Introduction
Renal cell carcinoma (RCC) is one of the most common
cancers of the urinary tract in the world, accounting for 2–3% of
adult cancers (1). It is reported
that there are ~209,000 new cases of RCC annually in the world and
the mortality rate is as high as 50% (2). The treatment effects of conventional
radiotherapy and chemotherapy are not satisfactory and the
treatment of RCC is mainly based on surgery (3). Therefore, it is essential to obtain a
better understanding of the RCC development mechanism and find new
therapeutic targets to reduce RCC mortality rate.
Human gene sequencing results have revealed that
>90% of human DNA sequences is involved in transcription, but
only 2% has protein encoding functions. RNA that does not have a
protein coding function is categorized as non-coding RNA (4). Non-coding RNA is divided into
long-chain non-coding RNA (lncRNA) and short-chain non-coding RNA
according to the size of RNA (4,5). Among
them, short-chain non-coding RNA, such as microRNA (miRNA), plays
an important role in regulating gene expression and cell function
(6). Previous research has revealed
that miRNA-22 (miR-22) is lowly expressed in breast cancer,
esophageal squamous cell carcinoma, gastric and colorectal cancer.
miR-22 inhibited cell growth, migration and epithelial-mesenchymal
transition in the above-mentioned cancers (7–11). It
was reported that miR-22 inhibited the invasion and migration of
gastric cancer cells by downregulating the expression of MMP-14 and
Snail (12). In addition, it has
been reported that miR-22 inhibited the proliferation and migration
of clear RCC cells by regulating the expression of PTEN, indicating
that miRNA-22 has an inhibitory effect in RCC (13).
Recent research has revealed that lncRNA has a
variety of biological functions in cancer progression (14,15).
lncRNAs, such as HOTAIR, TUG1, SPRY4-IT and metastasis-associated
lung adenocarcinoma transcript 1 lncRNA (lncR-MALAT1) act as
important regulators in the proliferation and metastasis of cancer
cells (16–18). lncR-MALAT1 is an lncRNA which is
located on the 11q13.1 chromosome (19). Previous research has demonstrated
that MALAT1 is highly expressed in a variety of cancers, such as
lung, breast, liver, pancreatic and colon cancer (20). Hirata et al (19) revealed that MALAT1 promoted the
mobility of RCC cells by influencing the interaction between Ezh2
and miR-205. Furthermore, MALAT1 was reported to affect the growth
of melanoma by targeting miR-22 (21). However, whether miR-22 is a target
of MALAT1 in the proliferation and migration of RCC cells has not
been reported.
In the present study, we identified that the
expression of MALAT1 was elevated, and the expression of miR-22-3p
was markedly decreased in RCC tumor tissues and cell lines. In
addition, silencing MALAT1 obviously inhibited the proliferation
and migration of tumor cells in vitro and in vivo. In
addition, miR-22-3p was demonstrated to be a target of MALAT1.
miR-22-3p inhibitor reversed the inhibitory effect of shRNA-MALAT1
on the progression of RCC and the PI3K/Akt signaling pathway.
Collectively, MALAT1 affected the development of RCC by targeting
miR-22-3p via the PI3K/Akt signaling pathway.
Materials and methods
Sample collection
Thirty pairs of RCC specimens and adjacent normal
tissues were obtained from patients who underwent surgical
treatment from October 2015 to August 2016 in Tai'an City Central
Hospital. Written informed consent was obtained prior to resection
from patients and the study was approved by the Ethics Committee of
Tai'an City Central Hospital.
Cell lines
RCC cell lines 786-O, Caki-1, Ketr-3, RT112 and T24,
as well as the normal renal epithelial cells Ect1/E6E7 were
purchased from the American Type Culture Collection (ATCC,
Gaithersburg MD, USA). The 786-O, Ketr-3, RT-112 and T24 cells were
cultured in RPMI-1640 medium (Gibco, Rockville, MD, USA) containing
10% fetal bovine serum (FBS; Gibco). The Caki-1 cells were cultured
in McCoy's 5A medium containing 10% FBS, and Ect1/E6E7 cells were
cultured in EMEM medium (Gibco) containing 10% FBS. All cells were
cultured in incubators at 37°C in a 5% CO2
atmosphere.
Quantitative real-time polymerase
chain reaction (qRT-PCR)
Total RNA was extracted from cells and tissues using
the TRIzol kit (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) according to the manufacturer's instructions and
cDNA was synthesized using Reverse Transcription kit (Thermo Fisher
Scientific). Quantitative analysis of cDNA (Thermo Fisher
Scientific) was performed using a Fluorescence Quantitative PCR kit
(Sangon Biotech Co., Ltd., Shanghai, China). The amplification
primers used were as follows: MALAT1 sense,
5′-CTCACTAAAGGCACCGAAGG-3′ and antisense,
5′-GGCAGAGAAGTTGCTTGTGG-3′; miR-22-3p sense,
5′-AAGCTGCCAGTTGAAGAACTGTA-3′ and antisense,
5′-CTCGCTTCGGCAGCACA-3′.
Cell transfection
Caki-1 cells were seeded in 24-well plates at
1×105 cells/well and were transfected with specified
fragments for 24 h, when the cell aggregation rate was over 80%.
Mimics/inhibitors specific for miR-22-3p and short hairpin RNA
(shRNA)/scramble fragments targeting MALAT1 were designed and
purchased from Hanheng Biotechnology Co., Ltd. (Shanghai, China).
Recombinant lentiviral vector for knocking out MALAT1 was
constructed and named LV-MALAT1 shRNA and the control group was
named LV-MALAT1 scramble group.
Cell viability
Caki-1 cells were transfected with shRNA- MALAT1 or
MALAT1 scramble, and cell viability was evaluated according to the
instructions of the CCK-8 assay kit (Sigma-Aldrich, St. Louis, MO,
USA).
Wound healing assay
Five parallel lines were evenly drawn on the back of
the 6-well plate using marker pens and the Caki-1 cells were grown
in 6-well plates at a concentration of 2×105 cells/ml.
After incubation for 24 h, the tip was scratched perpendicularly to
the horizontal line on the back of the 6-well plate. Destroyed
cells were rinsed off with phosphate-buffered saline (PBS) and
cultured in serum-free medium for another 24 h. Cell migration was
observed and imaged at 0 and 24 h with a digital camera (Olympus
Corp., Tokyo, Japan).
Western blot analysis
Protein samples were extracted from cells and
tissues with protein lysate, and the protein concentration was
assessed using a BCA kit (Pierce, Rockford, IL, USA). The protein
was separated by SDS-PAGE and the isolated protein was transferred
to the polyvinylidene fluoride (PVDF) membrane by semi-dry method
and then was blocked in 5% skim milk at room temperature, for 2 h.
The membrane was incubated with the primary antibodies (anti-Ki-67,
cat. no. ab16667; anti-PCNA, cat. no. ab29; anti-MMP3, cat. no.
ab53015; anti-MIIP, cat. no. ab167197; p-PI3K, cat. no. ab182651;
anti-p-AKT, cat. no. ab131443; anti-AKT, cat. no. ab179463; all at
the dilution 1:1,000) and the corresponding HRP-conjugated
secondary antibodies (cat. nos. ab6728 and ab6721 at the dilution
1:5,000; obtained from Abcam, Cambridge, UK) and was extensively
washed several times with PBST. The proteins were detected using a
ChemiDoc XRS imaging system and were analysed through Quantity One
analysis software (Bio-Rad Laboratories, San Francisco, CA, USA).
GAPDH was used as the internal control.
Luciferase activity assays
The binding site of MALAT1 on miR-22-3p was
predicted through bioinformatic analysis (TargetScan, http://www.targetscan.org/). PCR amplification of
MALAT1 fragments contained a fragment of the miR-22-3p binding
site, which was inserted into the pMIR-REPORT vector. The mutant
plasmid was used as the control group. Caki-1 cells were
co-transfected with MALAT1 WT/MALAT1-MUT and/or miR-22-3p mimic for
24 h. Fluorescence intensity was evaluated according to the
Dual-Luciferase Reporter kit (Promega Corporation, Madison, WI,
USA).
Immunofluorescence
Caki-1 cells were treated with 4% paraformaldehyde
for 30 min and 0.5% Triton X-100 for 30 min and then were blocked
with 5% goat serum for 2 h at room temperature. The cells were then
incubated with the primary antibody (Akt, 1:200; cat. no. ab179463;
Abcam) overnight at 4°C. On the second day, the cells were
incubated with the corresponding FITC-conjugated secondary
antibodies (1:2,000; cat. no. ab6785; Abcam) in the dark for 50
min, and then were stained with DAPI. Cell fluorescence was
observed under a fluorescence microscope (Olympus Corp.).
RCC xenografts
Sixteen male specific-pathogen-free (SPF) nude mice
(~20 g) were purchased from the Beijing Experimental Animal Center
(Beijing, China). The animal experiments were performed according
to the guidelines of the National Institute of Health (NIH). All
animal experiments were approved by the Medical Ethics Committee of
The Second Hospital Affiliated to Tianjin Medical University
(Tianjin, China). An RCC xenograft mouse model was created by
subcutaneous injection of 1×107 Caki-1 cells transfected
with LV-MALAT1 shRNA or LV-MALAT1 scramble to SPF nude mice. When
sacrificed, the weight of mice was ~25 g. The tumor volume was
evaluated every 5 days until 30 days after the development of a
palpable tumor according to the formula (a × b2)/2,
where a and b are the largest diameter and the perpendicular
diameter, respectively.
Statistical analysis
All data were analyzed using the SPSS 19.0 software
(IBM Corp., Armonk, NY, USA) and the results were expressed as the
mean ± standard deviation (SD). The Student's t-test was used to
assess comparisons between two groups. Multiple-group comparisons
were made using one-way analysis of variance (ANOVA). P<0.05 was
considered to indicate a statistically significant difference.
Results
MALAT1 is upregulated and miR-22-3p is
downregulated in the RCC tissues and cell lines
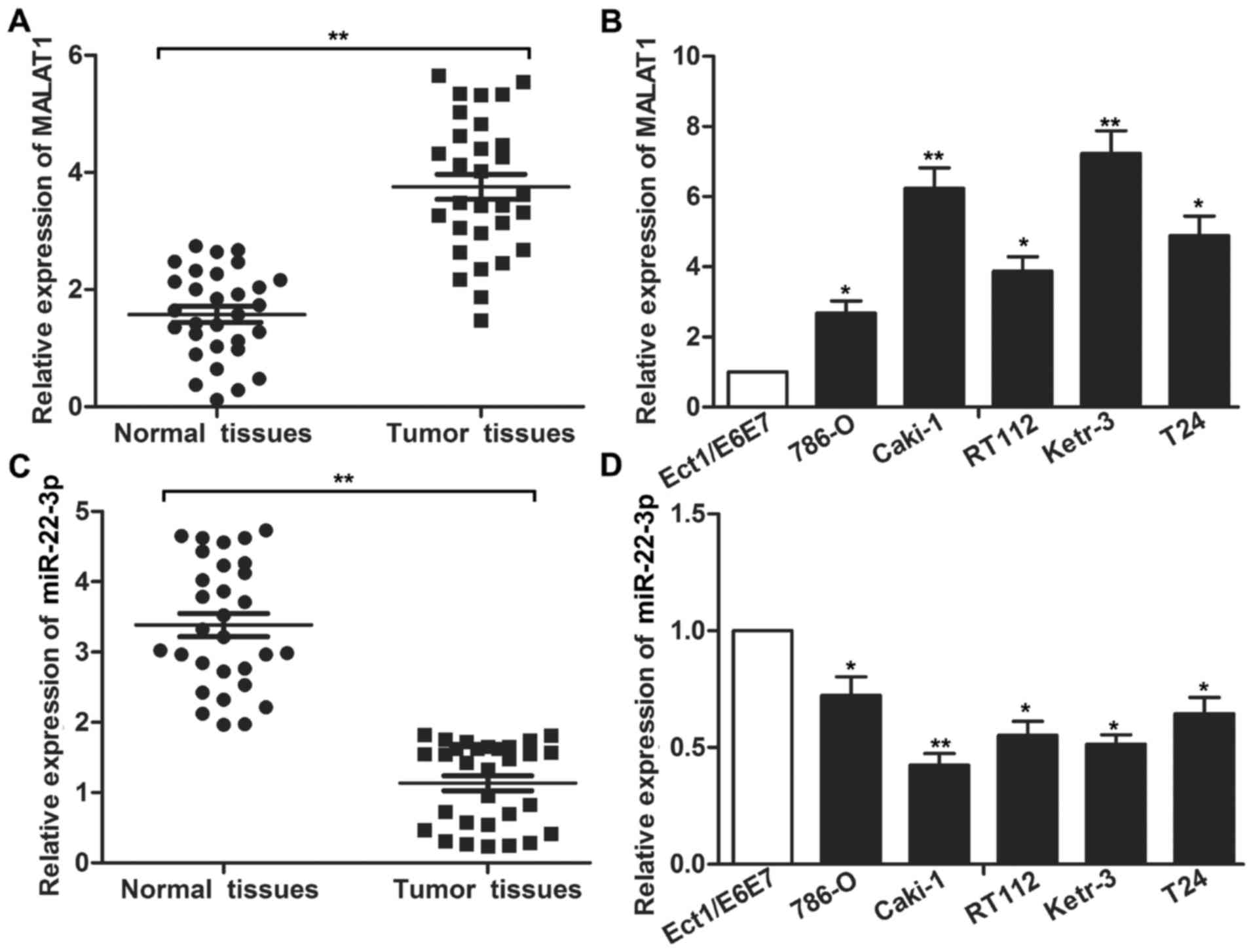
The expression of MALAT1 and miR-22-3p in RCC
tissues and cell lines was detected through qRT-PCR. As displayed
in Fig. 1A, the expression level of
MALAT1 in the RCC tumor tissue was significantly higher than that
in adjacent normal tissues (P<0.01). Concurrently, the
expression of MALAT1 in the RCC cell lines was significantly
increased compared with the normal renal epithelial cells Ect1/E6E7
(Fig. 1B; P<0.05 and P<0.01).
In addition, the expression level of miR-22-3p in the RCC tumor
tissues and cell lines was significantly decreased compared with
the control tissues and cell line (Fig.
1C and D; P<0.05 and P<0.01). The aberrant expression of
MALAT1 and miR-22-3p compared with normal renal epithelial cells
indicated a systematic predictable link between MALAT1/miR-22-3p
and the progression of RCC.
Effect of MALAT1 in RCC cell
proliferation and metastasis
Previous research has demonstrated that MALAT1
regulates the proliferation and migration of a variety of cancer
cells. In the present study, we selected the Caki-1 cell line for
the following experiments as it has the relatively highest
expression of MALAT1 and the lowest expression level of miR-22-3p
in the related cell lines. The expression of MALAT1 was
significantly downregulated through transfection with MALAT1 shRNA
in Caki-1 cells compared with the scramble group and the control
group (Fig. 2A; P<0.01).
Transfection with MALAT1 shRNA suppressed cell proliferation
compared with MALTA1 scramble group (Fig. 2B; P<0.05 and P<0.01).
Furthermore, the wound healing assay exhibited that the migration
ability of cells in the MALAT1 shRNA group was much lower than that
of the control group (Fig. 2C;
P<0.05). The results of the western blot analysis revealed that
MALAT1 shRNA significantly suppressed the expression of Ki-67 and
proliferating cell nuclear antigen (PCNA) compared with the
scramble and the control group (P<0.05) (Fig. 2D). As expected, MALAT1 shRNA
significantly inhibited the expression of matrix
metalloproteinase-3 (MMP-3) and promoted the expression of
migration and invasion inhibitory protein (MIIP) (Fig. 2E; P<0.05). The above-mentioned
results indicated that MALAT1 shRNA significantly inhibited the
proliferation and migration of RCC cells.
MALAT1 regulates cell viability and
mobility by targeting miR-22-3p in RCC
To investigate the related mechanism by which MALAT1
regulates the progression of RCC cells, we found a binding site for
MALAT1 on miR-22-3p through bioinformatic analysis (TargetScan,
http://www.targetscan.org/) (Fig. 3A). Concurrently, there were marked
negative relevant relations between the expression of MALAT1 and
miR-22-3p in RCC tissues (Fig. 3B).
As displayed in Fig. 3C, MALAT1
shRNA significantly elevated the expression of miR-22-3p in RCC
cells (P<0.05), but then this effect was reversed through the
co-transfection with miR-22-3p inhibitor (P<0.05). Furthermore,
the inhibiting effect of MALAT1 shRNA on the expression of MALAT1
was partially reversed by the co-transfection of miR-22-3p
inhibitor, identifying that miR-22-3p inhibitor elevated the level
of MALAT1 in turn (Fig. 3D;
P<0.05 and P<0.05, respectively). Subsequently, we used the
luciferase reporter assay to further examine the targeting
relationship between MALAT1 and miR-22-3p. The results demonstrated
that the luciferase signal in the lncR-MALAT1 MUT and the
lncR-MALAT1 WT group remained at the same level, but the addition
of miR-22-3p mimic significantly inhibited the luciferase activity
by binding to MALAT1 WT fragments. However, the regulatory
relationship disappeared when binding to the MALAT1 MUT fragments,
indicating that there existed a targeting relationship between
MALAT1 and miR-22-3p (Fig. 3E;
P<0.01). Furthermore, miR-22-3p inhibitor significantly reversed
the inhibitory effect of MALAT1 shRNA on the proliferation and
migration of Caki-1 cells (Fig. 3F and
G; P<0.05, P<0.01 and P<0.05). These results indicated
that MALAT1 may promote the proliferation and migration of RCC
cells by targeting miR-22-3p.
MALAT1 shRNA restrains the activation
of the PI3K/AKT pathway
Recent research has indicated that PI3K/Akt
signaling pathway is closely related to the proliferation and
migration of cancer cells (24,25).
We have also demonstrated that MALAT1 shRNA inhibited the
proliferation and migration of Caki-1 cells in the above-mentioned
experiments. Therefore, we further investigated the effect of
MALAT1 shRNA on the PI3K/Akt pathway. As displayed in Fig. 4A, MALAT1 shRNA significantly
inhibited the expression of p-PI3K and p-Akt, but the addition of
miR-22-3p inhibitor into the MALAT1 shRNA-treated Caki-1 cells
significantly reversed the inhibitory effect of MALAT1 shRNA on the
expression of p-PI3K and p-Akt (P<0.01, P<0.05). In addition,
the expression of Akt in the MALAT1 shRNA group was much lower than
that in the control group, indicating that MALAT1 shRNA inhibited
the activation of Akt (Fig. 4B).
Concurrently, miR-22-3p inhibitor reversed the inhibitory effect of
MALAT1 shRNA on Akt accumulation on the cell membrane (Fig. 4B). All these results revealed that
MALAT1 shRNA inhibited the activation of PI3K/Akt signaling pathway
through the upregulation of the expression of miR-22-3p.
MALATI shRNA suppresses tumor growth
and metastasis in vivo
To further validate the above results in
vivo, we replicated the xenograft animal model of RCC and
explored the effect of MALAT1 shRNA on tumor growth and metastasis.
The results demonstrated that MALAT1 shRNA inhibited the growth of
RCC tumor and elevated the expression of miR-22-3p in vivo
(Fig. 5A and B; P<0.05,
P<0.01). Concurrently, the expression of PCNA and MMP-3 in the
shRNA-MALAT1 group was significantly lower compared with the
control group. The ratio of p-Akt/Akt was also significantly lower
than that of the control group (Fig. 5C
and D, P<0.01). These results further indicated that MALAT1
shRNA can also inhibit the growth and metastasis of RCC tumors by
inhibiting the PI3K/Akt signaling pathway activation in
vivo.
Discussion
lncRNA is a class of transcripts with more than 200
nucleotides which do not have protein-coding abilities (4). Recent research has shown that lncRNA
plays a pivotal role in the development of many tumors, but the
related mechanism remains to be investigated (22). In the present study, we first
investigated the interaction of MALAT1 and miR-22-3p in the
development of RCC.
Lu et al (23) identified that the overexpression of
lncRNA BC032469 promoted the proliferation of gastric cancer cells
through the downregulation of miR-1207-5p. A previous study
reported that the overexpression of lncRNA ATB in hepatocellular
carcinoma competed with miR-200, leading to an upregulation of the
expression of ZEB1 and ZEB2, which promoted the metastasis of
hepatocellular carcinoma (24).
MALAT1 was firstly discovered in small cell lung cancer and is
often used as a prognostic indicator of lung cancer metastasis
(25). Numerous studies have
revealed that MALAT1 is closely related to multiple cancers by
regulating cell viability and mobility (26). Consistent with previous studies, in
the present study, we revealed that MALAT1 was highly expressed in
RCC tissues and cell lines. The CCK-8 and wound healing assay
further revealed that the knockdown of MALAT1 successfully
inhibited cell proliferation and migration. The result of western
blot analysis also demonstrated the inhibiting effect of MALAT1
shRNA on cell proliferation and migration in RCC cell lines by
regulating the expression of cell proliferation and migration
marker proteins. These results revealed that MALAT1 shRNA
suppressed the progression of RCC.
A previous study has demonstrated that MALAT1 is a
class of ceRNAs which promotes cancer development by binding to
miRNAs (27). As previously
reported, MALAT1 binds to miR-1 and inhibits its binding to
messenger RNA, thus promoting the invasion and metastasis of breast
cancer cells (28). MALAT1
regulates the development of RCC by binding to miR-200 and miR-205
(19,29). Recent research has also revealed
that miR-22 functions as a ‘double-edged sword’ in human cancer
(30). The overexpression of miR-22
can promote cancer cell proliferation and inhibit cancer cell
apoptosis in prostate cancer and chronic lymphocytic leukemia
(31,32). However, the expression of miR-22 is
suppressed in gastric cancer, melanoma, glioma, RCC and other types
of cancer. Zhang et al (33)
found that miR-22 expression in RCC was reduced and it played an
inhibiting role in RCC progression. In addition, it has been
demonstrated that MALAT1 competed with miR-22-3p for endogenous RNA
to protect endothelium from ox-LDL-induced endothelial dysfunction
(34). Similarly, suppressed
expression of miR-22-3p in RCC tumor tissues and cell lines was
detected in the present study. In addition, bioinformatic analysis
was used to predict the targeting relationship between MALAT1 and
miR-22-3p. The luciferase assay further confirmed the targeting
relationship between MALAT1 and miR-22-3p. Concurrently, miR-22-3p
inhibitor significantly reversed the inhibitory effect of MALAT1
shRNA on the proliferation and migration of Caki-1 cells. In
summary, MALAT1 regulated the proliferation and migration of RCC
cells by targeting miR-22-3p.
The PI3K/Akt signaling pathway is an important
pathway for regulating cell cycle, proliferation and apoptosis
(35). Previous research has shown
that miR-22 regulates the activation of PI3K/Akt pathway (32). MALAT1 can promote the proliferation
and epithelial-mesenchymal transition of cholangiocarcinoma and
breast cancer cells by activating the PI3K/Akt signaling pathway
(36,37). Therefore, we considered whether
MALAT1 promoted the proliferation of RCC cells via the PI3K/Akt
pathway. We found that MALAT1 shRNA (the experiments were conducted
using the most effective among three pairs of shRNAs targeting
MALAT1) inhibited the phosphorylation of PI3K and p-Akt and
membrane metastasis of Akt in RCC cells, thus inhibiting the
PI3K/Akt pathway activation. The co-transfection of miR-22-3p
significantly reversed the inhibiting effect of MALAT1 shRNA on the
PI3K/Akt pathway. These results indicated that MALAT1 shRNA
suppressed the activation of the PI3K/Akt signaling pathway by
targeting miR-22-3p.
Following the observation that MALAT1 shRNA
regulated cell proliferation and migration in RCC cell lines, the
effect of MALAT1 shRNA in RCC tumor was further investigated. As
previously described, MALAT1 promoted tumor growth and metastasis
by inducing epithelial-mesenchymal transition in oral squamous cell
carcinoma (38). A previous study
has also demonstrated that MALAT1 promoted tumor growth and
metastasis in colorectal cancer through binding to SFPQ (39). Similarly, in the present study,
MALAT1 shRNA was found to suppress tumor growth in an RCC xenograft
animal model. Furthermore, MALAT1 shRNA elevated the expression of
miR-22-3p and inactivated the PI3K/Akt signaling pathway in
vivo. The present study revealed that MALAT1 shRNA suppressed
RCC progression in vivo.
In summary, the present study demonstrated that the
expression of lncR-MALAT1 was upregulated in RCC tissues and cell
lines. Cell proliferation and motility was suppressed by knocking
out the expression of MALAT1 by MALAT1 shRNA in RCC cell lines.
Further research revealed that miR-22-3p was a direct target of
MALAT1 and miR-22-3p inhibitor counteracted the effect of MALAT1
shRNA on cell viability and motility. Furthermore, we found that
MALAT1 exerted its regulatory role by inactivating the PI3K/Akt
signaling pathway. The in vivo experiments further
demonstrated that MALAT1 shRNA suppressed the progression of RCC.
This investigation further revealed the mechanism of RCC
development and provided a new potential target for the clinical
treatment of RCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZL, ZM and XX conceived, designed the study and
performed the experiments. XX wrote the paper and edited the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All experimental protocols were approved by the
Ethics Committee of Tai'an City Central Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MALAT1
|
metastasis-associated lung
adenocarcinoma transcript 1
|
|
lncRNA
|
long non-coding RNA
|
|
RCC
|
renal cell carcinoma
|
|
miRNA
|
microRNA
|
|
MMP-3
|
matrix metaloproteinase-3
|
|
PCNA
|
proliferating cell nuclear antigen
|
|
MIIP
|
migration and invasion inhibitory
protein
|
|
FBS
|
fetal bovine serum
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Saini S, Yamamura S, Majid S, Shahryari V,
Hirata H, Tanaka Y and Dahiya R: MicroRNA-708 induces apoptosis and
suppresses tumorigenicity in renal cancer cells. Cancer Res.
71:6208–6219. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu ZH, Zhang Q, Wang YD, Chen J, Jiang ZM,
Shi M, Guo X, Qin J, Cui GH, Cai ZM, et al: Overexpression of
cyclooxygenase-1 correlates with poor prognosis in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:3729–3734. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bertone P, Stolc V, Royce TE, Rozowsky JS,
Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et
al: Global identification of human transcribed sequences with
genome tiling arrays. Science. 306:2242–2246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Song SJ and Pandolfi PP: miR-22 in
tumorigenesis. Cell cycle. 13:11–12. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen B, Tang H, Liu X, Liu P, Yang L, Xie
X, Ye F, Song C, Xie X and Wei W: miR-22 as a prognostic factor
targets glucose transporter protein type 1 in breast cancer. Cancer
Lett. 356:410–417. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jafarzadeh-Samani Z, Sohrabi S,
Shirmohammadi K, Effatpanah H, Yadegarazari R and Saidijam M:
Evaluation of miR-22 and miR-20a as diagnostic biomarkers for
gastric cancer. Chin Clin Oncol. 6:162017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang H, Tang J, Li C, Kong J, Wang J, Wu
Y, Xu E and Lai M: MiR-22 regulates 5-FU sensitivity by inhibiting
autophagy and promoting apoptosis in colorectal cancer cells.
Cancer Lett. 356:781–790. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang C, Ning S, Li Z, Qin X and Xu W:
miR-22 is down-regulated in esophageal squamous cell carcinoma and
inhibits cell migration and invasion. Cancer Cell Int. 14:1382014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fan W, Huang J, Xiao H and Liang Z:
MicroRNA-22 is downregulated in clear cell renal cell carcinoma,
and inhibits cell growth, migration and invasion by targeting PTEN.
Mol Med Rep. 13:4800–4806. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Orom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Szymanski M, Barciszewska MZ, Erdmann VA
and Barciszewski J: A new frontier for molecular medicine:
noncoding RNAs. Biochim Biophys Acta. 1756:65–75. 2005.PubMed/NCBI
|
|
16
|
Han Y, Liu Y, Gui Y and Cai Z: Long
intergenic non-coding RNA TUG1 is overexpressed in urothelial
carcinoma of the bladder. J Surg Oncol. 107:555–559. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xie HW, Wu QQ, Zhu B, Chen FJ, Ji L, Li
SQ, Wang CM, Tong YS, Tuo L, Wu M, et al: Long noncoding RNA
SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and
associated with poor prognosis. Tumour Biol. 35:7743–7754. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long Noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
Interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tian X and Xu G: Clinical value of lncRNA
MALAT1 as a prognostic marker in human cancer: Systematic review
and meta-analysis. BMJ Open. 5:e0086532015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luan W, Li L, Shi Y, Bu X, Xia Y, Wang J,
Djangmah HS, Liu X, You Y and Xu B: Long non-coding RNA MALAT1 acts
as a competing endogenous RNA to promote malignant melanoma growth
and metastasis by sponging miR-22. Oncotarget. 7:63901–63912. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang H, Chen Z, Wang X, Huang Z, He Z and
Chen Y: Long non-coding RNA: A new player in cancer. J Hematol
Oncol. 6:372013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu
YY, Wang SM, He FT and Yang SM: Long noncoding RNA BC032469, a
novel competing endogenous RNA, upregulates hTERT expression by
sponging miR-1207-5p and promotes proliferation in gastric cancer.
Oncogene. 35:3524–3534. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei Y and Niu B: Role of MALAT1 as a
prognostic factor for survival in various cancers: A systematic
review of the literature with meta-analysis. Dis Markers.
2015:1646352015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gutschner T, Hammerle M and Diederichs S:
MALAT1 - a paradigm for long noncoding RNA function in
cancer. J Mol Med. 91:791–801. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yu F, Lu Z, Cai J, Huang K, Chen B, Li G,
Dong P and Zheng J: MALAT1 functions as a competing endogenous RNA
to mediate Rac1 expression by sequestering miR-101b in liver
fibrosis. Cell Cycle. 14:3885–3896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu H, He Y, Lin L, Qi Z, Ma L, Li L and Su
Y: Long non-coding RNA MALAT1 modulates radiosensitivity of HR-HPV+
cervical cancer via sponging miR-145. Tumour Biol. 37:1683–1691.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xiao H, Tang K, Liu P, Chen K, Hu J, Zeng
J, Xiao W, Yu G, Yao W, Zhou H, et al: LncRNA MALAT1 functions as a
competing endogenous RNA to regulate ZEB2 expression by sponging
miR-200s in clear cell kidney carcinoma. Oncotarget. 6:38005–38015.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiong J: Emerging roles of microRNA-22 in
human disease and normal physiology. Curr Mol Med. 12:247–258.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Budd WT, Seashols-Williams SJ, Clark GC,
Weaver D, Calvert V, Petricoin E, Dragoescu EA, O'Hanlon K and
Zehner ZE: Dual action of miR-125b as a tumor suppressor and
OncomiR-22 promotes prostate cancer tumorigenesis. PLoS One.
10:e01423732015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Palacios F, Abreu C, Prieto D, Morande P,
Ruiz S, Fernández-Calero T, Naya H, Libisch G, Robello C, Landoni
AI, et al: Activation of the PI3K/AKT pathway by microRNA-22
results in CLL B-cell proliferation. Leukemia. 29:115–125. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang S, Zhang D, Yi C, Wang Y, Wang H and
Wang J: MicroRNA-22 functions as a tumor suppressor by targeting
SIRT1 in renal cell carcinoma. Oncol Rep. 35:559–567. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang Y, Jin X, Xiang Y, Chen Y, Shen CX,
Zhang YC and Li YG: The lncRNA MALAT1 protects the endothelium
against ox-LDL-induced dysfunction via upregulating the expression
of the miR-22-3p target genes CXCR2 and AKT. FEBS Lett.
589:3189–3196. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang D, Chen J, Chen H, Duan Z, Xu Q, Wei
M, Wang L and Zhong M: Leptin regulates proliferation and apoptosis
of colorectal carcinoma through PI3K/Akt/mTOR signalling pathway. J
Biosci. 37:91–101. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang C, Mao ZP, Wang L, Wu GH, Zhang FH,
Wang DY and Shi JL: Long non-coding RNA MALAT1 promotes
cholangiocarcinoma cell proliferation and invasion by activating
PI3K/Akt pathway. Neoplasma. 64:725–731. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Dong Y, Liang G, Yuan B, Yang C, Gao R and
Zhou X: MALAT1 promotes the proliferation and metastasis of
osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol.
36:1477–1486. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou X, Liu S, Cai G, Kong L, Zhang T, Ren
Y, Wu Y, Mei M, Zhang L and Wang X: Long non coding RNA MALAT1
promotes tumor growth and metastasis by inducing
epithelial-mesenchymal transition in oral squamous cell carcinoma.
Sci Rep. 5:159722015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han
Z, Sui H, Tang Y, Wang Y, Liu N, et al: Long non-coding RNA MALAT1
promotes tumour growth and metastasis in colorectal cancer through
binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2
complex. Br J Cancer. 111:736–748. 2014. View Article : Google Scholar : PubMed/NCBI
|