Introduction
Head and neck squamous cell carcinoma (HNSCC) is the
sixth most common cancer type with poor prognosis globally from
2005–2015 (1). Despite advances in
its diagnosis and treatment, the 5-year survival rate (1983–2002)
of patients with late stage HNSCC globally was 30–50% and has
improved slightly in recent decades (2). Therefore, development of novel
therapies with high efficacy and low toxicity is required.
Maternal embryonic leucine zipper kinase (MELK) has
been reported to serve critical roles in cancer cell proliferation
and maintenance of stemness (3). It
was previously reported that MELK is a promising target for cancer
therapy and we developed a potent MELK inhibitor, OTS167, which
exhibited effective growth suppression in mice xenograft models of
a number of cancer types, including breast, lung, prostate, and
pancreas cancer (4,5). At present, the therapeutic potential
of OTS167 is being evaluated in clinical trials, including
NCT01910545 (6), NCT02795520
(7) and NCT02926690 (8).
Cancer stem cells (CSCs) are more resistant to
chemotherapy/radiation than non-stem cancer cells and are
considered to serve important roles in cancer recurrence and/or
metastasis (9). Numerous studies
for targeting CSCs have purported pluripotency-associated
transcription factor (TF) SRY-box 2 (SOX2) to be a therapeutic
target due to its gene amplification and/or overexpression in
cancer cells (10–12). The SOX family is a group of TFs that
have been demonstrated to have critical roles in developmental and
stem cell biology (13). The SOX2
gene is located on chromosome 3q26.3-q27, belongs to the SOXB1
group and encodes for a protein consisting of 317 amino acids
(14). The aberrant expression of
SOX2, which promotes cellular proliferation, invasion, migration,
metastasis and evading apoptotic signals, has been reported in a
number of human cancer types, including lung, esophageal,
pancreatic, breast, ovarian and hepatocellular carcinoma, as well
as head and neck cancer (HNSCC) (15–19).
In HNSCC, SOX2 was demonstrated to regulate
self-renewal and tumorigenicity of HNSCC stem-like cells (20). Ectopic expression of SOX2 induced
cell proliferation and enhanced stemness-associated features
(20). Knockdown of SOX2 in HNSCC
CSCs attenuated their self-renewal capacity, chemoresistance,
invasiveness and in vivo tumorigenicity (18). Meta-analysis to examine associations
between SOX2 expression levels and clinicopathological/prognostic
parameters in HNSCC of 7 studies (9 cohorts) indicated that the
high SOX2 expression was strongly associated with unfavorable
5-year overall survival [hazard ratio (HR), 1.54; 95% confidence
interval (CI), 1.09–2.18] and disease-free survival rate (HR, 1.54;
95% CI, 1.13–2.10) of patients (21–27).
Additionally, increased SOX2 expression was also significantly
associated with a high tumor grade and an advanced TNM stage as
well as metastatic lymph-node status and distant metastasis
(28).
In the present study, the effect of MELK knockdown
and inhibition of MELK enzymatic activity by its inhibitor, OTS167,
on SOX2 expression in HNSCC cells was reported. The data indicated
that the MELK inhibitor may be a promising modality for the
treatment of HNSCC.
Materials and methods
Cell lines
The HNSCC cell lines UD-SCC-2, UT-SCC-40,
HN-SCC-151, FaDu, JSQ-3, HN-5, HN-6 and HN13 were provided by Dr
Tanguy Y. Seiwert (Department of Medicine, University of Chicago,
Chicago, IL, USA). The YD-10B cell line was purchased from the
Korean Cell Line Bank (KCLB; KCLB no. 60503; Korean Cell Line
Research Foundation, Seoul, Korea). UD-SCC-2, HN-SCC-151, JSQ-3 and
UT-SCC-40 cells were cultured in Dulbecco's modified Eagle's medium
(DMEM)/F12 (Life Technologies; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% FBS (Gemini Bio-Products, West
Sacramento, CA, USA) and 1X antibiotic-antimycotic solution
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). HN13, HN-5, HN-6
and YD-10B cells were cultured in DMEM (Life Technologies; Thermo
Fisher Scientific, Inc.) with 10% FBS and 1X antibiotic-antimycotic
solution. FaDu was cultured in RPMI-1640 media (Life Technologies;
Thermo Fisher Scientific, Inc.) with 10% FBS and 1X
antibiotic-antimycotic solution. All cells were maintained at 37°C
in humidified air in an atmosphere containing 5%
CO2.
Sphere formation
For the sphere formation assay, 1×103
cells of 6 individual HNSCC cell lines (HN13, FaDu, UD-SCC-2, HN-6,
YD-10B and HN-5) with a high or undetectable expression level of
MELK were seeded onto Ultra-Low attachment 60 mm plates (Corning
Incorporated, Corning, NY, USA) and cultured in corresponding
serum-free medium for two weeks at 37°C in humidified air in an
atmosphere containing 5% CO2. A total of 500 µl fresh
medium corresponding to each cell line was added every 3–4 days.
The spheres were observed and recorded with an Invitrogen EVOS FL
Auto 2 Cell Imaging System under an Evos FL Auto 2 light microscope
(×40; Invitrogen; Thermo Fisher Scientific, Inc.).
Western blot analysis and
antibodies
A total of 9 cell lines (FaDu, HN13, UD-SCC-2, HN-6,
UT-SCC-40, HN-SCC-151, JSQ-3, YD10-B and HN-5) were lysed on ice
with IP lysis buffer (Thermo Fisher Scientific, Inc.) containing
protease inhibitor cocktail set III (1:1,000) and phosphatase
inhibitor cocktail set I (1:100) (EMD Millipore, Billerica, MA,
USA) to evaluate the MELK expression level. HN13 and UT-SCC-40 were
used to evaluate the change of SOX2 expression level after MELK
inhibition following the same lysis protocol aforementioned.
Protein concentration was measured using a BCA assay. A total of 20
µg lysate protein were loaded and separated by electrophoresis
using an Any kD precast polyacrylamide gel (Bio-Rad Laboratories,
Inc., Hercules, CA, USA) and transferred onto a polyvinylidene
fluoride (PVDF) membrane. Following blocking with 5% skim milk
(Thermo Fisher Scientific, Inc.) in TBS with 0.1% Tween-20 (TBST)
buffer for 1 h at room temperature, membranes were incubated with
the primary antibodies, anti-MELK monoclonal antibody [1:5,000;
produced in-house, as previously described in (29)] or anti-SOX2 monoclonal antibody
(1:1,000; cat. no. MA1-014; Thermo Fisher Scientific, Inc.)
overnight at 4°C. Anti-β-actin antibody (1:10,000; cat. no. A5441;
Sigma-Aldrich; Merck KGaA) was used as a loading control and
incubated with membranes overnight at 4°C. After washing three
times with TBST buffer, membranes were incubated with rabbit
anti-mouse IgG-horseradish peroxidase (HRP) antibody as the
secondary antibody (1:1,000; cat. no. Sc358917; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) for 1 h at room temperature.
The expression level of MELK in each cancer cell line was
quantified with the ImageJ 1.51 h software (National Institutes of
Health, Bethesda, MD, USA) and normalized with that of β-actin with
GraphPad Prism version 7.01 (GraphPad Software, Inc., La Jolla, CA,
USA).
Growth suppressive effect of OTS167 in
HNSCC cells
MTT was conducted using Cell Counting Kit-8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan), according to the
manufacturer's protocols, to calculate the half-maximum inhibitory
concentration (IC50) value of OTS167 on cell lines. In
brief, 4×103 cells were seeded into 96-well flat plates.
Cells treated with graded concentrations of OTS167 were cultured
for 72 h at 37°C in an atmosphere containing 5% CO2.
OTS167 was provided by OncoTherapy Science, Inc. (Kawasaki, Japan).
UD-SCC2-2, HN13 and FaDu cells, which represent cell lines with
high MELK expression level, were treated with OTS167 at
concentrations of 0, 0.39, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50,
100, 500 or 1,000 nM. UT-SCC-40 cells and JSQ-3 cells represent
cell lines with moderate MELK expression level. UT-SCC-40 cells
were treated with OTS167 at concentrations of 0, 1.2, 2.4, 4.9,
9.8, 19.5, 39, 78, 156, 312, 625 or 1,000 nM. JSQ-3 cells were
treated with OTS167 at concentrations of 0, 0.6, 1.2, 2.4, 4.9,
9.8, 19.5, 39, 78, 156, 312 or 625 nM. YD-10B and HN-5 cells, which
represent cell lines with undetectable MELK expression level, were
treated with OTS167 at concentrations of 0, 0.78, 1.56, 3.12, 6.25,
12.5, 25, 50, 100, 200 or 1,000 nM. To quantify cell viability, the
96-well plate was measured at a wavelength of 450 nm in an iMark
microplate absorbance reader (Bio-Rad Laboratories, Inc.) following
reaction for 1–4 h at 37°C until the maximum absorbance reached
around 1. All these experiments were conducted in triplicate. The
IC50 values of OTS167 were calculated with GraphPad
Prism version 7.01.
Oligo small interfering RNA (siRNA)
and transfection
To knockdown MELK gene expression, HN13 and
UT-SCC-40 cells were transfected with 200 pmol oligonucleotide
siRNA using Lipofectamine® RNAiMAX (Invitrogen; Thermo
Fisher Scientific, Inc.), following the manufacturer's protocols.
The target sequence of oligo siMELK (200 pmol; Sigma-Aldrich, Merck
KGaA) was 5′-GACAUCCUAUCUAGCUGCA-3′ for MELK. Western blot analysis
was conducted to evaluate the efficiency of MELK knockdown with
siRNA at the 70% cell confluency condition and optimize the
transfection efficiency, as aforementioned. SIC001 Mission siRNA
Universal from Sigma-Aldrich (200 pmol; Merck KGaA) was used as a
control (siControl). After 48 h of transfection, the subsequent
analysis was performed.
MELK inhibition by OTS167
treatment
To inhibit MELK in HNSCCs, HN13 and UT-SCC-40 cells
were treated with OTS167 dissolved in dimethyl sulfoxide (DMSO) at
different concentrations (equivalent to 1X, 2X, 4X and 8X
IC50 concentrations) for 48 h at 37°C, cells treated by
equal amounts of DMSO were used as control.
In vitro kinase assay
MELK recombinant protein was obtained from
OncoTherapy Science, Inc. Histone H3 recombinant protein (EMD
Millipore, Burlington, MA) was used as a positive control in the
in vitro kinase assay. ATP (10 mM; cat. no. 9804) and kinase
buffer (10X; cat. no. 9802) were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA). In each reaction, 0.15 µM SOX2
recombinant protein (0.26 µg; cat. no. LS-G62-25; LifeSpan
Biosciences, Inc., Seattle, WA, USA) or 0.15 µM Histone H3
recombinant protein (0.13 µg; cat. no. 14-494; EMD Millipore) were
mixed with 0.15 µM MELK recombinant protein (0.29 µg) in 50 µl
kinase buffer and incubated for 2 h at 30°C. The reaction was
terminated by the addition of 1% SDS sample buffer (Thermo Fisher
Scientific, Inc.) and boiled at 70°C for 5 min. Finally, the total
reacted 50 µl samples were electrophoresed on an Any kD precast
polyacrylamide gel, transferred onto a PVDF membrane and followed
with the standard western blot analysis protocols, as
aforementioned. The primary antibody (anti-phospho-Ser/Thr/Tyr
antibody; 1:1,000; cat. no. ab15556) was purchased from Abcam
(Cambridge, MA, USA) and incubated with membranes overnight at 37°C
Rabbit anti-mouse IgG-HRP antibody was used as the secondary
antibody (1:1,000; cat. no. Sc358917; Santa Cruz Biotechnology) and
incubated for 1 h at room temperature.
Promoter binding TF profiling plate
array
A promoter binding TF profiling plate array
(Promoter-Binding TF Profiling Array II; Signosis, Inc., Santa
Clara, CA, USA) was performed according to the manufacturer's
protocols. Briefly, 96 biotin-labeled TF probes were mixed with
nuclear extract of HN13 cells with or without the SOX2 promoter
DNA. SOX2 promoter DNA was amplified by Q5®
High-Fidelity DNA Polymerase (New England Biolab, Inc., Ipswich,
MA, USA) with SOX2 promoter primers (Sigma-Aldrich, Merck KGaA)
detailed in Table I and PCR was
performed under the following thermocycling conditions: 95°C for 10
min, followed by 35 cycles of 95°C for 15 sec, 60°C for 30 sec and
72°C for 30 sec. PCR products was purified with a MinElute Gel
Extraction kit (Qiagen, Inc., Valencia, CA, USA). The TF-DNA
complex was separated from free probes using an isolation column,
and the TF-bound probes were eluted using elution buffer.
Hybridization of eluted probes was performed with hybridization
plate. The bound probe was detected using a
streptavidin-horseradish peroxidase conjugate and measured with a
SynergyH4 plate reader (BioTek Instruments, Inc., Winooski, VT,
USA). The decreased fold of signal was calculated.
 | Table I.Primers used in real time RT-PCR and
promoter-binding TFs assay. |
Table I.
Primers used in real time RT-PCR and
promoter-binding TFs assay.
| Primer | Forward | Reverse |
|---|
| MELK |
5′-GCTGCAAGGTATAATTGATGGA-3′ |
5′-CAGTAACATAATGACAGATGGGC-3′ |
| SOX2 |
5′-CCCCCGGCGGCAATAGCA-3′ |
5′-TCGGCGCCGGGGAGATACAT-3′ |
| C/EBP A |
5′-TGTATACCCCTGGTGGGAGA-3′ |
5′-TCATAACTCCGGTCCCTCTG-3′ |
| C/EBP B |
5′-GACAAGCACAGCGACGAGTA-3′ |
5′-AGCTGCTCCACCTTCTTCTG-3′ |
| Pbx1 |
5′-CAGATGCAGCTCAAGCAGAG-3′ |
5′-CTCTTTGGCTTCCTTCACTGG-3′ |
| Nkx2-5 |
5′-CTCAACAGCTCCCTGACTC-3′ |
5′-CTCATTGCACGCTGCATAAT-3′ |
| SMUC |
5′-GGCCACACACTGTCTCCAC-3′ |
5′-GTCGTTCAGGACACAGCAGA-3′ |
| SOX9 |
5′-TACGACTACACCGACCACCA-3′ |
5′-CTCCTCAAGGTCGAGTGAGC-3′ |
|
SOX2-promoter |
5′-TGAGAGAGTGTTGGCACCTG-3′ |
5′-GGGTTTCTAGCGACCAATCA-3′ |
| GAPDH |
5′-CGACCACTTTGTCAAGCTCA-3′ |
5′-GGTTGAGCACAGGGTACTTTATT-3′ |
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HN13 and UT-SCC-40
cells using a RNeasy Mini kit (Qiagen, Inc.), and then 2 µg RNA was
reverse transcribed using a SuperScript III First-Strand Synthesis
System (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocols. The reaction system was incubated for 60
min at 42°C. RT-qPCR was performed using primers (Sigma-Aldrich,
Merck KGaA) and SYBR®-Green Real-Time PCR Master mix
(Thermo Fisher Scientific, Inc.) in a ViiA 7 system (Life
Technologies; Thermo Fisher Scientific, Inc.). The PCR conditions
were as follows: 95°C for 10 min followed by 40 cycles of 95°C for
15 sec and 60°C for 1 min. The PCR primer sequences were listed in
Table I. The expression levels were
normalized with that of GAPDH with the ΔΔCq method (30).
Statistical analysis
Data analysis was conducted using GraphPad Prism
version 7.01. Data are presented as mean ± standard deviation.
Differences between two groups were calculated for significance
using Student's t-test and differences between multiple groups were
calculated for significance using the analysis of variance analysis
followed by Dunnett's multiple comparisons test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Increased MELK expression associates
with stronger sphere formation ability
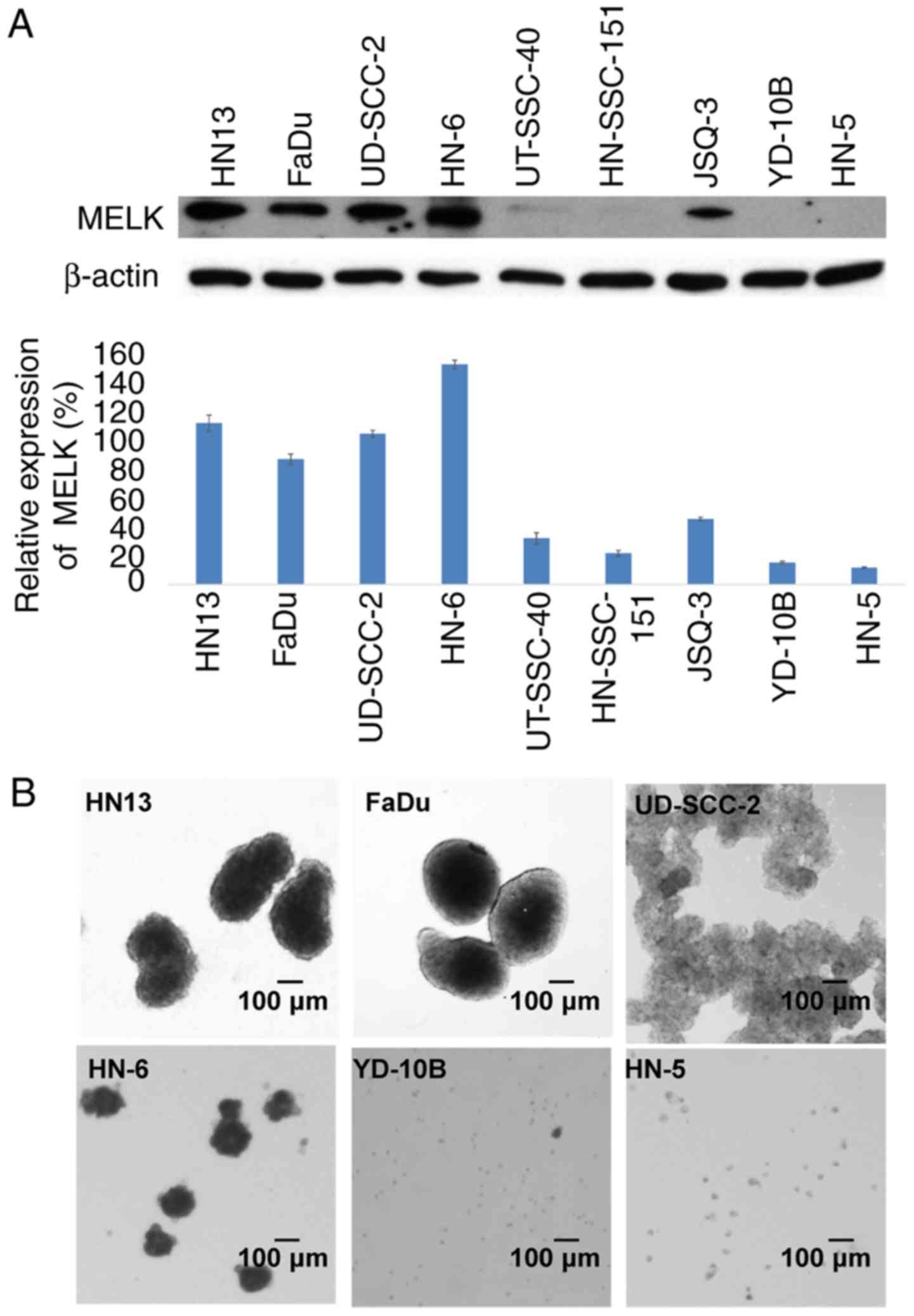
To investigate a role of MELK expression in the
stemness of HNSCC cells, the MELK expression levels was examined in
YD-10B, FaDu, HN-6, HN13, UT-SCC-40, HN-5, HN-SCC-151, JSQ-3 and
UD-SCC-2 cells by normalization with that of β-actin. The results
demonstrated that the relative expression levels of MELK in four
HNSCC cell lines, HN13, FaDu, UD-SCC-2 and HN-6, were notably high
(113, 87, 105 and 154% of the β-actin expression level,
respectively); whereas, in UT-SCC-40, HN-SCC-151, JSQ-3, YD-10B and
HN-5 cells, MELK expression levels were notably reduced (32, 21,
46, 15 and 11% of the β-actin expression level, respectively), as
depicted in Fig. 1A. Subsequent
sphere formation experiments of the two-week culture in serum-free
medium indicated that the four cell lines, HN13, FaDu, UD-SCC-2 and
HN-6, in which MELK had increased expression, formed large spheres,
compared with HN-5 and YD-10B cells with reduced MELK expression,
which formed small spheres. Additionally, UD-SCC-2 cells formed
numerous small spheres that stacked together (Fig. 1B).
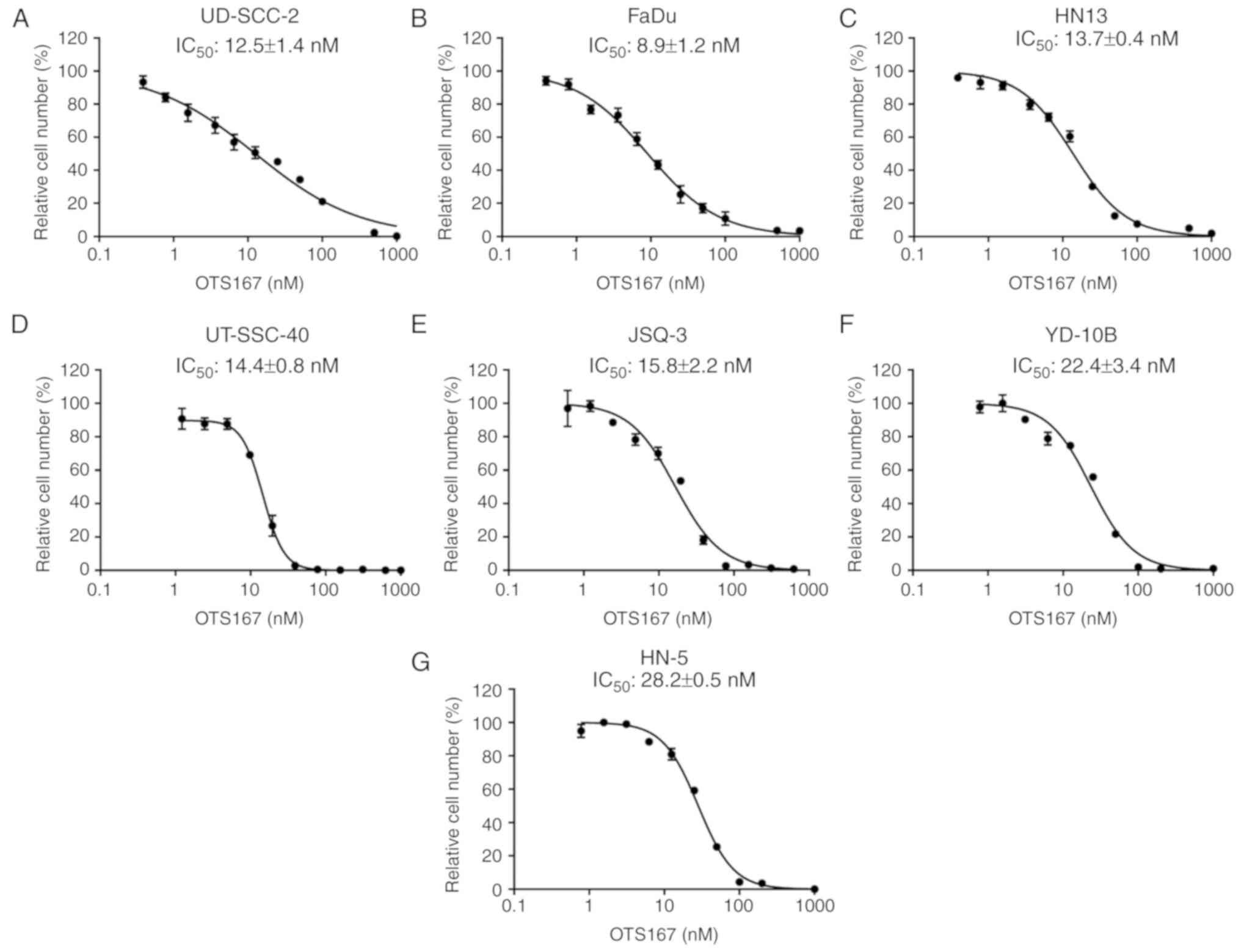
Growth suppressive effect of OTS167 in
HNSCC cancer cells
As previous reported, a potent MELK inhibitor,
OTS167, was developed, which exhibited a significant growth
suppressive effect on lung cancer cells (31), kidney cancer cells (32) as well as multiple myeloma cells
(33). In the present study, the
IC50 values of OTS167 were examined to measure its
growth inhibitory effect on seven HNSCC cell lines. This included
five HNSCC cell lines with relatively increased MELK expression
levels, UD-SCC-2, FaDu, HN13, UT-SCC-40 and JSQ-3 cells, which
exhibited relatively reduced IC50 values with a range of
8.9±1.2 to 15.8±2.2 nM, compared with the two cell lines with
reduced MELK expression, YD-10B and HN-5 cells, with 22.4±3.4 and
28.2±0.5 nM, respectively (Fig.
2).
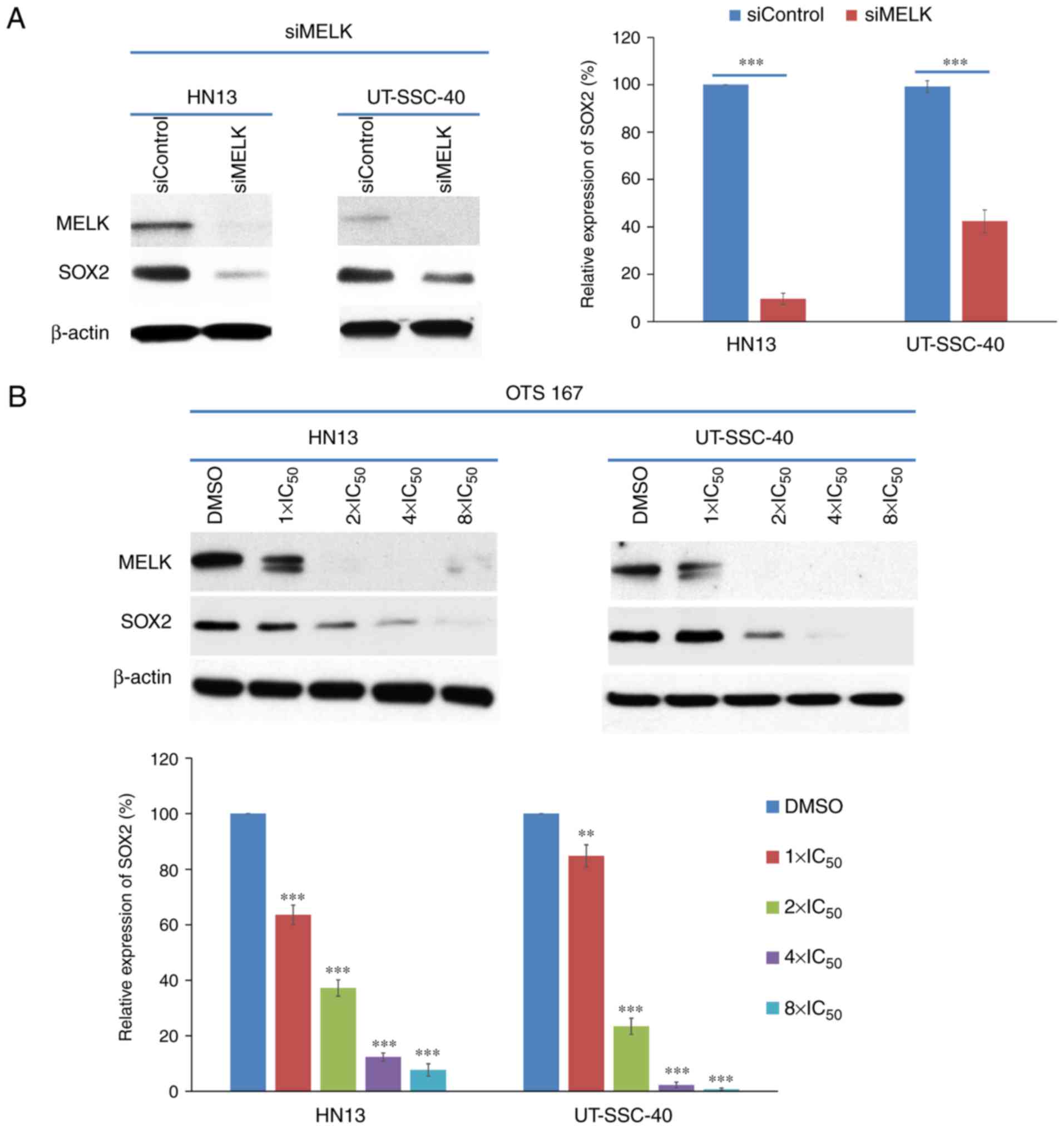
Knockdown with siRNA and OTS167
treatment could decrease SOX2 expression in HNSCC cells
Since SOX2 is considered as one of major
stemness-associated genes (10,11),
whether MELK expression would affect the SOX2 expression levels in
HNSCC cells was examined. Knockdown of MELK with siRNA decreased
SOX2 expression in HN13 and UT-SCC-40 cells. At 48 h after siMELK
transfection, the SOX2 expression level was significantly
decreased, to 9.6±2.4%, compared with HN13 cells transfected with
control siRNA (P<0.001). However, UT-SCC-40 cells, which had
reduced MELK expression, exhibited significant SOX2 downregulation
(decreased to 43.4±4.8%; P<0.001), compared with the cells
treated with control siRNA (Fig.
3A). These results indicated that SOX2 expression may be
regulated by multiple factors, including MELK, which may be a major
regulator of SOX2 expression in HN13 cells.
Additionally, HNSCC cells were treated with OTS167
at different concentrations (equivalent to 1X, 2X, 4X and 8X
IC50 concentrations) and it was identified that SOX2
expression was significantly downregulated in an OTS167
dose-dependent manner in the cell lines treated with OTS167
(P<0.0001, compared with the cells treated with DMSO), as
depicted in Fig. 3B. Due to MELK
being a kinase and serving a critical role through the
phosphorylation pathway (3–5), whether MELK could directly
phosphorylate SOX2 was examined with an in vitro kinase
assay using recombinant proteins.
In the in vitro kinase assay, without
co-incubation with MELK recombinant protein, Histone H3 alone also
exhibited a slight phosphorylation signal (Fig. 4; lane 3), which may be due to the
contamination of unknown protein kinase(s) during the purification
of recombinant Histone H3 protein. However, co-incubation of
Histone H3 protein with MELK exhibited a strong phosphorylation
signal (Fig. 4; lanes 6),
indicating that the MELK recombinant protein could effectively
phosphorylate the Histone H3 protein, which confirmed the
efficiency of the in vitro kinase assay strategy.
Furthermore, co-incubation of SOX2 with MELK demonstrated no
phosphorylation signal (Fig. 4;
lane 5), implying that SOX2 may not be a direct substrate of MELK.
Due to SOX2 gene expression being suppressed at a similar level in
two cell lines, OTS167 is considered to decrease SOX2 expression in
UT-SCC-40 cells by another mechanism, such as an off-target effect
(34).
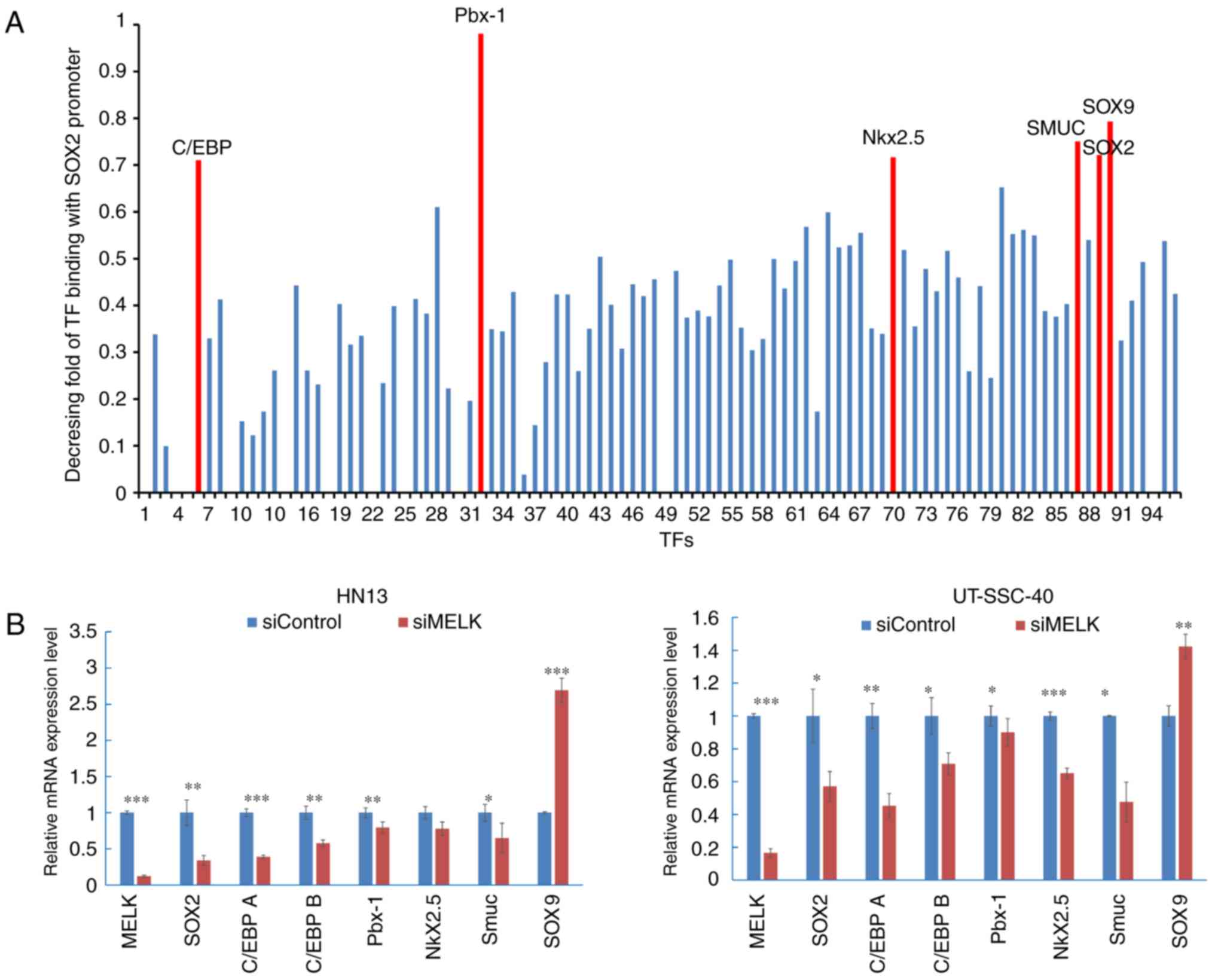
Knockdown of MELK decreases a series
of TFs of SOX2
To investigate a mechanism for SOX2 downregulation
by MELK inhibition, possible TFs of SOX2 were examined. SOX2
expression is reported to be exquisitely controlled in mammals and
it may be regulated by a large number of TFs, including serine
racemase 1 (SRR1) and SRR2 and forkhead box O1 (35). Therefore, a promoter-binding TF
profiling assay was performed to screen TFs that could bind to the
promoter region of SOX2 in HN13 cells. In the assay, the
synthesized DNA corresponding to a SOX2 promoter region was mixed
with a set of 96 biotin-labelled oligonucleotides corresponding to
96 TFs along with nuclear extract of cells. When the SOX2 promoter
DNA competes with the biotin-labelled oligonucleotide for the
binding to TFs in the nuclear extract, it causes no or reduced
detection of luminescent signals when the TF plate hybridized with
eluted probes. The decreasing fold of luminescent signals with or
without the SOX2 promoter DNA fragment was calculated and TFs with
a greater decreasing fold (>0.7) were selected. CCAAT enhancer
binding protein (C/EBP), Pbx1, Nkx2.5, snail family transcription
repressor 3 (SMUC), SOX2 and SOX9 were considered to bind strongly
to the SOX2 promoter region in the cancer cells examined (Fig. 5A). RT-qPCR was then performed to
quantify the expression levels of TF candidates in MELK-knockdown
cells and the control cells, and it was determined that following
knockdown of MELK in HN13 cells, SOX2 was downregulated to
0.34±0.06 (P<0.01), compared with the siControl group (1±0.17);
C/EBP A was downregulated to 0.39±0.02 (P<0.001), compared with
the siControl group (1±0.05); C/EBP B was downregulated to
0.58±0.04 (P<0.01), compared with the siControl group (1±0.09);
Pbx1 was downregulated to 0.79±0.08 (P<0.01), compared with the
siControl group (1±0.06); Nkx2.5 was downregulated to 0.78±0.09
(P>0.05), compared with the siControl group (1±0.08); and SMUC
were downregulated to 0.65±0.02 (P<0.05), compared with the
siControl group (1±0.12). In UT-SCC-40 cells, SOX2 was
downregulated to 0.57±0.09 (P<0.05), compared with the siControl
group (1±0.16); C/EBP A was downregulated to 0.45±0.07 (P<0.01),
compared with the siControl group (1±0.07); C/EBP B was
downregulated to 0.71±0.06 (P<0.05), compared with the siControl
group (1±0.06); Pbx1 was downregulated to 0.90±0.08 (P<0.05),
compared with the siControl group (1±0.08); Nkx2.5 was
downregulated to 0.65±0.03 (P<0.001), compared with the
siControl group (1±0.03); and SMUC were downregulated to 0.48±0.12
(P<0.05) compared with the siControl group (1±0.01). (Fig. 5B). However, SOX9 expression was
significantly increased in the cells, compared with the control
cells (P<0.001 in HN13, P<0.01 in UT-SCC40).
Discussion
It was previously reported that MELK could be a
promising target for cancer therapy and developed its potent
inhibitor, OTS167, which could effectively suppress MELK activity
and inhibit the phosphorylation of various MELK substrates,
including B-cell lymphoma 2 like 14, cell division cycle 25B,
mitogen-activated protein kinase kinase kinase 5 and zinc finger
protein 622 (4,5). It has to date been identified that
OTS167 exhibited significant suppressive effects on various cancer
cell lines, including breast cancer, kidney cancer, small cell lung
cancer (SCLC) and multiple myeloma (4,5,29,31–33).
Additionally, it was demonstrated that OTS167 could target CSCs and
suppress the mammosphere formation of cancer stem-like cells
(4,5). In a number of SCLC cell lines, it was
determined that OTS167 treatment could induce cytokinetic defects
with intercellular bridges and the formation of neuronal
protrusions accompanied with an increase of a neuronal
differentiation markerx (cycle of differentiation 56), indicating
that this compound may induce differentiation of cancer stem-like
cells to neuron-like cells (33).
In the present study, it was demonstrated that HNSCC
cells with increased MELK expression had an improved ability to
produce large spheres under the serum-free conditions. It was then
revealed that MELK inhibition by siRNA and OTS167 could
significantly downregulate SOX2 expression in HNSCC cells and it
was determined that the expression levels of a set of SOX2
transcriptional factors were decreased following MELK inhibition.
This indicated that OTS167 may target CSCs through the
downregulation of SOX2. Notably, it was previously reported in a
breast cancer MDA-MB-231 cell line, which has undifferentiated,
cancer stem-like characteristics, that snail family transcriptional
repressor 2 (Slug), a CSC marker, was determined to be reduced with
OTS167 treatment in a dose-dependent manner (5). Slug expression increased SOX2 and
Nanog expression and promoted the progression of hepatocellular
cancer. Knockdown of Slug with siRNA notably reduced SOX2 and Nanog
expression and resulted in inhibition of hepatocellular carcinoma
cell migration in vitro (36). These data indicated that there may
be an association between MELK, Slug and SOX2.
However, the present study has a number of
limitations, which should be addressed in the future. First of all,
although it was demonstrated that MELK inhibition affected the
expression levels of transcriptional factors for SOX2, the detailed
mechanism was not clarified. It would be beneficial to examine the
TF profiles with a chromatin immunoprecipitation assay to further
define the SOX2 TFs in these cell lines. The commercial
phosphorylated (p)-SOX2 or p-histone generally only targets a
number of specific sites of the proteins. However, in the present
study, there was no information available regarding the sites of
SOX2 that could be phosphorylated by MELK. Phosphorylation
frequently occurs at Ser/Thr or Tyr sites; therefore, the universal
antibody (anti-phospho-Ser/Thr/Tyr antibody) was used to include as
many phosphorylation sites as possible. Furthermore, additional
analyses regarding how MELK inhibition would affect the stemness,
migration and invasion of these cancer cells should also be
performed to obtain comprehensive information for the clinical
application of OTS167.
Although further investigation is necessary, the
present data demonstrates that MELK inhibition could target cancer
stemness through the suppression of SOX2 expression. It was
reported that partial suppression of SOX2 expression reduced the
cell viability, sphere formation, clonal growth as well as
tumorigenicity in multiple cancer types, including glioblastoma,
small-cell lung cancer and numerous forms of squamous cell
carcinoma (37–39). Considering the ability to target
CSCs as well as cancer cells, the MELK inhibitor may be a promising
drug in clinical applications for HNSCC.
Acknowledgements
The authors would like to thank Dr Tanguy Y. Seiwert
(Department of Medicine, University of Chicago, Chicago, IL, USA),
who provided the HNSCC cell lines (UD-SCC-2, UT-SCC-40, HN-SCC-151,
FaDu, JSQ-3, HN-5, HN-6 and HN13) for this research.
Funding
This study was supported by a research grant from
OncoTherapy Science Inc. and Science and Technology Planning
Project of Guangdong Province (grant no. 2016A020215207).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
YN designed and supervised the project and edited
the manuscript. LR performed the majority of the experiments and
wrote the manuscript. BD assisted in the experiments and data
analysis. VS set up and assisted with the sphere formation
experiment and the culture of cancer cell lines. JHP directed and
supervised the techniques, designed the project and was involved in
the data analysis. All authors read and approved the manuscript and
agree to be accountable for all aspects of the research in ensuring
that the accuracy or integrity of any part of the work are
appropriately investigated and resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
YN is a stock holder and a scientific advisor of
OncoTherapy Science, Inc. JP is a scientific advisor of OncoTherapy
Science, Inc. Oncotherapy Science, Inc., also provided financial
support and certain of the reagents for this study. No potential
conflicts of interest were disclosed by the other authors.
Glossary
Abbreviations
Abbreviations:
|
MELK
|
maternal embryonic leucine zipper
kinase
|
|
HNSCC
|
head and neck squamous cell
carcinoma
|
|
CSC
|
cancer stem cell
|
References
|
1
|
Global Burden of Disease Cancer
Collaboration, ; Fitzmaurice C, Allen C, Barber RM, Barregard L,
Bhutta ZA, Brenner H, Dicker DJ, Chimed-Orchir O, Dandona R, et al:
Global, regional, and national cancer incidence, mortality, years
of life lost, years lived with disability, and disability-adjusted
life-years for 32 cancer groups, 1990 to 2015: A systematic
analysis for the global burden of disease study. JAMA Oncol.
3:524–548. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chaturvedi AK, Anderson WF,
Lortet-Tieulent J, Curado MP, Ferlay J, Franceschi S, Rosenberg PS,
Bray F and Gillison ML: Worldwide trends in incidence rates for
oral cavity and oropharyngeal cancers. J Clin Oncol. 31:4550–4559.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ganguly R, Hong CS, Smith LG, Kornblum HI
and Nakano I: Maternal embryonic leucine zipper kinase: Key kinase
for stem cell phenotype in glioma and other cancers. Mol Cancer
Ther. 13:1393–1398. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chung S, Suzuki H, Miyamoto T, Takamatsu
N, Tatsuguchi A, Ueda K, Kijima K, Nakamura Y and Matsuo Y:
Development of an orally-administrative MELK-targeting inhibitor
that suppresses the growth of various types of human cancer.
Oncotarget. 3:1629–1640. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chung S and Nakamura Y: MELK inhibitor,
novel molecular targeted therapeutics for human cancer stem cells.
Cell Cycle. 12:1655–1656. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
simpleClinicalTrials.govPhase 1 study of OTS167 in
patients with solid tumors. https://clinicaltrials.gov/ct2/show/NCT01910545July
29–2013
|
|
7
|
simpleClinicalTrials.govPharmacological study of
intravenous OTS167 in patients with refractory or relapsed acute
myeloid leukemia, acute lymphoblastic leukemia, advanced
myelodysplastic syndromes, advanced myeloproliferative neoplastic
disorders, or advanced chronic myelogenous leukemia. https://clinicaltrials.gov/ct2/show/NCT02795520June
10–2016
|
|
8
|
simpleClinicalTrials.govSafety study of MELK inhibitor
to treat patients with advanced breast cancer and triple negative
breast cancer. https://clinicaltrials.gov/ct2/show/NCT02926690October
6–2016
|
|
9
|
Prince ME, Sivanandan R, Kaczorowski A,
Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles
LE: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Weina K and Utikal J: SOX2 and cancer:
Current research and its implications in the clinic. Clin Transl
Med. 3:192014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Herreros-Villanueva M, Zhang JS, Koenig A,
Abel EV, Smyrk TC, Bamlet WR, de Narvajas AA, Gomez TS, Simeone DM,
Bujanda L and Billadeau DD: SOX2 promotes dedifferentiation and
imparts stem cell-like features to pancreatic cancer cells.
Oncogenesis. 2:e612013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bareiss PM, Paczulla A, Wang H, Schairer
R, Wiehr S, Kohlhofer U, Rothfuss OC, Fischer A, Perner S, Staebler
A, et al: SOX2 expression associates with stem cell state in
human ovarian carcinoma. Cancer Res. 73:5544–5555. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prior HM and Walter MA: SOX genes:
Architects of development. Mol Med. 2:405–412. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stevanovic M, Zuffardi O, Collignon J,
Lovell-Badge R and Goodfellow P: The cDNA sequence and chromosomal
location of the human SOX2 gene. Mamm Genome. 5:640–642. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai YR, Zhang HQ, Qu Y, Mu J, Zhao D, Zhou
LJ, Yan H, Ye JW and Liu Y: Expression of MET and SOX2 genes in
non-small cell lung carcinoma with EGFR mutation. Oncol Rep.
26:877–885. 2011.PubMed/NCBI
|
|
16
|
Gen Y, Yasui K, Nishikawa T and Yoshikawa
T: SOX2 promotes tumor growth of esophageal squamous cell carcinoma
through the AKT/mammalian target of rapamycin complex 1 signaling
pathway. Cancer Sci. 104:810–816. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang Z, Pan X, Gao A and Zhu W: Expression
of Sox2 in cervical squamous cell carcinoma. J BUON. 19:203–206.
2014.PubMed/NCBI
|
|
18
|
Sun C, Sun L, Li Y, Kang X, Zhang S and
Liu Y: Sox2 expression predicts poor survival of hepatocellular
carcinoma patients and it promotes liver cancer cell invasion by
activating slug. Med Oncol. 30:5032013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lengerke C, Fehm T, Kurth R, Neubauer H,
Scheble V, Müller F, Schneider F, Petersen K, Wallwiener D, Kanz L,
et al: Expression of the embryonic stem cell marker SOX2 in
early-stage breast carcinoma. BMC Cancer. 11:422011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee SH, Oh SY, Do SI, Lee HJ, Kang HJ, Rho
YS, Bae WJ and Lim YC: SOX2 regulates self-renewal and
tumorigenicity of stem-like cells of head and neck squamous cell
carcinoma. Br J Cancer. 111:2122–2130. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo W, Li S, Peng B, Ye Y, Deng X and Yao
K: Embryonic stem cells markers SOX2, OCT4 and Nanog expression and
their correlations with epithelial-mesenchymal transition in
nasopharyngeal carcinoma. PLoS One. 8:e563242013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ge N, Lin HX, Xiao XS, Guo L, Xu HM, Wang
X, Jin T, Cai XY, Liang Y, Hu WH, et al: Prognostic significance of
Oct4 and Sox2 expression in hypopharyngeal squamous cell carcinoma.
J Transl Med. 8:942010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang X, Liang Y, Chen Q, Xu HM, Ge N, Luo
RZ, Shao JY, He Z, Zeng YX, Kang T, et al: Prognostic significance
of SOX2 expression in nasopharyngeal carcinoma. Cancer Invest.
30:79–85. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Du L, Yang Y, Xiao X, Wang C, Zhang X,
Wang L, Zhang X, Li W, Zheng G, Wang S, et al: Sox2 nuclear
expression is closely associated with poor prognosis in patients
with histologically node-negative oral tongue squamous cell
carcinoma. Oral Oncol. 47:709–713. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dai W, Tan X, Sun C and Zhou Q: High
expression of SOX2 is associated with poor prognosis in
patients with salivary gland adenoid cystic carcinoma. Int J Mol
Sci. 15:8393–8406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tang XB, Shen XH, Li L, Zhang YF and Chen
GQ: SOX2 overexpression correlates with poor prognosis in laryngeal
squamous cell carcinoma. Auris Nasus Larynx. 40:481–486. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gonzalez-Marquez R, Llorente JL, Rodrigo
JP, Garcia-Pedrero JM, Alvarez-Marcos C, Suarez C and Hermsen MA:
SOX2 expression in hypopharyngeal, laryngeal, and sinonasal
squamous cell carcinoma. Hum Pathol. 45:851–857. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong Z, Liu G, Huang B, Sun J and Wu D:
Prognostic significance of SOX2 in head and neck cancer: A
meta-analysis. Int J Clin Exp Med. 7:5010–5020. 2014.PubMed/NCBI
|
|
29
|
Chung S, Kijima K, Kudo A, Fujisawa Y,
Harada Y, Taira A, Takamatsu N, Miyamoto T, Matsuo Y and Nakamura
Y: Preclinical evaluation of biomarkers associated with antitumor
activity of MELK inhibitor. Oncotarget. 7:18171–18182. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Inoue H, Kato T, Olugbile S, Tamura K,
Chung S, Miyamoto T, Matsuo Y, Salgia R, Nakamura Y and Park JH:
Effective growth-suppressive activity of maternal embryonic
leucine-zipper kinase (MELK) inhibitor against small cell lung
cancer. Oncotarget. 7:13621–13633. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kato T, Inoue H, Imoto S, Tamada Y,
Miyamoto T, Matsuo Y, Nakamura Y and Park JH: Oncogenic roles of
TOPK and MELK, and effective growth suppression by small molecular
inhibitors in kidney cancer cells. Oncotarget. 7:17652–17664. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stefka AT, Park JH, Matsuo Y, Chung S,
Nakamura Y, Jakubowiak AJ and Rosebeck S: Anti-myeloma activity of
MELK inhibitor OTS167: Effects on drug-resistant myeloma cells and
putative myeloma stem cell replenishment of malignant plasma cells.
Blood Cancer J. 6:e4602016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin A, Giuliano CJ, Sayles NM and Sheltzer
JM: CRISPR/Cas9 mutagenesis invalidates a putative cancer
dependency targeted in on-going clinical trials. Elife.
6:e241792017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wuebben EL and Rizzino A: The dark side of
SOX2: Cancer - A comprehensive overview. Oncotarget. 8:44917–44943.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao X, Sun B, Sun D, Liu T, Che N, Gu Q,
Dong X, Li R, Liu Y and Li J: Slug promotes hepatocellular cancer
cell progression by increasing sox2 and nanog expression. Oncol
Rep. 33:149–156. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jia X, Li X, Xu Y, Zhang S, Mou W, Liu Y,
Liu Y, Lv D, Liu CH, Tan X, et al: SOX2 promotes tumorigenesis and
increases the anti-apoptotic property of human prostate cancer
cell. J Mol Cell Biol. 3:230–238. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tian Y, Jia X, Wang S, Li Y, Zhao P, Cai
D, Zhou Z, Wang J, Luo Y and Dong M: SOX2 oncogenes amplified and
operate to activate AKT signaling in gastric cancer and predict
immunotherapy responsiveness. J Cancer Res Clin Oncol.
140:1117–1124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sanada Y, Yoshida K, Ohara M, Oeda M,
Konishi K and Tsutani Y: Histopathologic evaluation of stepwise
progression of pancreatic carcinoma with immunohistochemical
analysis of gastric epithelial transcription factor S:
Comparison of expression patterns between invasive components and
cancerous or nonneoplastic intraductal components. Pancreas.
32:164–170. 2006. View Article : Google Scholar : PubMed/NCBI
|