Introduction
Pathological complete response (pCR) is defined by
the rate of the absence of residual invasive breast cancer disease
after preoperative neoadjuvant chemotherapy (NeoCh), and pCR has
been used as the primary end point in many neoadjuvant trials
(1). NeoCh constitutes an important
standard approach for locally advanced breast cancer, and it takes
place before surgical extraction of tumors with the objective of
reducing high tumor size (2). This
procedure aims to render locally advanced cancers operable, to
facilitate the removal of tumors, to allow breast-conserving
surgery, and to improve postoperative recovery and long-term
outcome for the patients (3,4). In
addition, neoadjuvant trials provide the opportunity to test new
drugs preoperatively in patients with locally invasive breast
cancer accordingly to subtypes and hormonal receptor status
(5–7). Large multi-centric studies have
stablished that patients who better benefit from NeoCh are those
who achieve a successful pCR which has been associated with both
improved disease-free survival (DFS) and overall survival (OS)
rates (8). For example, the CTNeoBC
study in a large cohort of women with breast cancer identified a
good association between pCR and DFS/OS (9). However, no specific and sensitive
biomarkers to predict the clinical response to NeoCh in breast
cancer have yet been defined.
MicroRNAs (miRNAs) are evolutionarily conserved
single-stranded tiny non-coding RNAs of 21–25 nucleotides in length
that function as negative post-transcriptional regulators of gene
expression (10). The mechanism of
action of miRNAs relies in the partially complementary binding with
the 3′ untranslated region (UTR) of specific target mRNAs resulting
in either translation inhibition or deadenylation-dependent
degradation of encoding protein transcripts in the cytoplasmic
P-bodies. Thus, these small RNAs function as guide molecules in
post-transcriptional gene silencing. miRNAs have normal functions
in eukaryotic cells including cell growth, differentiation,
survival and metabolism. However, alterations in the
transcriptional and epigenetic mechanisms leading to aberrant
expression of miRNAs and its target genes have been frequently
observed in breast cancer. To date, there is sufficient
experimental evidence which strongly links miRNAs with the
development and progression of breast cancer, as they function as
oncomiRs through the regulation of tumor-suppressor genes and
cellular oncogenes (11). Moreover,
changes in the abundance of miRNAs have been associated with
clinical and pathological features of patients. Notably, miRNAs
have also been recently investigated as potential predictors of
clinical response to cytotoxic therapy in diverse types of cancers
(12–19). Previously, we reported a miRNA
expression signature associated with pathological complete response
to NeoCh in triple negative breast cancer patients (19). In the present study, we focused on
the clinical and molecular analysis of miR-145-5p, as its
relationships with response to therapy have not been addressed in
triple negative breast cancer. Our data strongly suggest that
miR-145-5p could be a predictor of pCR to NeoCh in breast cancer
patients. Moreover, the present experimental findings demonstrated
a potential function for miR-145-5p as a regulator of cell
proliferation and apoptosis in breast cancer cells.
Materials and methods
Statement of ethics
The Breast Cancer Foundation (FUCAM) of Mexico
provided the breast tumors and normal tissue collection. The Ethics
Committee of the Breast Cancer Foundation (FUCAM) of Mexico
approved the protocols using human tissues. Signed informed consent
forms were obtained from the participants prior to release for
research use. This study was carried out in accordance with the
ethical standards of the committee and in accordance with the
Helsinki Declaration of 1975.
Tissue samples
Formalin-fixed paraffin-embedded (FFPE) tissues from
triple-negative breast cancer patients (n=32) who received
neoadjuvant cisplatin/doxorubicin-based chemotherapy at FUCAM
between November 2008 and August 2017 were collected. Tumor samples
were classified as with or without pathological complete response
(pCR) to neoadjuvant therapy. Patients were aged between 28 and 65
years; mean age was 46 years in the pCR group and 53 in the no-pCR
group. Pathologist confirmed the existence of at least 80% tumor
cells in the clinical specimens.
Cell lines
Human breast cancer cell lines were obtained from
the American Type Culture Collection (ATCC; Manassas, VA, USA).
MCF-7 (ATCC: HTB-22), MDA-MB-231 (ATCC: HTB-26), SKBR3 (ATCC:
HTB-30), BT-20 (ATCC: HTB19) and no-tumorigenic MCF-10A (ATCC:
CRL-10317) were routinely grown in Dulbecco's modified Eagle's
minimal essential medium (DMEM; Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), supplemented with 10% fetal bovine serum
(FBS) and penicillin-streptomycin (50 U/ml; Invitrogen; Thermo
Fisher Scientific, Inc.) in 5% CO2 atmosphere at
37°C.
Reverse transcription and real-time
polymerase chain reaction
Five serial 20-µm-thick sections of FFPE tissue
specimens were used for total RNA isolation using the RNeasy FFPE
kit (Qiagen Inc., Valencia, CA, USA) with modifications to the
manufacturer's protocol. Briefly, the sections were incubated twice
in xylene for 1 h at 63°C for deparaffinization followed by
purification of total RNA using TRIzol protocol (Ambion; Thermo
Fisher Scientific, Inc.). RNA concentration and purity were
analyzed for spectrophotometry (NanoDrop Technologies; Thermo
Fisher Scientific, Inc.), and integrity was evaluated by 1% agarose
gel electrophoresis. Quantitative RT-PCR (qRT-PCR) assay of
individual miR-145 was performed using MicroRNA assays (cat. no.
4427975; Thermo Fisher Scientific, Inc.). Briefly, 10 ng total RNA
were reverse transcribed using a stem looped-RT specific primer,
0.15 µl dNTPs (100 mM), 1.0 µl reverse transcriptase MultiScribe
(Thermo Fisher Scientific, Inc.) (50 U/µl), 1.5 µl 10X buffer, 0.19
µl RNase inhibitor (20 U/µl) and 4.16 µl RNase-free water. Then,
retrotranscription reaction (1:15 dilution) was mixed with 10 µl
TaqMan Universal PCR Master Mix, No AmpErase UNG 2X, 7.67 µl
RNase-free water and 1.0 µl PCR probe. PCR reaction was performed
in a GeneAmp System 9700 (Applied Biosystems; Thermo Fisher
Scientific, Inc.) as follows: 95°C for 10 min, and 40 cycles at
95°C for 15 sec and 60°C for 1 min. Tests were normalized using
RNU44 as internal control. Experiments were performed three times
in triplicate, and the results were expressed as mean ± SD.
Relative quantification was referred as ΔΔCq as previously
described (20). P<0.05 was
considered to indicate statistical significance.
miR-145-5p restoration in breast
cancer cells
miR-145-5p mimics (60 nM, GUCCAGUUUUCCCAGGAAUCCCU;
Thermo Fisher Scientific, Inc.) and scramble sequence used as a
negative control (60 nM, AM17110; Thermo Fisher Scientific, Inc.)
were individually transfected into MDA-MB-231 cells using siPORT
amine transfection agent (Ambion; Thermo Fisher Scientific, Inc.).
Briefly, miR-145-5p and scramble were added to wells containing
1×107 cells and incubated for 48 h. Then, total RNA was
extracted using TRIzol and miR-145-5p restoration was evaluated by
qRT-PCR using specific stem-looped RT oligonucleotide and TaqMan
probe (4427975; Thermo Fisher Scientific, Inc.) as implemented in
the TaqMan MicroRNA Assay protocol. Experiments were performed
three times in triplicate and the results are expressed as mean ±
SD. P<0.05 was considered to indicate statistical
significance.
Cell proliferation assays
Cell proliferation was measured using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay (5 mg/ml; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). MTT
reagent was added to the MDA-MB-231 cells (1×105/well)
transfected with mimic miR-145-5p or scramble and mock and
incubated for 3.5 h at 37°C. Then, dissolution buffer (99%
isopropanol) was added to the cells and incubated for an additional
15 min. Absorbance was recorded at 12 h using a spectrophotometer
(570–630 nm). Data were analyzed using BioStat software
(AnalystSoft, Inc., Walnut, CA, USA). Experiments were performed
three times in triplicate and the results are expressed as mean ±
SD. P<0.05 was considered to indicate statistical
significance.
Fluorescence-activated cell sorting
assays (FACS)
MDA-MB-231 cells (2×105) were treated for
48 h with siPORT transfection agent (mock), scramble (60 nM) and
mimic miR-145-5p (60 nM). Cisplatin (56 µM, IC50) was
added to the miR-145-5p mimic-transfected cells and to the
non-transfected cells and incubated by 24 h. Subsequently, cells
were harvested, washed twice with phosphate-buffered saline (PBS)
1X and resuspended in 100 µl buffer (10 mM HEPES, 140 mM NaCl and
2.5 mM CaCl2), and processed for apoptosis assays and
FACS following the manufacturer's instructions (Annexin V-FLUOS
staining kit; Roche Diagnostics, Basel, Switzerland). Briefly, the
cells were stained with 2 µl Annexin V-FITC and 2 µl propidium
iodide (PI) mixed with 100 µl incubation buffer for 15 min, washed
with 500 µl binding buffer and resuspended in 300 µl PBS 1X.
Apoptosis events were evaluated using the FACSCalibur flow
cytometer [BD Immunocytometry Systems (BDIS); BD Biosciences,
Franklin Lakes, NJ, USA). Briefly, Annexin V and PI emissions were
detected in the FL-1 and FL-2 channels, respectively. For each
sample, data from 20,000 cells were acquired in list mode on
logarithmic scales. Data were analyzed using the Summit V4.3
software (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA)
and the results are represented as the total percentage of
apoptotic cells as the sum of both early and late phases of
apoptosis (Annexin V-FITC-positive). Assays were performed in
triplicate and data are expressed as mean ± SD. P<0.05 was
considered to indicate statistical significance.
Bioinformatic prediction of miR-145-5p
gene targets
miR-145 target genes were predicted using TargetScan
v.7.2 (http://www.targetscan.org/vert_72/), miRWalk v.2.0
(http://mirwalk.umm.uni-heidelberg.de/) and PicTar
(https://pictar.mdc-berlin.de/) software.
Only those gene targets predicted by the three algorithms were
included in downstream analysis. Cellular pathways and processes
potentially affected by miR-145-5p were predicted using DAVID v.6.7
software (https://david.ncifcrf.gov/).
Kaplan-Meier analysis
Kaplan-Meier method was used to evaluate the
disease-free survival (DFS) associated with miR-145-5p expression
in breast cancer patients. The significance of the survival
differences was determined by the log-rank test with 95% confidence
intervals (CI).
Western blot analysis
Whole protein extracts from MDA-MB-231 cells
transfected with miR-145-5p (60 nM) mimic, scramble (60 nM) or mock
were obtained using TNTE buffer (50 mM TRIS-HCl pH 7.4, 150 mM
NaCl, 0.5% Triton X-100 and 5 mM EDTA) supplemented with complete
protease inhibitor cocktail (Roche Molecular Biochemicals,
Penzberg, Upper Bavaria, Germany). Protein extracts (40 µg) were
separated by 10% SDS-PAGE and electrotransferred to nitrocellulose
membrane (Bio-Rad Laboratories, Hercules, CA, USA). After blocking
with 5% non-fat dry milk and 0.05% Tween-20 in PBS pH 7.4 overnight
at 4°C, the membranes were probed with the TGFβR2 antibody
(dilution 1:500; cat. no. ab78419; Abcam, Cambridge, UK) overnight
at 4°C. For detection, the membranes were incubated with
peroxidase-conjugated goat anti-mouse secondary antibodies
(dilution 1:2,000; cat. no. G-21040; Molecular Probes; Thermo
Fisher Scientific, Inc.) in 5% non-fat dry milk and 0.05% Tween-20
in PBS pH 7.4 and immunocomplexes were developed using the ECL
chemiluminescence system (Amersham Pharmacia Biotech, Little
Chalfont, UK). Membranes were subjected to striping and re-blotting
with GAPDH monoclonal antibodies (dilution 1:2,000; cat. no.
sc-47724; Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Densitometric analysis of immunodetected bands in western blots
assays was performed using the public domain ImageJ software
(https://imagej.nih.gov/ij/index.html).
Statistical analysis
A t-test was used to identify significant
differences in miRNA expression between patients with pathological
complete response to chemotherapy treatment in comparison to the
non-responder group. For parametric data we used one-way analysis
of variance (ANOVA) to compare between groups. A P<0.05 was
considered as statistically significant. GraphPad Prism v.5 was
used for statistical anlaysis (https://www.graphpad.com/scientific-software/prism/).
Experiments were performed three times in triplicate and the
results were represented as mean ± standard deviation (SD).
Results
Low miR-145-5p levels are associated
with response to chemotherapy and higher disease-free survival
Breast cancer tumors were collected from a cohort of
patients (n=32) diagnosed with locally advanced triple-negative
breast cancer. Tissues were tested by immunohistochemistry to
confirm the triple-negative status. To evaluate whether changes in
miR-145-5p expression levels could identify the breast cancer
patients that achieved pCR from no-responder individuals, we set up
stem-loop reverse transcription-quantitative PCR (RT-PCR)
experiments. Our data from the 2−ΔΔCq analyses showed
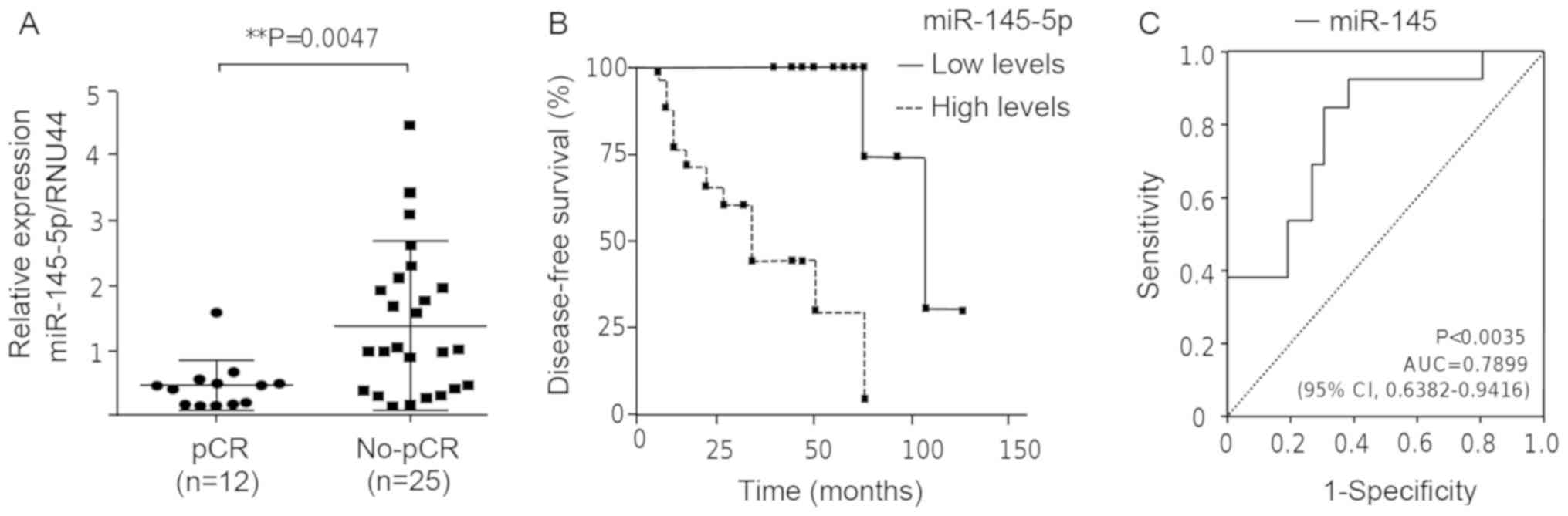
that miR-145-5p was differentially expressed between both groups
(Fig. 1A). miR-145-5p exhibited a
significantly low expression (P<0.0047) in patients that
achieved a pathological complete response (pCR) to chemotherapy
treatment in comparison to the non-responder group. Kaplan-Meier
survival analysis for miR-145-5p expression was performed to
estimate the disease-free survival (DFS) in the triple-negative
breast cancer patients. Patients were dichotomized at their median
into two groups with low and high miR-145-5p expression according
to quantile expression. The log-rank (Mantel-Cox) test identified
significant differences (P<0.0007) between the groups of
patients with a median survival of 104 months in the pCR group (95%
CI, 0.03462–0.2513). Breast cancer patients with low levels of
miR-145-5p had a higher DFS in comparison to the patients with high
levels of miR-145-5p (Fig. 1B). In
contrast, breast cancer patients with high levels of miR-145-5p did
not respond to the chemotherapy regimen and had a worst outcome.
Moreover, receiver operating characteristic (ROC) curve analysis
suggested that miR-145-5p could be a predictor of pCR. The area
under the curve (AUC) was 0.7899 (P<0.0035, 95% CI,
0.6382–0.9416) (Fig. 1C).
miR-145-5p is downregulated in breast
cancer cell lines and impairs cell proliferation
To investigate the biological relevance of
miR-145-5p, its expression was evaluated in breast cancer cells
using stem-loop RT-PCR assays. Data showed that miR-145-5p
expression was significantly downregulated in BT-20, MDA-MB-231,
MCF-7 and SK-BR-3 breast cancer cell lines in comparison to MCF-10A
normal mammary cells (Fig. 2A). We
next aimed to ascertain whether forced expression of miR-145-5p has
negative effects on cell proliferation using MTT assays. BT-20
cells were initially used for miR-145-5p analysis. However, after
RNA mimic transfection, we repeatedly observed a large decrease in
cell viability (80%) indicating an effect on this cell line which
impeded to continue with further characterization (data not shown).
Thus, we decided to use the MDA-MB-231 cells as a model for
functional assays as also it exhibited a significant and important
downregulation of miR-145-5p expression. Transfection of miR-145-5p
mimics (30 nM) was effective to restore the expression by 20-fold
in MDA-MB-231 cells in comparison to the non-transfected and
scramble negative control transfected cells (Fig. 2B). Cell proliferation assay data
indicated that the growth rate of triple-negative MDA-MB-231 cells
transfected with precursor miR-145-5p (60 nM) was significantly
(P<0.001) decreased at an early (12 h) to late (48 h) time of
incubation in comparison with the mock and scramble control cells
(Fig. 2C).
miR-145-5p restoration induces
apoptosis and sensitizes breast cancer cells to cisplatin
therapy
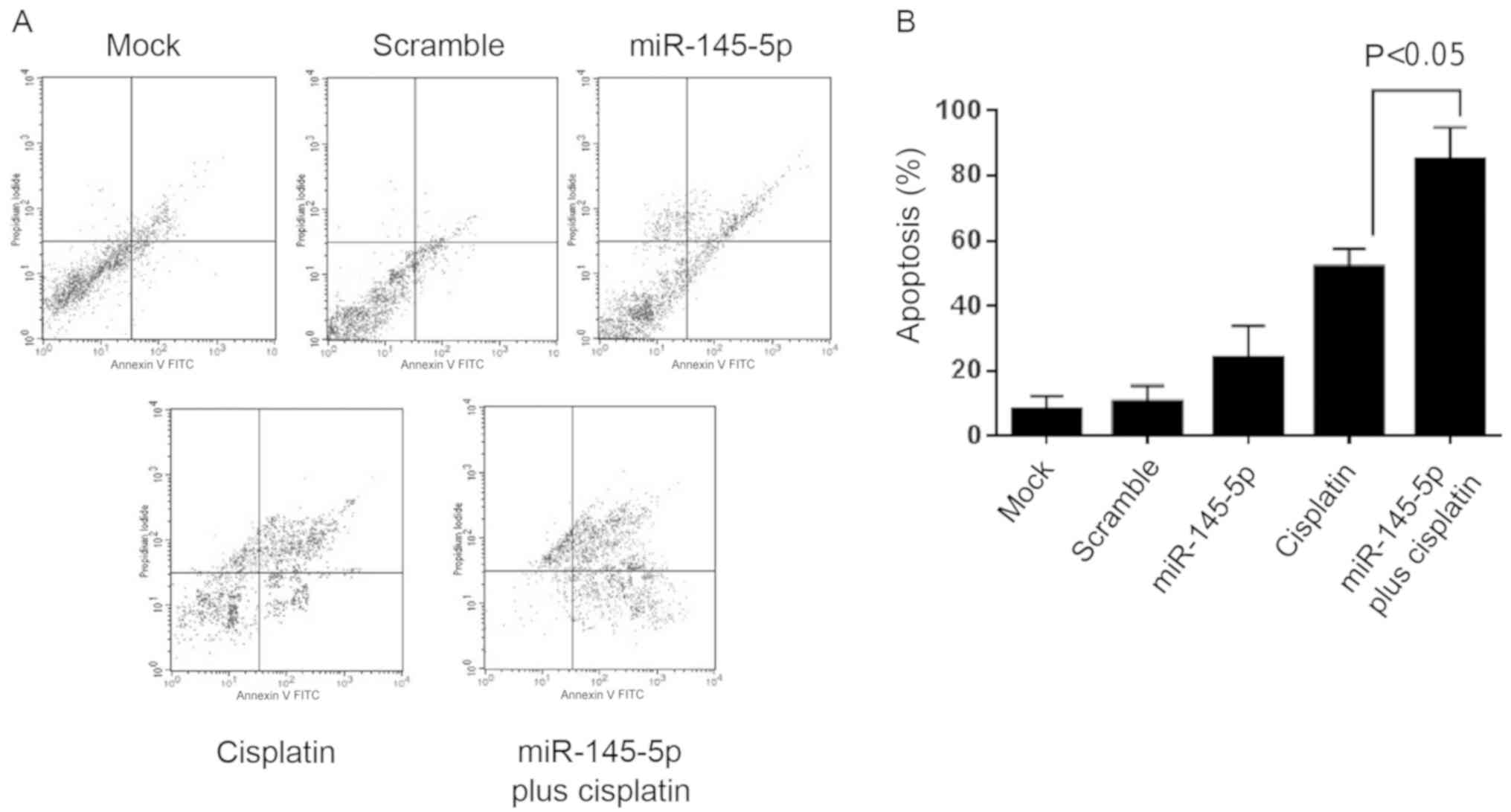
It was next evaluated whether miR-145-5p
overexpression results in apoptosis activation. For this purpose
Annexin V assays and fluorescence-activated cell sorting were
performed. Results showed that treatment with miR-145-5p mimics was
able to induce a modest but significant increase in apoptosis of
MDA-MB-231 cells relative to the mock and scramble-transfected
controls (Fig. 3A). Then, we
evaluated the effect of miR-145-5p in the response to cisplatin in
MDA-MB-231 breast cancer cells. Both precursor
miR-145-5p-transfected and non-transfected cells were submitted to
cisplatin (IC50 55 µM) monotherapy for 48 h. Notably,
combined dual therapy using miR-145-5p plus cisplatin induced a
synergistic increase (P<0.05) in the early and late apoptosis of
MDA-MB-231 cells in comparison to cisplatin alone (Fig. 3B). These data indicate that
miR-145-5p sensitizes breast cancer cells to cisplatin therapy.
miR-145-5p modulates diverse oncogenic
signaling pathways
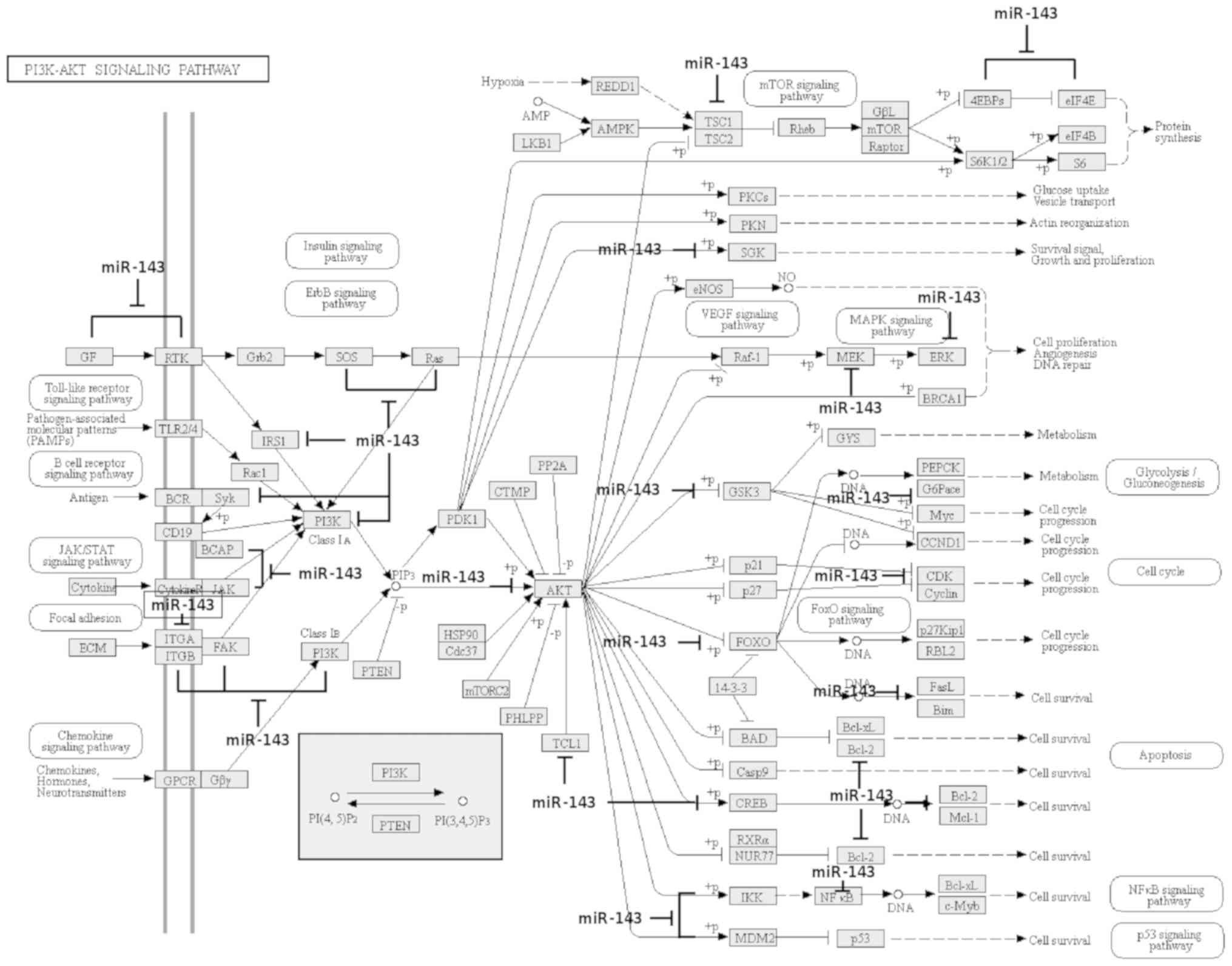
In order to obtain insight concerning the molecular
mechanisms of miR-145-5p in breast cancer, a bioinformatic analysis
of potential target genes was performed using TargetScan 7.2,
miRWalk v.2.0 and PicTar softwares as described in Materials and
methods. Our computational analysis identify 1,007 potential gene
targets of miR-145-5p many of them involved in the regulation of
diverse transducers with pivotal functions in oncogenic signaling
pathways including TGFβ, PI3K/AKT, ErbB, VEGF/MAPK, FOXO, FAK,
JAK/STAT and mTOR (Fig. 4).
Collectively these signaling transduction pathways may regulate
apoptosis, cell cycle, migration, metastasis and survival of tumor
cells which highlights the potential tumor-suppressor functions of
miR-145-5p. Of these pathways, we focused on the study of TGFβ due
to its role as an oncogenic pathway at the advanced stages of
disease.
miR-145-5p downregulates TGFβR2
protein
TGFβ signaling functions as a function of tumor
suppressors or oncogenes during early and late stages of
carcinogenesis, respectively (21).
Our bioinformatic analyses identified two potential miR-145-5p
binding sites in the 3UTR of the TGFβR2 gene (Fig. 5A). Given its importance in the
positive regulation of cancer hallmarks, we decided to ascertain
whether miR-145-5p exerts a potential posttranscriptional
repression of TGFβR2. For this purpose, western blot assays were
performed using specific antibodies and whole protein extracts from
MDA-MB-231 cells transfected with precursor miR-145-5p and
non-transfected mock and scramble-transfected controls. Our data
revealed that miR-145-5p mimics resulted in a significant
(P<0.05) and severe downregulation of TGFβR2 protein levels in
comparison to mock and scramble controls (Fig. 5B and C). GADPH levels used as
control did not show significant changes after miR-145-5p treatment
(Fig. 5B). These data indicate that
miR-145-5p negatively regulates TGFβR2 by a direct or indirect
mechanism.
Discussion
Currently, there are no effective methods to
identify breast cancer patients who would benefit from neoadjuvant
therapy. Thus, there is a need to identify novel biomarkers that
may predict clinical response to therapy and will achieve a
pathological complete response. Recent studies revealed that
changes in miRNA expression could be associated with successful
pathological complete response (pCR) after neoadjuvant treatment in
different types of human cancers (5–7). In
order to contribute to the scarce list of potential predictors of
clinical response to chemotherapy in breast cancer, in the present
study, analysis of miR-145-5p was focused on as a potential
predictor for pCR to NeoCh in triple-negative breast cancer
patients. First, it was shown that low miR-145-5p levels were
significantly associated with pCR to neoadjuvant therapy. Receiver
operating characteristic (ROC) curve analysis suggested that
miR-145-5p could be a predictor of pCR (P<0.0035, AUC=0.7899;
95% CI, 0.6382–0.9416). Moreover, low levels of miR-145-5p also
predicted a higher disease-free survival (Fig. 1). These data are relevant and
highlight the potential of miR-145-5p as a biomarker of response to
therapy.
It is important to note that during chemotherapy
treatments, miRNA dynamics is a complex event. Before NeoCh, we
distinguish two groups of patients: i) a group with very low
miR-145 (low miR-145-5p); and ii) a group with relative high
expression (high miR-145-5p). Remarkably, both groups of cancer
patients had lower expression of miR-145-5p in comparison to normal
cell lines (Fig. 2A) and tissues,
as expected for a tumor-suppressor gene. In our cohort of patients,
relative high expression of miR-145-5p before the therapy was
associated with a worse response to chemotherapy (no-pCR) (Fig. 1A) and low disease-free survival
(DFS) in comparison to the miR-145-5p low group (Fig. 1B). These data confirm our previous
global miRNA profiling findings in pCR and no-pCR patients treated
with a novel chemotherapeutic regimen (19). In addition, it has been reported
that several miRNAs with known oncogenic or tumor-suppressor
functions frequently exhibit significant up- and down-variations in
their expression levels before, during and after chemotherapy,
indicating the profound effect of drugs in miRNA regulation.
Therefore, alterations in miRNA abundance are not always associated
with its functions in carcinogenesis studied in vitro with
cell lines, although the possibility of an unknown dual function
cannot be ruled out (e.g. at early stages of tumorigenesis TGFβ is
a tumor suppressor, but at the late stages of disease it acts as a
potent oncogene). These observations may reflect the complexity of
therapy resistance observed in vivo with cancer patients,
which also have been pointed out by other authors investigating
miRNAs as potential predictors of clinical response to neoadjuvant
therapies. Thus, the fact that low levels of miR-145-5p were
associated with a good response to neoadjuvant therapy may be
inconsistent with the tumor-suppressive role reported for
miR-145-5p. Since miR-145-5p targets many genes, its functions may
differ by diverse biological and therapeutic events (e.g. in
vivo vs. in vitro; adjuvant chemotherapy vs.
chemo-radiotherapy; the percentage of patients achieving pCR vs.
patients without response, neoadjuvant vs. no-neoadjuvant regimen).
Indeed, consistent with our findings, recent studies have shown a
potential role of miR-145 associated with resistance to
chemo-radiotherapy. For instance, similar data were reported in
locally advanced rectal cancer patients in whom low levels of
miR-145 and miR-143 predicted pCR to neoadjuvant chemo-radiotherapy
(22), indicating that miR-145 and
miR-143 levels may be novel, non-invasive predictive markers of
response to therapy in cancer patients. Thus, we propose that in
vivo variations in miR-145 levels before and after therapy may
not always reflect the functions observed in isolated cancer cell
lines. Further analyses are needed to confirm the miR-145 levels in
large cohorts of patients and its biological roles associated with
resistance to therapy.
It was also found that miR-145-5p expression was
significantly repressed in breast tumors and in four breast cancer
cell lines relative to normal mammary tissues and non-tumorigenic
cells, respectively (Fig. 2A). At
the functional level, miR-145 regulates tumor growth, cell
proliferation, apoptosis, cell migration and invasion in breast
cancer cells (23–28). In agreement with its
tumor-suppressor functions reported in vitro, here it was
revealed that ectopic restoration of miR-145-5p inhibited cell
proliferation in triple-negative MDA-MB-231 cells and sensitized
tumor cells to cisplatin treatment reinforcing the notion that
miR-145-5p is a bona fide tumor suppressor associated with therapy
response (Fig. 2). Intriguingly,
miR-145-5p RNA mimic transfection in BT-20 cells induced a large
decrease in cell viability. We hypothesized that the BT-20 cell
genetic background may influence and exacerbate the cell response
to miR-495. Although this cell line is also triple negative it
contains important sequence variations in master genes controlling
cell proliferation and survival including PI3KCA, CDKN2A, EGFR and
p53 (29), that may exert an
unknown effect in response to miR-145-5p restoration; however,
further investigation is needed to support these assumptions.
Novel data concerning the miR-145-5p mechanisms
associated with cancer hallmark inhibition is provided as the
findings elucidated that miR-145 directly or indirectly targets
TGFβ signaling in breast cancer cells. During the preparation of
this manuscript a recent study reported that miR-145 inhibits cell
proliferation by targeting TGFβ1 in breast cancer cells (30). In the present study, we added a
piece in the puzzle of miR-145-5p functions and demonstrated that
it also targets TGFβR2 which strengthens the notion that this tiny
non-coding RNA has a profound impact on the suppression of cancer
hallmarks through modulation of TGFβ signaling in breast cancer. In
summary, our data suggest that miR-145-5p could be a potential
predictor of response to neoadjuvant therapy, and also elucidate
the molecular functions of miR-145-5p in breast cancer cells.
Acknowledgements
We acknowledge Consejo Nacional de Ciencia y
Tecnología CONACyT, México and Universidad Autónoma de la Ciudad de
México for support.
Funding
The present study was funded by the Consejo Nacional
de Ciencia y Tecnología CONACyT México, Fondo SSA/IMSS/ISSSTE
(grant no. 233370 and 222335).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
CLC, LAM, SRC and ERG conceived and designed the
study. SRC provided the tissues collection. FGG, YMSV, RGV and ACR
performed the experiments. CLC, CPP, RRP, YMSV, RGV and MAM
analyzed the data. CLC, FGG and LAM wrote the paper. ERG, RRP, MAM
and JSLG reviewed and edited the manuscript. JSLG, ACR, CPP and SRC
provided the additional materials and reagents. All authors read
and approved the manuscript and agree to be accountable for all
aspects of the research in ensuring that the accuracy or integrity
of any part of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
The Breast Cancer Foundation (FUCAM) of Mexico
provided the breast tumors and normal tissue collection. The Ethics
Committee of the Breast Cancer Foundation (FUCAM) of Mexico
approved the protocols using human tissues. Signed informed consent
forms were obtained from the participants prior to release for
research use. This study was carried out in accordance with the
ethical standards of the committee and in accordance with the
Helsinki Declaration of 1975.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Prowell TM and Pazdur R: Pathological
complete response and accelerated drug approval in early breast
cancer. N Engl J Med. 366:2438–2441. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Von Minckwitz G, Untch M, Blohmer JU,
Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich
J, Huober J, et al: Definition and impact of pathologic complete
response on prognosis after neoadjuvant chemotherapy in various
intrinsic breast cancer subtypes. J Clin Oncol. 30:1796–1804. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, et al: Response to neoadjuvant therapy and long-term
survival in patients with triple negative breast cancer. J Clin
Oncol. 26:1275–1281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kong X, Moran MS, Zhang N, Haffty B and
Yang Q: Meta-analysis confirms achieving pathological complete
response after neoadjuvant chemotherapy predicts favourable
prognosis for breast cancer patients. Eur J Cancer. 47:2084–2090.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tanioka M, Sasaki M, Shimomura A,
Fujishima M, Doi M, Matsuura K, Sakuma T, Yoshimura K, Saeki T,
Ohara M, et al: Pathologic complete response after neoadjuvant
chemotherapy in HER2-overexpressing breast cancer according to
hormonal receptor status. Breast. 23:466–472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang P, Yin Y, Mo H, Zhang B, Wang X, Li
Q, Yuan P, Wang J, Zheng S, Cai R, et al: Better pathologic
complete response and relapse-free survival after carboplatin plus
paclitaxel compared with epirubicin plus paclitaxel as neoadjuvant
chemotherapy for locally advanced triple-negative breast cancer: A
randomized phase 2 trial. Oncotarget. 7:60647–60656.
2016.PubMed/NCBI
|
|
7
|
Luangdilok S, Samarnthai N and Korphaisarn
K: Association between pathological complete response and outcome
following neoadjuvant chemotherapy in locally advanced breast
cancer patients. J Breast Cancer. 17:376–385. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ring AE, Smith IE, Ashley S, Fulford LG
and Lakhani SR: Oestrogen receptor status, pathological complete
response and prognosis in patients receiving neoadjuvant
chemotherapy for early breast cancer. Br J Cancer. 91:2012–2017.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cortazar P, Zhang L, Untch M, Mehta K,
Costantino JP, Wolmark N, Bonnefoi H, Cameron D, Gianni L,
Valagussa P, et al: Pathological complete response and long-term
clinical benefit in breast cancer: The CTNeoBC pooled analysis.
Lancet. 384:164–172. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kolacinska A, Morawiec J, Fendler W,
Malachowska B, Morawiec Z, Szemraj J, Pawlowska Z, Chowdhury D,
Choi YE, Kubiak R, et al: Association of microRNAs and pathologic
response to preoperative chemotherapy in triple negative breast
cancer: Preliminary report. Mol Biol Rep. 41:2851–2857. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hummel R, Hussej DJ and Haier J:
MicroRNAs: Predictors and modifiers of chemo- and radiotherapy in
different tumor types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gasparini P, Cascione L, Fassan M, Lovat
F, Guler G, Balci S, Irkkan C, Morrison C, Croce CM, Shapiro CL and
Huebner K: microRNA expression profiling identifies a four microRNA
signature as a novel diagnostic and prognostic biomarker in triple
negative breast cancers. Oncotarget. 5:1174–1184. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ohzawa H, Miki A, Teratani T, Shiba S,
Sakuma Y, Nishimura W, Noda Y, Fukushima N, Fujii H, Hozumi Y, et
al: Usefulness of miRNA profiles for predicting pathological
responses to neoadjuvant chemotherapy in patients with human
epidermal growth factor receptor 2-positive breast cancer. Oncol
Lett. 13:1731–1740. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Raychaudhuri M, Bronger H, Buchner T,
Kiechle M, Weichert W and Avril S: MicroRNAs miR-7 and miR-340
predict response to neoadjuvant chemotherapy in breast cancer.
Breast Cancer Res Treat. 162:511–521. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pedroza-Torres A, Fernández-Retana J,
Peralta-Zaragoza O, Jacobo-Herrera N, Cantú de Leon D, Cerna-Cortés
JF, Lopez-Camarillo C and Pérez-Plasencia C: A microRNA expression
signature for clinical response in locally advanced cervical
cancer. Gynecol Oncol. 142:557–565. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Petrillo M, Zannoni GF, Beltrame L,
Martinelli E, DiFeo A, Paracchini L, Craparotta I, Mannarino L,
Vizzielli G, Scambia G, et al: Identification of high-grade serous
ovarian cancer miRNA species associated with survival and drug
response in patients receiving neoadjuvant chemotherapy: A
retrospective longitudinal analysis using matched tumor biopsies.
Ann Oncol. 27:625–634. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
García-Vazquez R, Ruiz-García E, Meneses
García A, Astudillo-de la Vega H, Lara-Medina F, Alvarado-Miranda
A, Maldonado-Martínez H, González-Barrios JA, Campos-Parra AD,
Rodríguez Cuevas S, et al: A microRNA signature associated with
pathological complete response to novel neoadjuvant therapy regimen
in triple-negative breast cancer. Tumour Biol.
39:10104283177028992017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Syed V: TGF-β signaling in cancer. J Cell
Biochem. 117:1279–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hiyoshi Y, Akiyoshi T, Inoue R, Murofushi
K, Yamamoto N, Fukunaga Y, Ueno M, Baba H, Mori S and Yamaguchi T:
Serum miR-143 levels predict the pathological response to
neoadjuvant chemoradiotherapy in patients with locally advanced
rectal cancer. Oncotarget. 8:79201–79211. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao H, Kang X, Xia X, Wo L, Gu X, Hu Y,
Xie X, Chang H, Lou L and Shen X: miR-145 suppresses breast cancer
cell migration by targeting FSCN-1 and inhibiting
epithelial-mesenchymal transition. Am J Transl Res. 8:3106–3114.
2016.PubMed/NCBI
|
|
24
|
Gao M, Miao L, Liu M, Li C, Yu C, Yan H,
Yin Y, Wang Y, Qi X and Ren J: miR-145 sensitizes breast cancer to
doxorubicin by targeting multidrug resistance-associated protein-1.
Oncotarget. 7:59714–59726. 2016.PubMed/NCBI
|
|
25
|
Zheng M, Wu Z, Wu A, Huang Z, He N and Xie
X: MiR-145 promotes TNF-α-induced apoptosis by facilitating the
formation of RIP1-FADD caspase-8 complex in triple-negative breast
cancer. Tumour Biol. 37:8599–8607. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yan X, Chen X, Liang H, Deng T, Chen W,
Zhang S, Liu M, Gao X, Liu Y, Zhao C, et al: miR-143 and miR-145
synergistically regulate ERBB3 to suppress cell proliferation and
invasion in breast cancer. Mol Cancer. 13:2202014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Götte M, Mohr C, Koo CY, Stock C, Vaske
AK, Viola M, Ibrahim SA, Peddibhotla S, Teng YH, Low JY, et al:
miR-145-dependent targeting of junctional adhesion molecule A and
modulation of fascin expression are associated with reduced breast
cancer cell motility and invasiveness. Oncogene. 29:6569–6580.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng
L, Zhou H and Zhao RC: miR-145 inhibits breast cancer cell growth
through RTKN. Int J Oncol. 34:1461–1466. 2009.PubMed/NCBI
|
|
29
|
Saunus JM, Smart CE, Kutasovic JR,
Johnston RL, Kalita-de Croft P, Miranda M, Rozali EN, Vargas AC,
Reid LE, Lorsy E, et al: Multidimensional phenotyping of breast
cancer cell lines to guide preclinical research. Breast Cancer Res
Treat. 167:289–301. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding Y, Zhang C, Zhang J, Zhang N, Li T,
Fang J, Zhang Y, Zuo F, Tao Z, Tang S, et al: miR-145 inhibits
proliferation and migration of breast cancer cells by directly or
indirectly regulating TGF-β1 expression. Int J Oncol. 50:1701–1710.
2017. View Article : Google Scholar : PubMed/NCBI
|