorn et al
(10) demonstrated that in a
co-culture system, cancer cells produced numerous inflammatory
mediators in a tumor microenvironment and further induced autophagy
in adjacent fibroblasts via oxidative stress and nuclear factor
(NF) κB-activation (10). This
indicated that inflammation mediators in the tumor microenvironment
contribute to tumor progression by activating the autophagy
response (10,11).
Hence, the present study established an oral cancer
mouse model with 4-nitroquinoline-1-oxide (4NQO) and assessed the
expression of autophagy markers dihydrosphingosine 1-phosphate
phosphatase LCB3 (LC3B), p62/SQSTM1 (p62) and Beclin 1 at various
stages of tongue lesions of these mice. Furthermore, the number of
Gr-1+CD11b+ myeloid derived suppressor cells
(MDSCs) and CD4+ Foxp3+ regulatory T cells
(Tregs) during oral carcinogenesis and the association with
autophagy status was also examined. The data will help to address
the dynamic change of autophagy activity during multistage oral
carcinogenesis and its association with inflammation.
Materials and methods
Ethics statement
All procedures involving mice were approved by the
Subcommittee on Research and Animal Care of Sichuan University
(WCHSIRB-D-2016-149, Chengdu, China). The written informed consents
were obtained from participants through their signatures. The use
of human tissue samples and clinical data was approved by the
Institutional Ethics Committee of the West China Medical Center,
Sichuan University (WCHSIRB-D-2012-097).
Experimental model
A total of 38 female wild-type C57BL/6 mice, eight
weeks old, weighing 20–25 g were purchased from the Chengdu Dashuo
Biological Technology Co., Ltd. (Sichuan, China). The mice were
housed in State Key Laboratory of Oral Diseases West China Hospital
of Stomatology (Sichuan University), five per cage, maintained at
22±1°C and within the range of 30–70% relative humidity with a 12-h
light/dark cycle. The animals were permitted free access to a
normal chow (LabDiet with constant nutrition, Dashuo Co. Ltd.,
Chengdu, China) and drinking water provided in water bottles. A
stock solution of 4-nitroquinoline-1-oxide (4NQO; Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) was prepared at 0.5 mg/ml and added
to the drinking water to obtain the working concentration. To avoid
the decomposition of 4NQO, light avoidance precaution was taken.
After a week of acclimation, mice were randomly assigned into three
separate experimental groups: Control group (n=10), low-4NQO group
(n=18), high-4NQO group (n=20). In the experimental groups, the
mice were given 100 µg/ml 4NQO (low-4NQO group) or 200 µg/ml 4NQO
(high-4NQO group) in drinking water. Then, mice in low-4NQO and
high-4NQO group were divided into 6 weeks, 10 weeks, 14 weeks or 18
weeks, as presented in Table I.
Following corresponding time of 4NQO exposure, the animals in
4NQO-treated groups were switched to distilled water for 8 weeks.
The animals in the control group received only distilled water.
 | Table I.Incidence of tongue lesions in
various 4-NQO-treated mice groups. |
Table I.
Incidence of tongue lesions in
various 4-NQO-treated mice groups.
|
|
|
|
| Dysplasia |
|
|---|
|
|
|
|
|
|
|
|---|
| Group | No. of mice | Lesion
numbera | Lesion size
(mm)b | Mild-moderate
dysplasia (%) | Severe dysplasia
(%) | Total (%) | Carcinoma (%) |
|---|
| 14 weeks | 11 | 0.7 | 1.3 | 2/11
(18) | 4/11
(36.4) | 6/11
(54.4) | 5/11
(45.5) |
| Low-dose 4NQO | 5 | 0.8 | 1.0 | 1/5
(20) | 1/5
(20) | 2/5
(40) | 3/5
(60) |
| High-dose 4NQO | 6 | 0.7 | 1.6 | 1/6
(17.7) | 3/6
(50) | 4/6
(67.7) | 2/6
(33.3) |
| 18 weeks | 8 | 1.6 | 1.3 | 4/8
(50) | 2/8
(25) | 6/8
(75) | 2/8
(25) |
| Low-dose 4NQO | 4 | 1.7 | 1.5 | 1/4
(25) | 1/4
(25) | 2/4
(50) | 2/4
(50) |
| high-dose 4NQO | 4 | 1.5 | 1.1 | 3/4
(75) | 1/4
(25) | 4/4
(100) | 0/4
(0) |
| 22 weeks | 9 | 2.0 | 1.6 | 1/9
(11.1) | 2/9
(22.2) | 3/9
(33.3) | 6/9
(66.6) |
| Low-dose 4NQO | 4 | 1.8 | 1.4 | 1/4
(25) | 1/4
(25) | 2/4
(50) | 2/4
(50) |
| Hgh-dose 4NQO | 5 | 2.2 | 1.7 | 0/5
(0) | 1/5
(20) | 1/5
(20) | 4/5
(80) |
| 26 weeks | 10 | 1.9 | 2.1 | 0/10
(0) | 2/10
(20) | 2/10
(20) | 8/10
(80) |
| Low-dose 4NQO | 5 | 1.6 | 1.8 | 0/5
(0) | 1/5
(20) | 1/5
(20) | 4/5
(80) |
| High-dose 4NQO | 5 | 2.2 | 2.4 | 0/5
(0) | 1/5
(20) | 1/5
(20) | 4/5
(80) |
| Total | 38 | 1.5 | 1.9 | 7/38
(18.4) | 10/38 (26.3) | 17/38 (44.7) | 21/38 (55.3) |
At the end of the experimental period, mice were
anesthetized using isoflurane, and blood was collected by
vacutainer vials containing heparin. Tongue and spleen tissues were
collected in each group and tongue tissues were then longitudinally
bisected. One part of each tongue tissue was fixed in 10% buffered
formalin at room temperature for 24 h and then embedded with
paraffin. The other part was immediately snap-frozen and stored at
−80°C for western blotting or frozen with OCT for
immunofluorescence staining.
Histopathological analysis
Sections (4-µm) of tongue tissue from different
groups were processed for hematoxylin and eosin (H&E) staining.
After deparaffinization and rehydration, the sections were stained
with hematoxylin (OriGene Technologies, Inc., Rockville, MD, USA)
at room temperature for 5 min. Then the sections were
differentiated in 1% hydrochloric acid alcohol for 2 sec followed
by incubation in ammonia water for 2 min, and stained with eosin
(OriGene Technologies, Inc.) at room temperature for 1 min.
Histopathological diagnosis was performed by an experienced oral
pathologist in a blind manner, and the samples were classified into
the following four types: Normal epithelium, mild-moderate
dysplasia, severe dysplasia, and carcinoma, according to the
criteria described by the World Health Organization (12). All histopathological examination was
conducted using a light microscope (Olympus BX46; Olympus
Corporation, Tokyo, Japan).
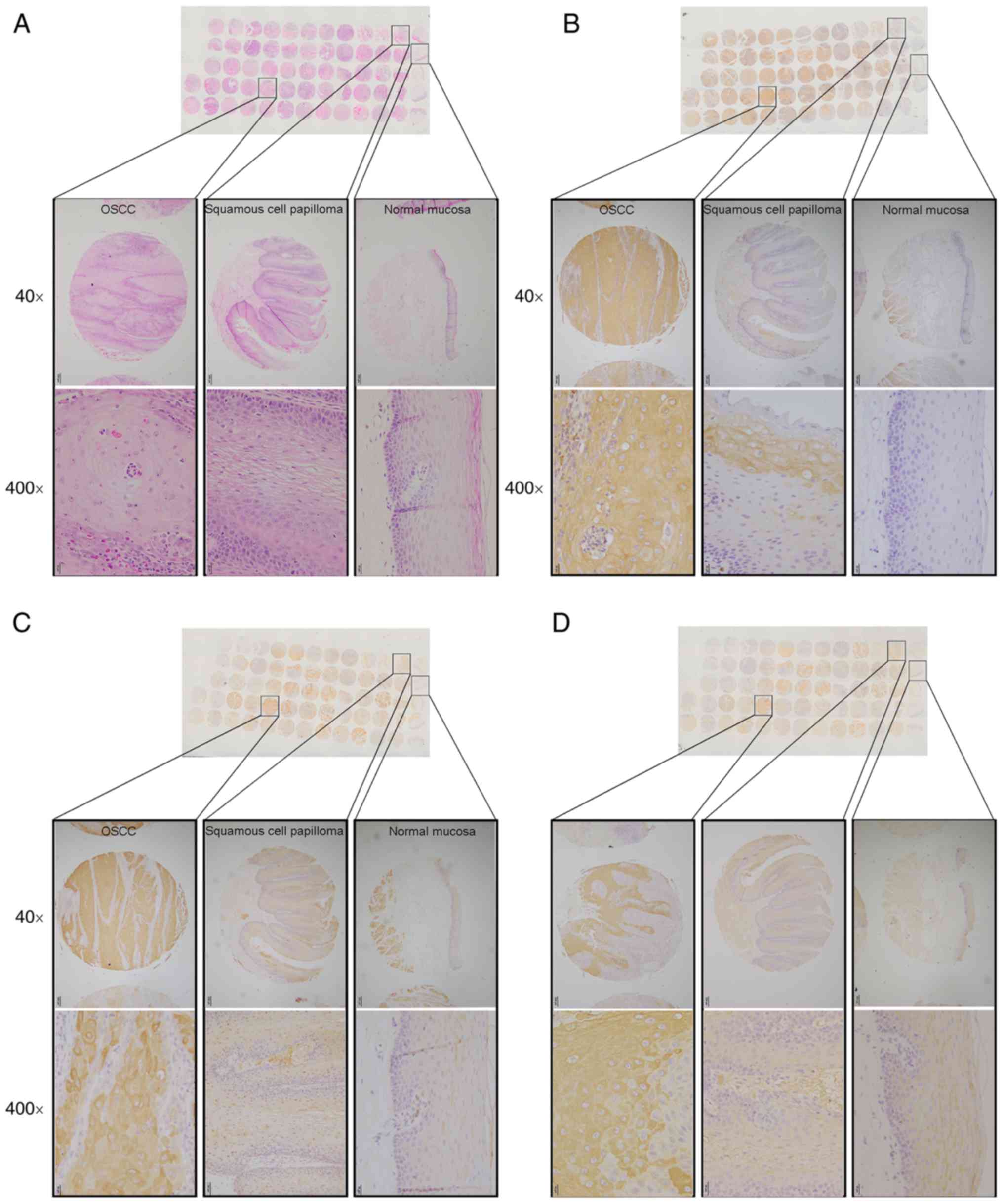
Tissue microarray (TMA)
construction
The tissue microarray used for the present study
included 52 oral squamous cell carcinoma (OSCC), 5 squamous cell
papilloma and 5 normal mucosa specimens from the Department of Oral
Pathology, West China Hospital of Stomatology, Sichuan University
between 2013 and 2014. The OSCC samples used in the study were
typical keratinizing-type, not verrucous or other variant types.
There were 32 male and 20 female patients, and their ages ranged
from 27 to 91 years, with a median age of 62 years. The diagnosis
of these specimens was confirmed by pathologic examination. The
tissue microarray slide was constructed as previously described
(13). Briefly, based on the
results of HE-stained tissue slides, formalin-fixed,
paraffin-embedded tissue blocks were punched to obtain tissue
cylinders with a 3 mm diameter containing representative tissue
areas. Then the punched tissue cores were placed on a recipient
block and arranged with an array pattern. Four-µm-thick sections of
TMA were produced for H&E and immunohistochemistry
staining.
Immunohistochemical analysis
Paraffin-embedded sections at 4-µm thickness were
deparaffinized and rehydrated in graded ethanol series and
distilled water. For antigen retrieval, slides were immersed in
0.01 M sodium citrate buffer, pH 6.0 in a water bath at 95°C for 30
min. Endogenous peroxidases were inhibited by treatment with 3%
hydrogen peroxide for 20 min. After blocking with goat serum
albumin (OriGene Technologies, Inc.) at room temperature for 20
min, sections were incubated overnight at 4°C with
anti-proliferating cell nuclear antigen (PCNA) monoclonal antibody
(1:200; catalog no. ZM-0213, OriGene Technologies, Inc.), anti-LC3B
antibody (1:200; catalog no. 14600-1-AP; Proteintech Group,
Chicago, IL, USA), anti-p62 antibody (1:100; catalog no. WL02385;
Wanleibio Co., Ltd., Shanghai, China) or anti-Beclin 1 mouse
monoclonal antibody (1:100; catalog no. WL02508; Wanleibio Co.,
Ltd.). After rinsing with PBS, the slides were then incubated with
biotinylated anti-mouse/rabbit IgG (catalog no. SP-9000; OriGene
Technologies, Inc.) at room temperature for 15 min, followed by
incubation with peroxidase-streptavidin (catalog no. SP-9000;
OriGene Technologies, Inc.) at room temperature for 15 min.
Finally, the slices were visualized with diaminobenzidine and
nuclei were counterstained with hematoxylin at room temperature for
2 min. As negative control specimens, PBS instead of the primary
antibody was incubated.
One hundred epithelial cells or tumor cells from
five representative fields of each section were randomly selected
and evaluated by two independent researchers (JW, SW) under a light
microscope (Olympus BX46; Olympus Corporation). The inconsistent
results were re-evaluated by discussion until consensus was
reached. The immunoreactivity of each protein was evaluated by
combined assessment of the intensity and the percentage of
positivity. The intensity was evaluated as: 0 (no staining), 1
(mild staining), 2 (moderate staining), 3 (strong staining). The
proportion of stained cells was graded as: 0 (negative), 1 (0–10%
positive), 2 (10–50% positive) or 3 (>50% positive). The two
variables were multiplied to provide a total score and the total
score was categorized as negative (0–4) or positive (>4) for
evaluation of the expression level in human patient samples.
Flow cytometry analysis
To prepare a single-cell suspension the spleen
tissues were disaggregated mechanically by a Medimachine (Dako;
Agilent Technologies GmbH, Waldbronn, Germany). Heparinized
peripheral blood was diluted with equal PBS. Peripheral blood
mononuclear cells (PBMCs) were isolated using Ficoll-Hypaque
(eBioscience; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
density gradient centrifugation. For MDSCs analysis, spleen cell
suspensions or PBMCs were incubated with 5 µl fluorescent
monoclonal antibodies against Gr-1-PE, CD11b-FITC or isotype
control (BD Biosciences, Franklin Lakes, NJ, USA) for 30 min in the
dark at 4°C. For Tregs analysis, cells were incubated with CD4-FITC
or isotype control (BD Biosciences) followed by intracellular
staining for Foxp3-PE (BD Biosciences). Finally, cells were washed
and resuspended in PBS, and cell sorting occurred with flow
cytometry (Becton Coulter, Inc., Brea, CA, USA) and analyzed using
Winmdi2.9 software (Scripps Institute, San Diego, CA, USA).
Immunofluorescence staining
Frozen tissue specimens in OCT were cut into 8-µm
cryostat sections, fixed with acetone for 10 min and then brought
to room temperature. The slides were incubated overnight at 4°C
with anti-LC3B polyclonal antibody (1:200; catalog no. 14600-1-AP,
Proteintech Group). The Rhodamine (TRITC) tagged anti-rabbit IgG
(1:100; catalog no. ZF-0316; OriGene Technologies, Inc.) were
diluted in 1% BSA in PBS and incubated with the sample for 1 h at
37°C in the dark, followed by counterstaining with DAPI to
visualize the nuclei. The slides were observed under laser scanning
confocal microscope (Olympus FluoView FV1000; Olympus Corporation)
at ×400 magnification, and excitation wavelength was 559 nm for
detection of Rhodamine.
Western blot analysis
Total protein lysates were extracted from the mice
tongue from each experimental group by using a Total protein lysate
kit (catalog no. KGP250; Nanjing KeyGen Biotech Co., Ltd., Nanjing,
China) according to the manufacturer's protocol. Protein
concentration was quantified using the bicinchoninic acid assay
(catalog no. KGP902; Nanjing KeyGen Biotech Co., Ltd., and 30 µg of
total protein was subjected to 12% SDS-PAGE. The fractionated
samples were transferred to polyvinylidene difluoride membranes
(EMD Millipore, Billerica, MA, USA), immersed in 5% blotto at room
temperature for 60 min to block non-specific binding and then
incubated with anti-GAPDH antibody (1:5,000; catalog no.
10494-1-AP; Proteintech Group), anti-LC3B antibody (1:1,000;
catalog no. 14600-1-AP; Proteintech Group), anti-P62 antibody
(1:500; catalog no. WL02385; Wanleibio Co., Ltd.) and anti-Beclin 1
antibody (1:500; catalog no. WL02508; Wanleibio Co., Ltd.)
overnight at 4°C. The membranes were then incubated with the
HRP-conjugated goat anti-rabbit IgG (1:5,000; catalog no.
SA00001-2; Proteintech Group) for 1 h at room temperature. The
specific bands were detected using the Immobilon Western
Chemiluminescent HRP Substrate detection kit (EMD Millipore). The
intensities of the protein bands were quantified with Quantity One
4.5.0 software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Data were analyzed using SPSS software, version 22.0
(IBM SPSS, Armonk, NY, USA) and GraphPad Prism (version 5.03;
GraphPad Software, Inc., La Jolla, CA, USA). The normally
distributed quantitative data were expressed as the mean ± standard
deviations and were compared using one-way analysis of variance.
The SNK test was performed for multiple testing among all groups.
Comparisons of the positive rates of autophagy marker expression in
human tissues were performed using Fisher's exact test and
Bonferroni correction was performed for multiple testing (data not
shown). P<0.05 was considered to indicate a statistically
significant difference. Pearson correlation analysis was performed
to evaluate the correlation between autophagy marker expression and
the number of MDSCs and Tregs. Immunohistochemical analysis, flow
cytometry analysis, immunofluorescence staining and western blot
analysis were repeated at least three times.
Results
Characteristics of mouse oral tumor
model in macroscopic examinations
In the present study, mice were exposed to low or
high doses of 4NQO for different time-periods and returned to
normal drinking water for 8 weeks. Visible and gross lesions of the
oral cavity were observed both in low-dose and high-dose 4NQO
groups after a period of 6 weeks 4NQO treatment and 8 weeks
observation. At 14 weeks, the average lesion number and lesion size
of low-dose 4NQO-treated mice were 0.8 and 1 mm respectively, and
0.7 and 1.6 mm in high-dose 4NQO-treated mice (Table I). At 26 weeks, the average lesion
number and lesion size of low-dose 4NQO-treated mice were 1.6 and
1.8 mm respectively, and 2.2 and 2.4 mm in high-dose 4NQO-treated
mice. This indicated that lesion sizes and number in each mouse
increased with the increase of 4NQO exposure duration. However,
none of the control mice exhibited visible changes during the
period of observation (n=10).
Histopathological analysis and
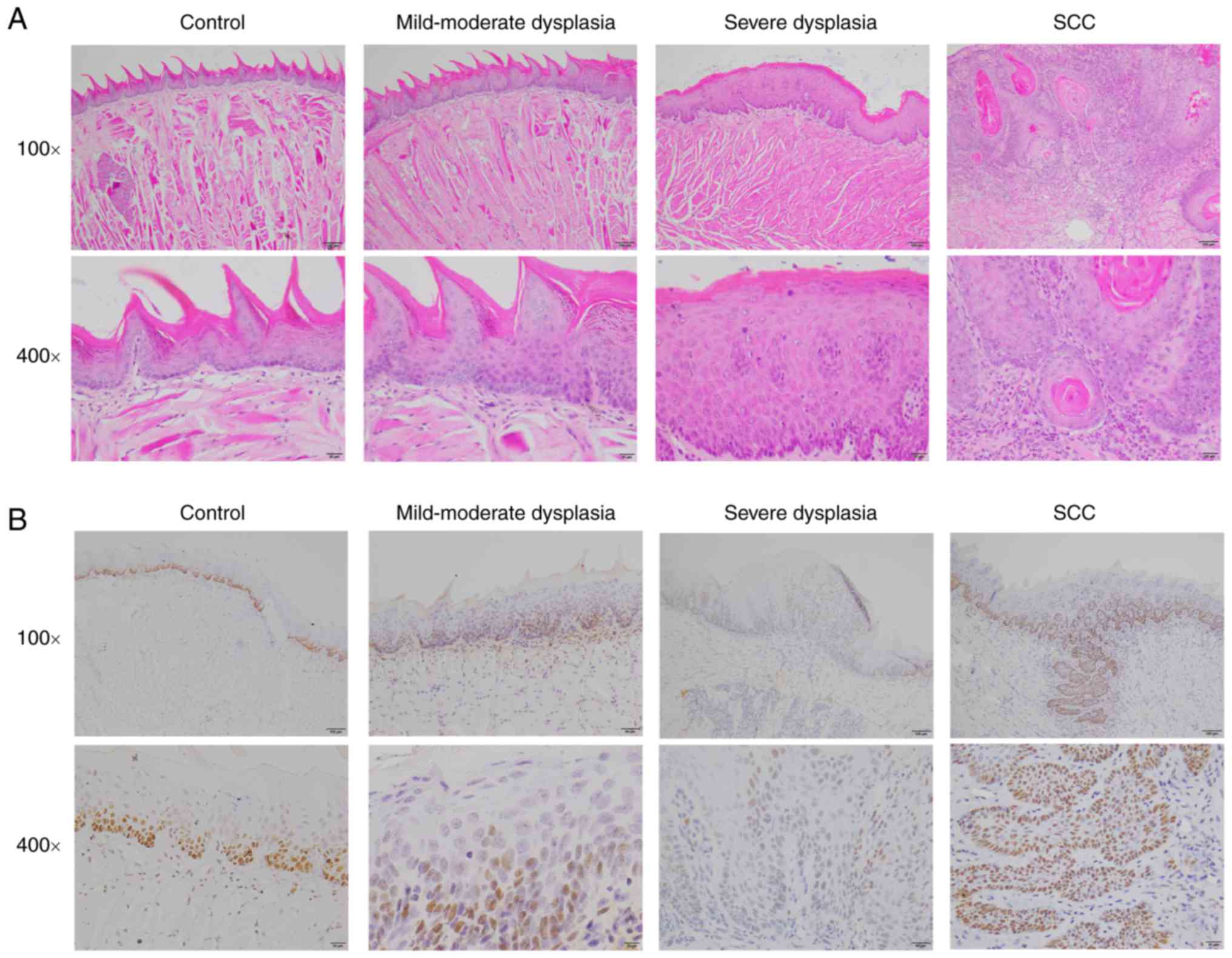
immunohistochemical staining of PCNA
Histological examination of 4NQO-induced lesion
tissues was conducted by a trained pathologist blinded to the
sample identities. As expected, 4NQO-treated mice induced oral
carcinogenesis with a well-defined progression from normal
epithelia, through dysplasia of different severity to early
invasive carcinoma (Fig. 1A).
Histopathologically, the tumor lesions were usually squamous cell
carcinoma of well-differentiated or moderately-differentiated type
with typical keratin pearl formation. A few of the tumor lesions
exhibited a papillary surface configuration and spread into the
submucosa and underlying muscle layer. At 14 weeks, 40% low-dose
4NQO-treated mice exhibited dysplasia, and 60% exhibited squamous
cell carcinoma (SCC), whereas 67.7% high-dose 4NQO-treated mice
exhibited dysplasia lesion, and 3.3% showed SCC. At 18 weeks, 50%
low-dose 4NQO-treated mice showed squamous cell carcinoma (SCC),
whereas no high-dose 4NQO-treated mice showed SCC. At 26 weeks, 80%
mice of both the low-dose 4NQO group and high-dose 4NQO group
showed SCC. The histopathological analysis was summarized in
Table I.
PCNA is a marker of cell proliferation and the PCNA
expression in mouse tongues was detected by immunohistochemistry.
In the control group, PCNA expression was weakly observed only in
the basal layer and suprabasal layers of tongue epithelium.
However, PCNA expression was markedly increased in the dysplasia
and SCC groups (Fig. 1B).
Expression of LC3B, p62 and Beclin 1
in different periods of 4NQO-induced oral carcinogenesis
The present study examined LC3B expression, a common
marker of autophagy, in tongue tissue of normal, mild-moderate
dysplasia, severe dysplasia, and SCC groups. Immunohistochemical
analysis demonstrated that LC3B was mainly expressed in the
cytoplasm of the cells and infrequently in the perinuclear membrane
and nucleus (Fig. 2A). By
semiquantitative assessment of IHC staining, LC3B was highly
expressed in dysplasia epithelia and SCC compared to control group,
and LC3B levels in SCC were increased compared with dysplasia
epithelia (P<0.05; Fig. 2B).
Additionally, immunofluorescence assessment revealed that LC3B
puncta (representing autophagosomes) formation also increased
during oral carcinogenesis (Fig.
2C). These results indicated that increased LC3B levels were
associated with tongue carcinogenesis progression. Then, the
present study further tested the isoform conversion of LC3, another
indicator of autophagosome formation during 4NQO-induced tongue
carcinogenesis. The result of the western blot analysis
demonstrated that the LC3B-II expression level was upregulated in
SCC compared with normal and dysplasia tissues (Fig. 2D). Taken together, the increased
LC3B expression suggested that autophagosome formation increased
during oral carcinogenesis.
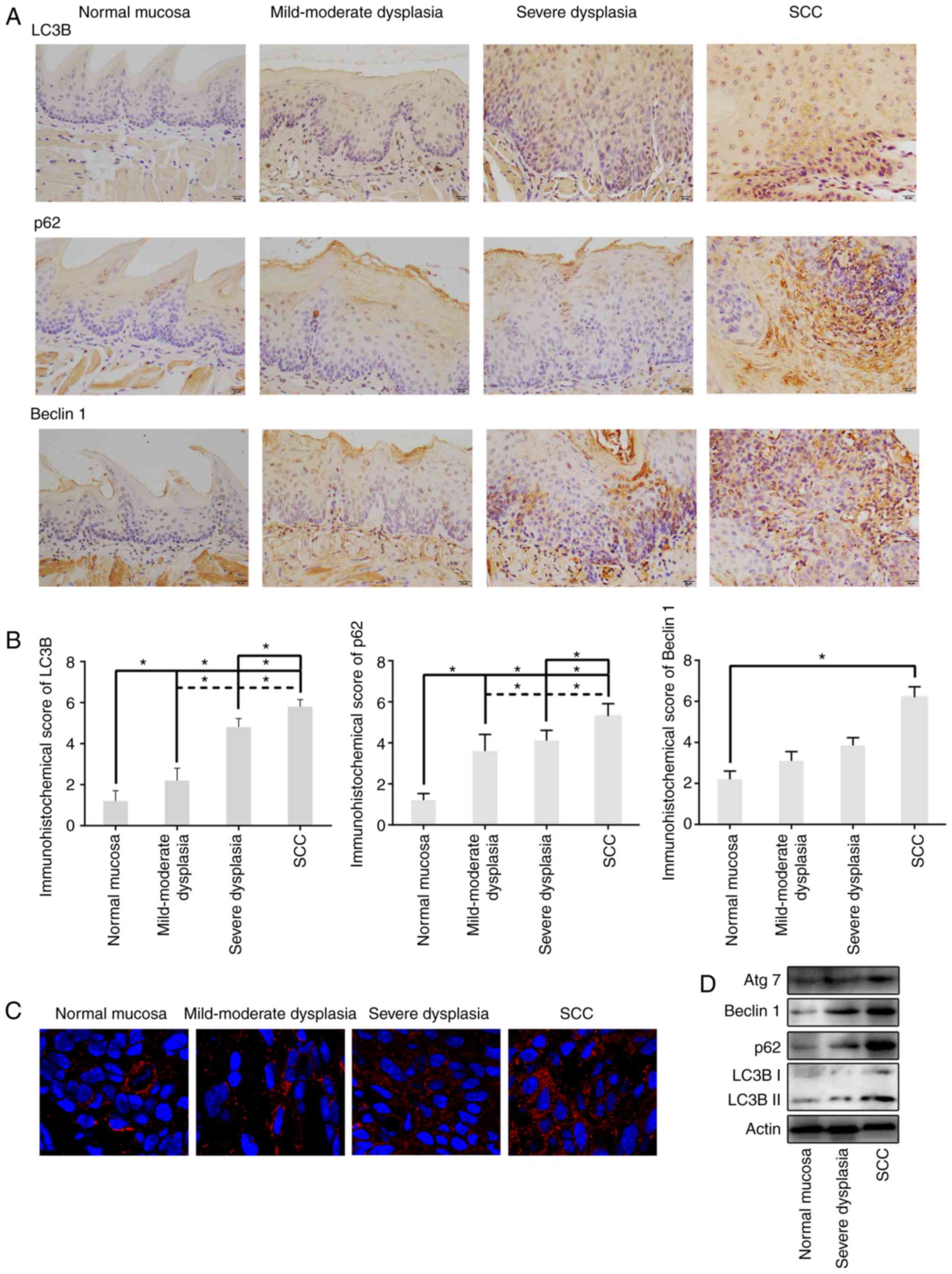 | Figure 2.Expression levels of LC3B, p62,
Beclin 1 and Atg7 during 4NQO-induced oral carcinogenesis. (A)
Representative immunohistochemical analysis of LC3B, p62 and Beclin
1 expression in sectioned tissue samples from the 4NQO-treated
mice. All images were taken at ×400 magnification with 20 µm scale
bars. (B) Semiquantitative staining analysis showed that the
expression of LC3B, p62 and Beclin 1 was gradually upregulated from
normal mucosa, mild-moderate dysplasia, severe dysplasia, and SCC.
(C) Immunofluorescent staining revealed LC3 puncta in tongue tissue
of 4NQO-treated mice accumulated during oral carcinogenesis. (D)
Representative western blot images displaying 30 µg of total
protein extracted from the tongue tissue of each experimental group
showed increased levels of LC3, p62, Beclin 1 and Atg7 in dysplasia
and SCC groups, compared with normal mucosa group. Data are
expressed as the mean ± standard deviation. *P<0.05. LC3B,
dihydrosphingosine 1-phosphate phosphatase LCB3; p62, p62/SQSTM1;
Atg7, autophagy related 7; SCC, squamous cell carcinoma; 4NQO,
4-nitroquinoline-1-oxide. |
p62, a ubiquitin-binding scaffold protein, serves as
an autophagy substrate and cargo adapter that can be selectively
degraded by autophagy. To identify the level of autophagy flux, p62
expression was assessed by immunohistochemistry and western blot
analysis at various stages of 4NQO-induced tongue carcinogenesis.
p62 was primarily expressed in the cytoplasm and infrequently in
the nucleus, consistent with the finding that p62 can shuttle
between nucleus and cytoplasm (14). Normal tongue epithelium exhibited
weak p62 expression, which was predominantly detected in the
stratum spinosum of stratified squamous epithelia. In dysplastic
epithelium and SCC, increased cytoplasmic expression of p62 was
observed. Furthermore, SCC exhibited increased cytoplasmic
expression of p62 compared with dysplastic epithelium (Fig. 2A and B). Western blot analysis of
tongue tissues from different groups also revealed that the levels
of p62 expression increased during oral carcinogenesis (Fig. 2D).
Beclin 1, an essential regulator of autophagy,
functions by interaction with LC3B and p62. Immunohistochemistry
results revealed that Beclin 1 was localized dominantly in the
cytoplasm, although perinuclear expression was also observed. In
normal tongue epithelium, Beclin 1 was weakly expressed, whereas
strong expression of Beclin 1 was observed in dysplastic epithelium
and SCC (Fig. 2A and B). Western
blot analysis of tongue tissues from different groups also revealed
that the expression levels of p62 increased during oral
carcinogenesis (Fig. 2D).
Furthermore, the present study also evaluated
expression of autophagy related 7 (Atg7), a key regulator of
autophagosome maturation, in 4NQO-induced mice tongue
carcinogenesis by western blotting. Consistent with other autophagy
markers, elevated Atg7 expression was also detected in dysplastic
epithelium and SCC (Fig. 2D). Taken
together, the autophagy level increased during the progressive
stages of 4NQO-induced tongue carcinogenesis.
Expression of autophagy markers in
human oral cancer
The present study investigated the protein level of
autophagy-associated genes in resection specimens from 5 normal
mucosa, 5 squamous cell papilloma and 52 OSCC patients by a tissue
microarray and immunohistochemical staining (Fig. 3A). Compared with negative expression
of autophagy markers in normal mucosa specimens, OSCC specimens
exhibited stronger immunohistochemical staining (Fig. 3B-D). Specifically, 86.3%, 70.6 and
78.4% OSCC specimens showed LC3B, p62 and Beclin 1 positive
expression, respectively. Notably, squamous cell papilloma showed a
higher positive rate of LC3B, p62 and Beclin 1 (20%, 40%, 20%
respectively) compared with normal mucosa, however this was
decreased compared with OSCC (data not shown). These results
suggested that autophagy was associated with human OSCC
progression.
Number of MDSCs and Tregs in
peripheral blood and spleen tissue of 4NQO-treated mice at
different stages of 4NQO-induced oral carcinogenesis
To examine the number of MDSCs and Tregs in
4NQO-treated mice, peripheral blood and spleen cells were screened
by flow cytometry. A substantial accumulation of MDSCs and Tregs
was observed in the peripheral blood and spleens during oral
carcinogenesis. It was revealed that the number of MDSCs in the
peripheral blood and spleens gradually increased in mild-moderate
dysplasia, severe dysplasia, and SCC groups compared with control
group (Fig. 4A and B). A similar
change in the Tregs number was observed in the peripheral blood and
spleens of the control, mild-moderate dysplasia, severe dysplasia,
and SCC groups (Fig. 4C and D).
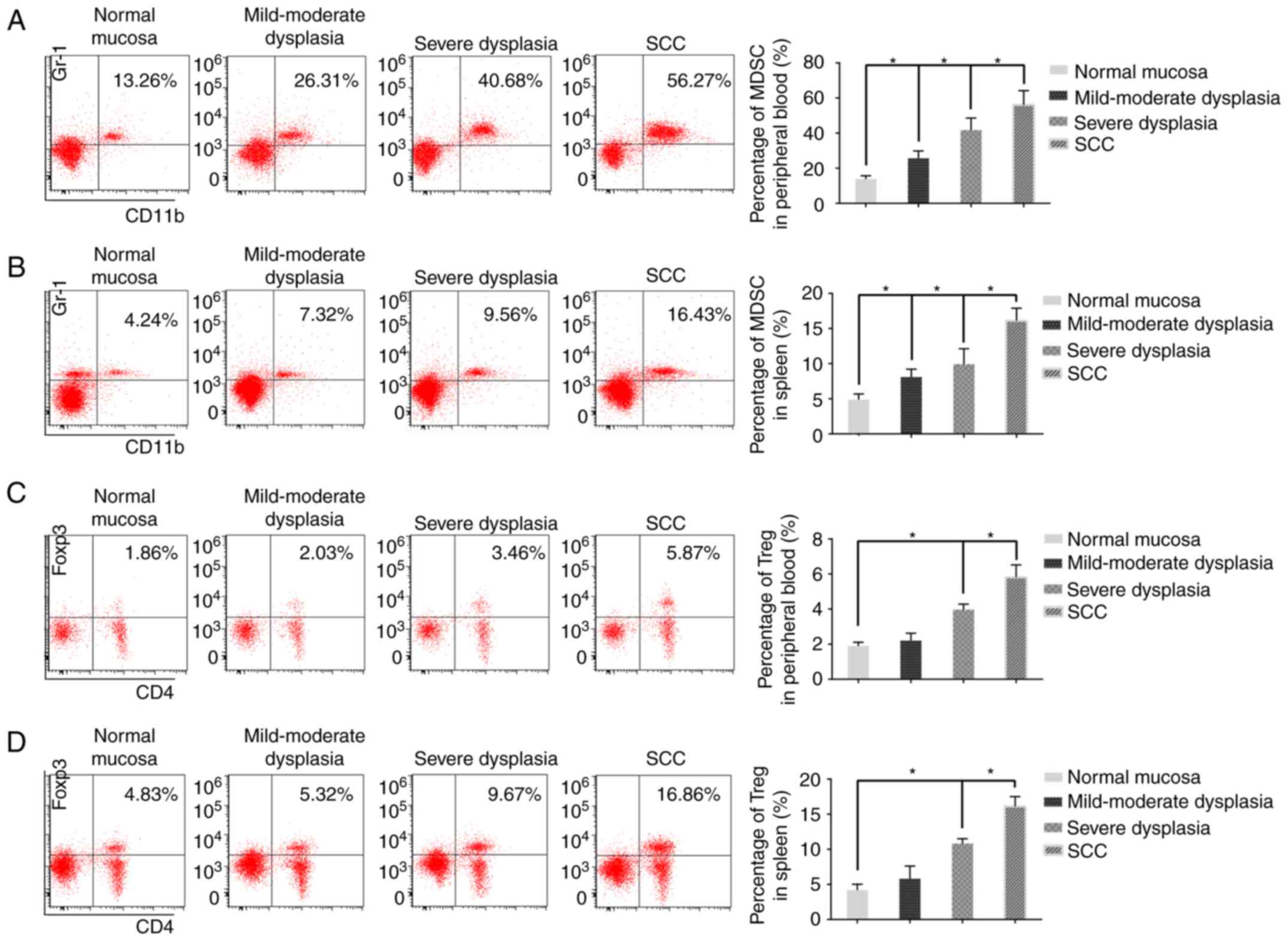 | Figure 4.Flow cytometric analysis of the
number of MDSCs and Tregs in 4NQO-treated mice. Representative flow
cytometry plots of MDSCs in (A) blood and (B) spleen tissues of
normal mice, mild-moderate dysplasia, severe dysplasia, and
tumor-bearing mice. Representative flow cytometry plots of Tregs
from (C) blood and (D) spleen tissues of normal mice (n=5),
mild-moderate dysplasia (n=6), severe dysplasia (n=6), and
tumor-bearing mice (n=12). Data are expressed as the mean ±
standard deviation. *P<0.05. MDSCs, Gr-1+CD11b+ myeloid derived
suppressor cells; Tregs, CD4+ Foxp3+ regulatory T cells. SCC,
squamous cell carcinoma; 4NQO, 4-nitroquinoline-1-oxide. |
Relationship between
autophagy-associated proteins and inflammation during oral
carcinogenesis
The present study used Pearson correlation analysis
to evaluate the correlation between autophagy marker expression and
the number of MDSCs and Tregs during oral carcinogenesis (data not
shown). The expression levels of LC3B and p62 were positively
associated with the number of MDSCs in the peripheral blood and
spleen tissues, whereas the expression of Beclin 1 was closely
associated with the number of Tregs in the peripheral blood and
spleen tissues. These results indicated that the expression of
autophagy-associated proteins appeared to be associated with tumor
inflammation during oral carcinogenesis.
Discussion
It has been recognized that autophagy is closely
associated with carcinogenesis (15). Previous evidence has revealed that
inflammation in the tumor microenvironment can induce autophagy,
which produces recycled nutrients to ‘feed’ anabolic cancer cells
and maintain tumor cell survival (10). In the present study, the results
revealed that the expression levels of LC3B, p62 and Beclin 1 and
the number of MDSCs and Tregs increased during oral carcinogenesis.
A close association was observed between the overexpression of
autophagy biomarkers and the accumulation of myeloid derived
suppressor cells. In addition, autophagy activation was also
observed in squamous cell papilloma and OSCC patient specimens.
These data indicated that autophagy may be activated by tumor
inflammation microenvironment during oral carcinogenesis. To the
best of the authors' knowledge, this is the first study to evaluate
the role of autophagy in 4NQO-induced oral carcinogenesis, which
was demonstrated to provide a favorable tumor microenvironment for
oral carcinogenesis.
The 4NQO-induced oral tumorigenesis mouse model has
been proven to provide an ideal model for studying tumor
pathobiology during sequential progression of human oral cancer
(16). Numerous studies have
established the 4NQO-induced oral cancer mouse model by adding
various does of 4NQO in drinking water, but the problems of
time-consuming and slow effect of this mice model remain unsolved
(17–19). In the present study, based on the
histological analysis, continuous 4NQO exposure in the mouse model
induced the multistage oral carcinogenesis lesions including
different grades of dysplasia and squamous cell carcinoma, similar
to human oral carcinogenesis. Consistent with previous studies, a
long period (26 weeks) of 4NQO exposure increased the mice oral
carcinogenesis rate compared with the short period (14 weeks)
(18,20,21).
However, 200 µg/ml 4NQO exposure does not further accelerate oral
carcinogenesis, although a similar lethality rate remains (22,23).
It therefore became evident that 100 µg/ml was an appropriate dose
with which to induce mice oral cancer in this model and a higher
dose cannot accelerate oral carcinogenesis. This is in line with
most previous studies, in which the 4NQO dose used to treat mice is
usually lower than 100 µg/ml (22,24,25).
However, this result may be attributed to the small number of mice
in the study, and further studies of larger sample size should be
conducted in the future.
LC3 is considered a well-established marker of
autophagy activity for monitoring autophagosome formation (26). Previous studies have reported that
LC3 expression is elevated and positively associated with poor
survival in many cancers (27–29).
The present study demonstrated that LC3B expression was increased
in premalignant lesion and squamous cell carcinoma of 4NQO-treated
mice. Similarly, increased LC3B expression was also detected in
human OSCC specimens. These were consistent with a previous study
in which LC3B puncta formation exhibited an increasing trend from
human normal oral mucosa, verrucous hyperplasia to OSCC (30). Additionally, high LC3 expression
status in OSCC is closely associated with lymph node metastasis,
advanced TNM stage and unfavorable outcome (31).
p62, as a selective substrate, is routinely used as
a biomarker to monitor autophagy flux and p62 accumulates when
autophagy is suppressed (32). The
results revealed that normal tongue epithelia exhibited limited
cytoplasmic p62 expression, whereas both 4NQO-induced squamous cell
carcinoma and human OSCC exhibited increased expression, suggesting
that p62 expression was upregulated during oral carcinogenesis
(30). Although a previous study
reported that no differences in p62 expression were observed among
normal oral mucosa, verrucous hyperplasia, and OSCC, p62 has been
found to be upregulated in several human cancers, including
colorectal carcinoma, epithelial ovarian cancer and gliomas
(30,33–35).
p62 is considered an autophagy substrate and can be degraded by
interaction with LC3 (32).
However, high levels of p62 protein have been revealed to induce
oncogenic transformation independent of its autophagy-associated
functions (36). Valencia et
al (37) demonstrated that in
the tumor microenvironment, p62 can promote the inflammatory
response via NF-κB activation and upregulation of c-Myc genes by
activating mechanistic target of rapamycin kinase complex 1
(37). Moreover, p62 also activates
the nuclear factor, erythroid 2 like 2-dependent anti-oxidant
response and thereby controls cell death and survival (38). Taken together, p62 may act as a
pro-oncogenic regulator in oral carcinogenesis but the underlying
mechanism is not clear.
Beclin 1 has been recognized as a central regulator
of autophagy. In the present study, Beclin 1 expression was
revealed to be upregulated during 4NQO-induced carcinogenesis.
Similarly, Beclin 1 overexpression was also observed in papilloma
and OSCC patient specimens. Consistent with the results of the
present study, the accumulation of Beclin 1 has been found in
breast, colon and ovarian cancer tissues (29,39,40).
Notably, increase of Beclin 1 has been reported to be a marker of
poor prognosis in colon cancer (40). However, Hu et al (41) reported that Beclin 1 expression is
decreased in oral tongue squamous cell carcinoma tissues relative
to the matched non-cancerous tissues (41). Moreover, Beclin 1+/−
mutant mice develop a large number of spontaneous tumors, including
lymphomas, lung cancer and hepatocarcinoma (42,43).
The reason for these controversial outcomes in a variety of cancers
may be that the role of autophagy depends upon intrinsic properties
of the tumor type and upon the specific tumor environment.
Accumulating evidence has indicated that
inflammation contributes to tumor initiation and progression
(44). MDSCs and Tregs, two
populations of inflammatory cells, notably increase in
tumor-bearing mice and cancer patients and contribute to an
immunosuppressive tumor microenvironment (17,23).
The present study observed a progressive increase in the proportion
of MDSCs and Tregs in the spleens and peripheral blood during oral
carcinogenesis. These results are consistent with previous studies
(17,23). Tregs are not considered a homogenous
population. The multiple subpopulations of Tregs are distinguished
by the expression of different cell surface markers, including
FoxP3, CD4, CD25 and CD127 (45,46).
The present study examined the level of CD4+
Foxp3+ Tregs, which has been widely reported in previous
studies (47–52). The multiple subpopulations of Tregs
may play different roles in oral carcinogenesis and the authors
hope to examine CD4+ CD25+ CD127 low
FoxP3+ Tregs in future studies.
Autophagy has been demonstrated to be activated by
inflammation in the tumor microenvironment (10). In the present study, it was
demonstrated that the expression of LC3B and p62 was positively
associated with MDSCs number, and that the expression of Beclin 1
was closely associated with an increase in Treg number. This
indicated that there was a correlation between autophagy and
inflammation in oral carcinogenesis. However, the study cannot
address how inflammation and autophagy act synergistically in
carcinogenesis. One hypothesis is that autophagy can be activated
in the inflammation microenvironment via activation of NLR Family
Pyrin Domain Containing 3 and inflammasomes or oxidative
stress-induced NF-κB activation (10). However, given that activated
autophagy can suppress excessive inflammation by regulating the
secretion of cytokines and chemokines and activate antitumor
immunity by presentation of tumor antigens, further study to
address this issue is necessary (9).
In conclusion, the study demonstrated that the
expression of LC3B, p62 and Beclin 1 was upregulated in 4NQO tongue
carcinogenesis, accompanied with MDSCs and Tregs accumulation. The
close correlation between autophagy markers and inflammatory cell
number suggested that autophagy may be activated by tumor
inflammation which provides a favorable tumor microenvironment for
cancer development. However, temporal and adjustable autophagy
inhibition studies in animal models are required in order to
further confirm the roles of autophagy in oral carcinogenesis and
its mechanisms in the tumor inflammation environment.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Program
on Key Research Project of China (grant no. 2016YFC0902700),
National Natural Science Foundation of China grants (grant nos.
81672672, 81572650, 81772891, 81502357 and 81621062), Natural
Science Foundation of Zhejiang Province (grant no. Q142114001),
Zhoushan Science and Technology Bureau Project (grant no.
2014C3106) and by State Key Laboratory of Oral Diseases Special
Funded Projects (2018).
Availability of data and materials
The majority of data generated or analyzed during
this study are included in this published article. The data of
expression of autophagy markers in human oral cancer and Pearson
correlation analysis were not shown.
Authors' contributions
JSW conducted the study, carried out most of
experiments and drafted the manuscript. XHL and MZ conceived the
study and participated in its design. JSW, LL, SRS and XP assisted
in development of methodology and acquisition of data. SSW and JBW
performed the analysis or interpretation of data and drafted the
manuscript. YJT and YLT participated in scoring of
immunohistochemistry, performed data interpretation and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures involving mice were approved by the
Subcommittee on Research and Animal Care (SRAC) of Sichuan
University. The written informed consents were obtained from
participants through their signatures. The use of human tissue
samples and clinical data was approved by the Institutional Ethics
Committee of the West China Medical Center, Sichuan University,
China (WCHSIRB-D-2012-097).
Patient consent for publication
Informed consent for the publication was obtained
from the participants.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
4NQO
|
4-nitroquinoline-1-oxide
|
|
OSCC
|
oral squamous cell carcinoma
|
|
MDSCs
|
Gr-1+CD11b+
myeloid derived suppressor cells
|
|
Tregs
|
CD4+ Foxp3+
regulatory T cell
|
|
p62
|
p62/SQSTM1
|
|
TMA
|
tissue microarray.
|
References
|
1
|
Petersen PE: Oral cancer prevention and
control-the approach of the World Health Organization. Oral Oncol.
45:454–460. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sarasin A: An overview of the mechanisms
of mutagenesis and carcinogenesis. Mutat Res. 544:99–106. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Danaei G, Vander Hoorn S, Lopez AD, Murray
CJ and Ezzati M: Comparative Risk Assessment collaborating group
(Cancers): Causes of cancer in the world: Comparative risk
assessment of nine behavioural and environmental risk factors.
Lancet. 366:1784–1793. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mizushima N and Klionsky DJ: Protein
turnover via autophagy: Implications for metabolism. Ann Rev Nutr.
27:19–40. 2007. View Article : Google Scholar
|
|
5
|
Green DR and Levine B: To be or not to be?
How selective autophagy and cell death govern cell fate. Cell.
157:65–75. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
White E: Deconvoluting the
context-dependent role for autophagy in cancer. Nat Rev Cancer.
12:401–410. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin S: p53, Autophagy and tumor
suppression. Autophagy. 1:171–173. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rosenfeldt MT, O'Prey J, Morton JP, Nixon
C, MacKay G, Mrowinska A, Au A, Rai TS, Zheng L, Ridgway R, et al:
p53 status determines the role of autophagy in pancreatic tumour
development. Nature. 504:296–300. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhong Z, Sanchez-Lopez E and Karin M:
Autophagy, inflammation, and immunity: A troika governing cancer
and its treatment. Cell. 166:288–298. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martinez-Outschoorn UE, Whitaker-Menezes
D, Lin Z, Flomenberg N, Howell A, Pestell RG, Lisanti MP and Sotgia
F: Cytokine production and inflammation drive autophagy in the
tumor microenvironment: Role of stromal caveolin-1 as a key
regulator. Cell Cycle. 10:1784–1793. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bonora M, Wieckowsk MR, Chinopoulos C,
Kepp O, Kroemer G, Galluzzi L and Pinton P: Molecular mechanisms of
cell death: Central implication of ATP synthase in mitochondrial
permeability transition. Oncogene. 34:16082015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kramer IR, Lucas RB, Pindborg JJ and Sobin
LH: Definition of leukoplakia and related lesions: An aid to
studies on oral precancer. Oral Surg Oral Med Oral Pathol.
46:518–539. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Vogel U: Overview on techniques to
construct tissue arrays with special emphasis on tissue
microarrays. Microarrays (Basel). 3:103–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pankiv S, Lamark T, Bruun JA, Øvervatn A,
Bjørkøy G and Johansen T: Nucleocytoplasmic shuttling of p62/SQSTM1
and its role in recruitment of nuclear polyubiquitinated proteins
to promyelocytic leukemia bodies. J Biol Chem. 285:5941–5953. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Galluzzi L, Pietrocola F, Bravo-San Pedro
JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J,
Gewirtz DA, Karantza V, et al: Autophagy in malignant
transformation and cancer progression. EMBO J. 34:856–880. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tang XH, Scognamiglio T and Gudas LJ:
Basal stem cells contribute to squamous cell carcinomas in the oral
cavity. Carcinogenesis. 34:1158–1164. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chu M, Su YX, Wang L, Zhang TH, Liang YJ,
Liang LZ and Liao GQ: Myeloid-derived suppressor cells contribute
to oral cancer progression in 4NQO-treated mice. Oral Dis.
18:67–73. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao J, Wang Z, Han J, Qiu X, Pan J and
Chen J: Increased frequency of CD4+ CD25+
FOXP3+ cells correlates with the progression of
4-nitroquinoline1-oxide-induced rat tongue carcinogenesis. Clin
Oral Investig. 18:1725–1730. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang M, Qiao X, Zhao L, Jiang L and Ren
F: Lactobacillus salivarius REN counteracted unfavorable
4-nitroquinoline-1-oxide-induced changes in colonic microflora of
rats. J Microbiol. 49:877–883. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kanojia D and Vaidya MM:
4-nitroquinoline-1-oxide induced experimental oral carcinogenesis.
Oral Oncol. 42:655–667. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Vitale-Cross L, Czerninski R,
Amornphimoltham P, Patel V, Molinolo AA and Gutkind JS: Chemical
carcinogenesis models for evaluating molecular-targeted prevention
and treatment of oral cancer. Cancer Prev Res (Phila). 2:419–422.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tang XH, Osei-Sarfo K, Urvalek AM, Zhang
T, Scognamiglio T and Gudas LJ: Combination of bexarotene and the
retinoid CD1530 reduces murine oral-cavity carcinogenesis induced
by the carcinogen 4-nitroquinoline 1-oxide. Proc Natl Acad Sci USA.
111:8907–8912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen MF, Kuan FC, Yen TC, Lu MS, Lin PY,
Chung YH, Chen WC and Lee KD: IL-6-stimulated CD11b+
CD14+ HLA-DR-myeloid-derived suppressor cells, are
associated with progression and poor prognosis in squamous cell
carcinoma of the esophagus. Oncotarget. 5:8716–8728. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang M, Wang F, Jiang L, Liu R, Zhang L,
Lei X, Li J, Jiang J, Guo H, Fang B, et al: Lactobacillus
salivarius REN inhibits rat oral cancer induced by 4-nitroquioline
1-oxide. Cancer Prev Res. 6:686–694. 2013. View Article : Google Scholar
|
|
25
|
Chen PT, Hsieh CC, Wu CT, Yen TC, Lin PY,
Chen WC and Chen MF: 1α,25-dihydroxyvitamin D3 inhibits esophageal
squamous cell carcinoma progression by reducing IL6 signaling. Mol
Cancer Ther. 14:1365–1375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kabeya Y, Mizushima N, Ueno T, Yamamoto A,
Kirisako T, Noda T, Kominami E, Ohsumi Y and Yoshimori T: LC3, a
mammalian homologue of yeast Apg8p, is localized in autophagosome
membranes after processing. EMBO J. 19:5720–5728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yoshioka A: LC3, an autophagosome marker,
is highly expressed in gastrointestinal cancers. Int J Oncol.
33:461–468. 2008.PubMed/NCBI
|
|
28
|
Schmitz KJ, Ademi C, Bertram S, Schmid KW
and Baba HA: Prognostic relevance of autophagy-related markers LC3,
p62/sequestosome 1, Beclin-1 and ULK1 in colorectal cancer patients
with respect to KRAS mutational status. World J Surg Oncol.
14:1892016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Choi J, Jung W and Koo JS: Expression of
autophagy-related markers beclin-1, light chain 3A, light chain 3B
and p62 according to the molecular subtype of breast cancer.
Histopathology. 62:275–286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu JL, Chen FF, Lung J, Lo CH, Lee FH, Lu
YC and Hung CH: Prognostic significance of p62/SQSTM1 subcellular
localization and LC3B in oral squamous cell carcinoma. Br J Cancer.
111:944–954. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang JY, Hsi E, Huang YC, Hsu NC, Chu PY
and Chai CY: High LC3 expression correlates with poor survival in
patients with oral squamous cell carcinoma. Hum Pathol.
44:2558–2562. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Klionsky DJ, Abdelmohsen K, Abe A, Abedin
MJ, Abeliovich H, Arozena Acevedo A, Adachi H, Adams CM, Adams PD,
Adeli K, et al: Guidelines for the use and interpretation of assays
for monitoring autophagy (3rd edition). Autophagy. 12:1–222. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Iwadate R, Inoue J, Tsuda H, Takano M,
Furuya K, Hirasawa A, Aoki D and Inazawa J: High expression of
SQSTM1/p62 protein is associated with poor prognosis in epithelial
ovarian cancer. Acta Histochem Cytochem. 47:295–301. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakayama S, Karasawa H, Suzuki T, Yabuuchi
S, Takagi K, Aizawa T, Onodera Y, Nakamura Y, Watanabe M, Fujishima
F, et al: p62/sequestosome 1 in human colorectal carcinoma as a
potent prognostic predictor associated with cell proliferation.
Cancer Med. 6:1264–1274. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao M, Xu H, Zhang B, Hong B, Yan W and
Zhang J: Impact of nuclear factor erythroid-derived 2-like 2 and
p62/sequestosome expression on prognosis of patients with gliomas.
Hum Pathol. 46:843–849. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Umemura A, He F, Taniguchi K, Nakagawa H,
Yamachika S, Font-Burgada J, Zhong Z, Subramaniam S, Raghunandan S,
Duran A, et al: p62, upregulated during preneoplasia, induces
hepatocellular carcinogenesis by maintaining survival of stressed
HCC-initiating cells. Cancer Cell. 29:935–948. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Valencia T, Kim JY, Abu-Baker S,
Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA,
Metallo CM, Diaz-Meco MT and Moscat J: Metabolic reprogramming of
stromal fibroblasts through p62-mTORC1 signaling promotes
inflammation and tumorigenesis. Cancer Cell. 26:121–135. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Moscat J and Diaz-Meco MT: p62 at the
crossroads of autophagy, apoptosis, and cancer. Cell.
137:1001–1004. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhao Y, Chen S, Gou WF, Xiao LJ, Takano Y
and Zheng HC: Aberrant Beclin 1 expression is closely linked to
carcinogenesis, differentiation, progression, and prognosis of
ovarian epithelial carcinoma. Tumour Biol. 35:1955–1964. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park JM, Huang S, Wu TT, Foster NR and
Sinicrope FA: Prognostic impact of Beclin 1, p62/sequestosome 1 and
LC3 protein expression in colon carcinomas from patients receiving
5-fluorouracil as adjuvant chemotherapy. Cancer Biol Ther.
14:100–107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hu Z, Zhong Z, Huang S, Wen H, Chen X, Chu
H, Li Q and Sun C: Decreased expression of Beclin-1 is
significantly associated with a poor prognosis in oral tongue
squamous cell carcinoma. Mol Med Rep. 14:1567–1573. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yue Z, Jin S, Yang C, Levine AJ and Heintz
N: Beclin 1, an autophagy gene essential for early embryonic
development, is a haploinsufficient tumor suppressor. Proc Natl
Acad Sci USA. 100:15077–15082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh
H, Troxel A, Rosen J, Eskelinen EL, Mizushima N, Ohsumi Y, et al:
Promotion of tumorigenesis by heterozygous disruption of the beclin
1 autophagy gene. J Clin Invest. 112:1809–1820. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Elinav E, Nowarski R, Thaiss CA, Hu B, Jin
C and Flavell RA: Inflammation-induced cancer: Crosstalk between
tumours, immune cells and microorganisms. Nat Rev Cancer.
13:759–771. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Romano M, Tung SL, Smyth LA and Lombardi
G: Treg therapy in transplantation: A general overview. Transpl
Int. 30:745–753. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Edozie FC, Nova-Lamperti EA, Povoleri GA,
Scottà C, John S, Lombardi G and Afzali B: Regulatory T-cell
therapy in the induction of transplant tolerance: The issue of
subpopulations. Transplantation. 98:370–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wei J, Long L, Yang K, Guy C, Shrestha S,
Chen Z, Wu C, Vogel P, Neale G, Green DR and Chi H: Autophagy
enforces functional integrity of regulatory T cells by coupling
environmental cues and metabolic homeostasis. Nat Immunol.
17:277–285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang XX, Qiao YC, Li W, Zou X, Chen YL,
Shen J, Liao QY, Zhang QJ, He L and Zhao HL: Human amylin induces
CD4+Foxp3+ regulatory T cells in the
protection from autoimmune diabetes. Immunol Res. 66:179–186. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yin E, Matsuyama S, Uchiyama M, Kawai K
and Niimi M: Graft protective effect and induction of
CD4+Foxp3+ cell by thrombomodulin on
allograft arteriosclerosis in mice. J Cardiothorac Surg. 13:482018.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun IH, Oh MH, Zhao L, Patel CH, Arwood
ML, Xu W, Tam AJ, Blosser RL, Wen J and Powell JD: mTOR complex 1
signaling regulates the generation and function of central and
effector Foxp3+ regulatory T cells. J Immunol.
201:481–492. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Oyarce K, Campos-Mora M, Gajardo-Carrasco
T and Pino-Lagos K: Vitamin C fosters the in vivo differentiation
of peripheral CD4+ Foxp3− T cells into
CD4+ Foxp3+ regulatory T cells but impairs
their ability to prolong skin allograft survival. Front Immunol.
9:1122018. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Olguin JE, Medina-Andrade I, Molina E,
Vázquez A, Pacheco-Fernández T, Saavedra R, Pérez-Plasencia C,
Chirino YI, Vaca-Paniagua F, Arias-Romero LE, et al: Early and
partial reduction in CD4+Foxp3+ regulatory T
cells during colitis-associated colon cancer induces
CD4+ and CD8+ T cell activation inhibiting
tumorigenesis. J Cancer. 9:239–249. 2018. View Article : Google Scholar : PubMed/NCBI
|