Introduction
Multidrug resistance (MDR) to cytostatics, whether
intrinsic or acquired, remains a barrier to successful therapy for
solid tumors. The MDR phenotype often correlates with high
expression of P-glycoprotein, which is the most commonly studied
ATP-binding cassette (ABC) transporter (1). The protein is encoded by the
ABCB1 gene [Online Mendelian Inheritance in Man (OMIM)
entry: 171050], which is located on chromosome 7q21.1, and
functions as a cellular efflux pump for numerous xenobiotics,
including anticancer drugs (2–4). ABCB1 is
mostly expressed in excreting organs (such as the liver and kidney)
and physiological barriers, including the blood-brain, -testis and
-placental barriers (5,6), as well as tumor tissues such as breast
and ovarian carcinomas (7–11). Correlations between ABCB1
expression and overall or disease-free survival and response to
chemotherapy have been reported in patients with breast carcinoma
(12–14). A previous study on ovarian carcinoma
has shown that ABCB1 gene/protein expression is associated
with MDR (15).
Epigenetic mechanisms (i.e. mainly DNA and histone
modifications) result in the regulation of genes without changes in
their coding sequence (16).
Epigenetic changes can be inherited (such as imprinting) and
relatively stable (such as chromosome X inactivation), but more
often reflect rapidly changing cell needs (17). Epigenetic changes can be induced by
DNA damage (18,19). It is therefore not surprising that
errors in DNA methylation are linked to a variety of effects,
including imprinting defect syndromes and cancer (17).
The contribution of DNA methylation to cancer
prognosis and progression has been extensively studied in recent
years. Methylation of the ABCB1 promoter occurs early during
breast tumorigenesis, and it has been detected in ductal carcinoma
in situ and invasive breast tumors (20–22). A
significant association between ABCB1 promoter methylation
and protein expression was observed in invasive ductal carcinomas
and paired serum samples (23). Other
authors have reported an association between ABCB1 promoter
methylation and treatment response and survival of patients with
breast carcinoma (21,24).
Taken together, previously published studies suggest
potential epigenetic effects of promoter methylation on
ABCB1 expression and prognosis of patients with breast
carcinoma, but the data are inconsistent. Different studies include
different regions of the ABCB1 gene and use non-validated
technologies. Furthermore, the clinical impact of ABCB1
promoter methylation on ovarian carcinoma prognosis or survival is
currently unknown. In particular, ovarian carcinoma patients are
predominantly treated with a combination of platinum derivatives
and taxanes. Taxanes are substrates of ABC membrane transporters
including P-glycoprotein and epigenetic regulation of the
ABCB1 promoter affecting its function may play an important
role in therapeutic efficiency and development of chemoresistance
to taxanes in ovarian carcinomas.
DNA methylation of the ABCB1 promoter region
can be detected and quantified by various technologies, including
pyrosequencing (20–22) and methylation-specific polymerase
chain reaction (PCR) mainly encompassing the binding SP-1 site in
the 3′-region of the ABCB1 gene (23–25).
The aim of the present study was to develop a novel,
rapid and simple method for ABCB1 promoter methylation
analysis overlapping a transcript site using gene-specific,
high-resolution melting (HRM) analysis. The present study also
evaluated the functional consequences of ABCB1 promoter
methylation by the analysis of correlation between the methylation
status with the expression levels of ABCB1 gene. Finally,
associations of ABCB1 promoter methylation with the
prognosis of patients with breast and ovarian carcinoma were
assessed. Such associations may have a clinical impact on prognosis
and individualized patient therapy, thus, offering potential
socio-economic benefits.
Patients and methods
Patients
This retrospective study utilized the following
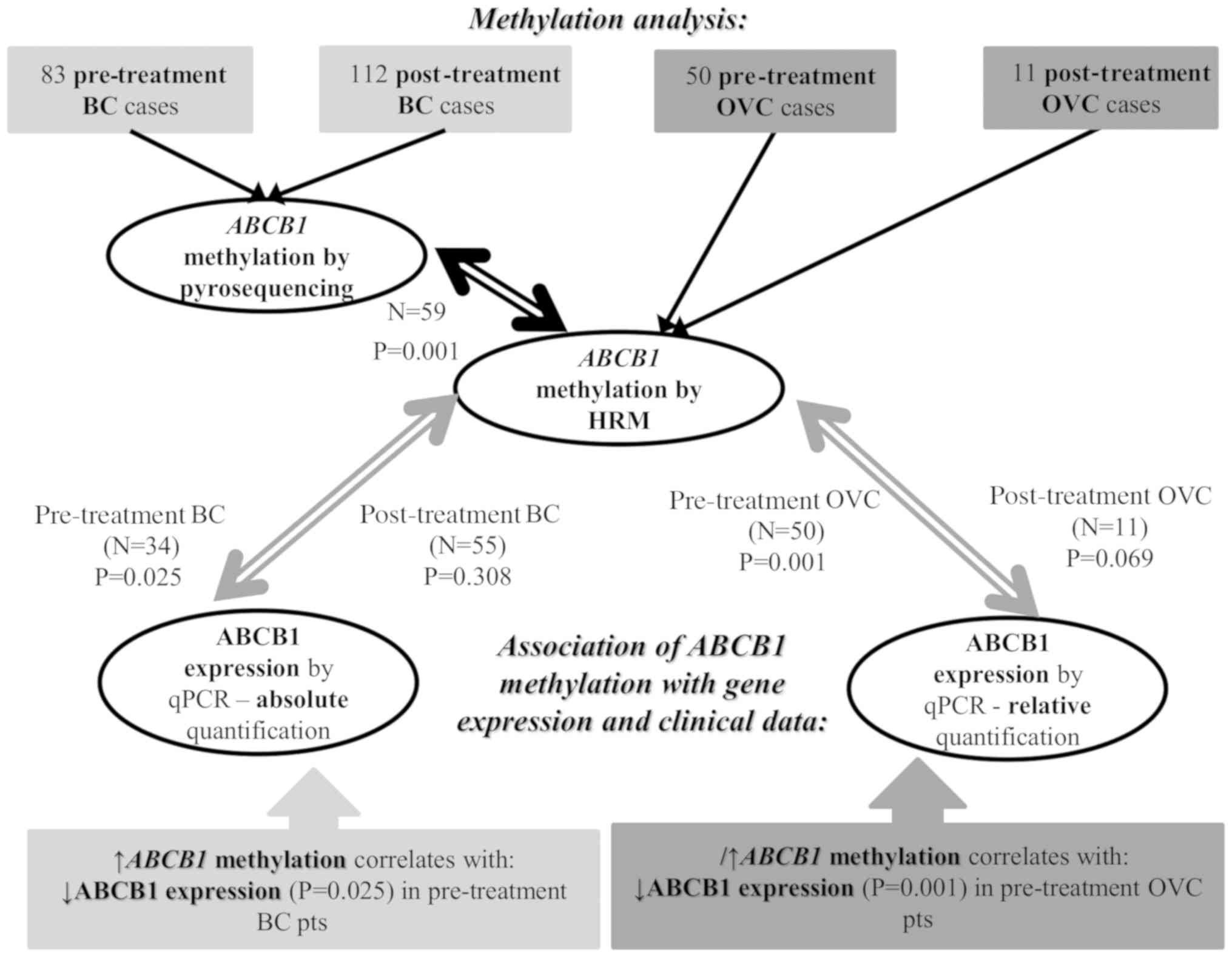
historical pre-treatment and post-treatment cohorts of patients
with breast and ovarian carcinoma (Fig.
1).
Breast tumor samples
Tumor tissues of 83 patients with breast carcinoma
collected prior to chemotherapy (pre-treatment set). The patients
were diagnosed at Motol University Hospital (Prague, Czech
Republic; n=71) and the General University Hospital (Prague, Czech
Republic; n=12) between February 2000 and December 2006. The
collection and handling of tissue samples was described in detail
elsewhere (8,26). The post-treatment set of patients with
breast carcinoma was used for validation of the HRM method, and the
samples were collected from hospitals in Oslo, Norway as described
previously (22,27). The post-treatment set of patients was
treated with 5-fluorouracil and mitomycin (n=34) or doxorubicin
(n=78). The patients were enrolled in an Institutional Review Board
approved protocol evaluating the drug response in a neoadjuvant
setting (28–30). Paired adjacent control tissues without
morphological signs of carcinoma were available for 6 patients.
Ovarian tumor samples
A total of 61 samples of patients with epithelial
ovarian carcinoma (EOC) diagnosed at Motol University Hospital
(Prague, Czech Republic) during the 2009–2013 period were used in
the present study. In total, 11 samples were collected upon
neoadjuvant treatment (post-treatment set) based on a combination
of paclitaxel and platinum derivatives, while the remaining samples
(n=50) were collected at the time of surgery prior to chemotherapy
treatment. A total of 11 samples of ovarian tissues without
morphological signs of carcinoma were used as controls, and were
obtained from patients who underwent surgery for reasons other than
ovarian malignancy at Motol University Hospital. The collection and
handling of these tissue samples has been described in detail
elsewhere (8,26,31,32).
All patients had given informed consent, and the
project was approved by the Ethics Commission of the National
Institute of Public Health in Prague (ethic codes: IGA no. 9799-4
of 30 January 2008, 14055-3 of 2 July 2012, and 14056-3 of 2 July
2012), and the Institutional Review Board of Norwegian
Radiumhospital (Oslo, Norway) in the frame of the Norwegian Cancer
Society (D-03067) and the Norwegian Research Council (163027/V409)
projects. The methods were carried out in accordance with
guidelines approved by the above Ethics Commissions.
ABCB1 methylation analysis by
pyrosequencing
The overall study design is described in Fig. 1. DNA was extracted from freshly frozen
breast tumor tissue samples by the standard phenol/chloroform
extraction method. The extracted DNA was bisulfite modified using
the EpiTect® Bisulfite kit (Qiagen GmbH, Hilden,
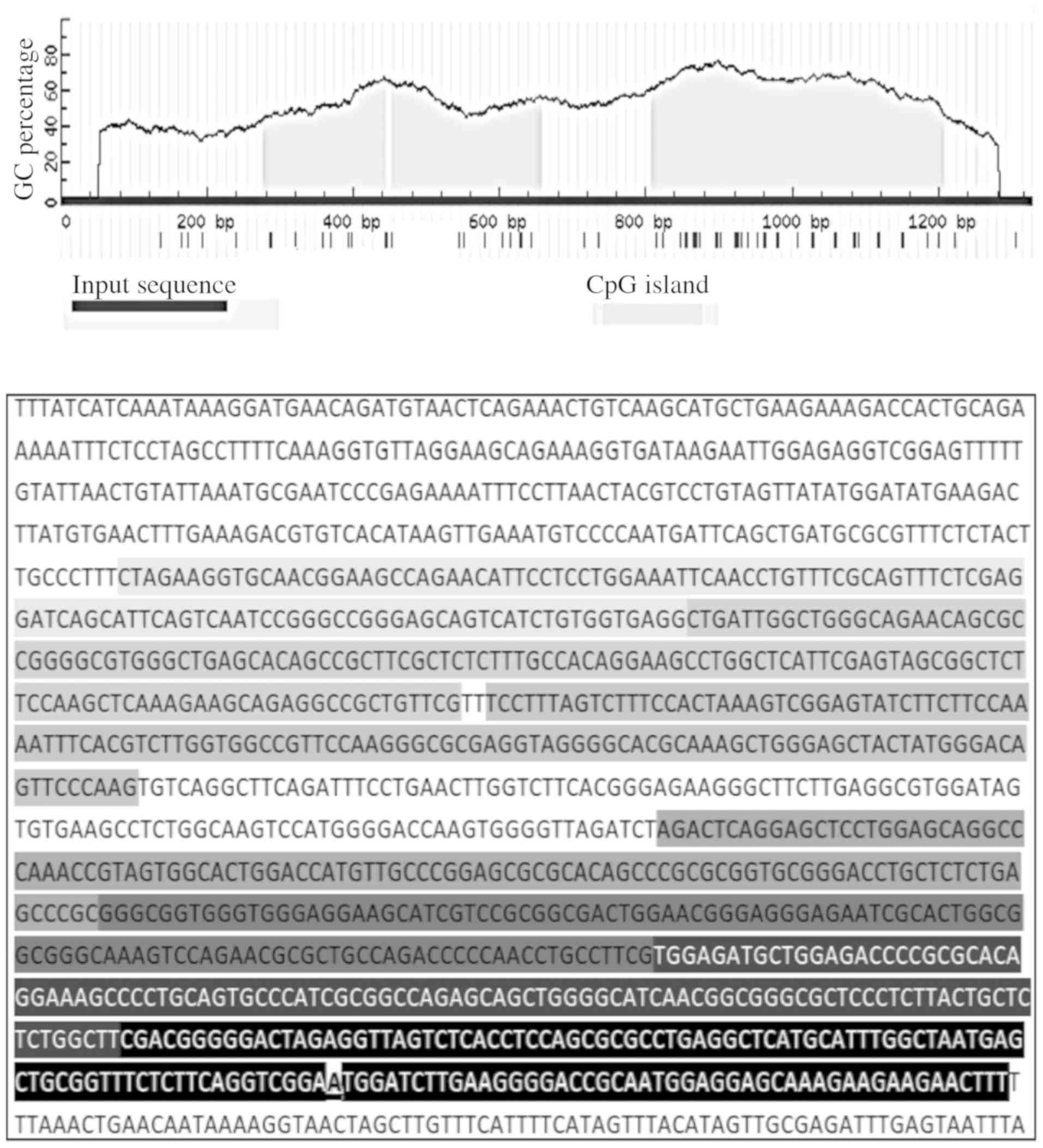
Germany) following the manufacturer's protocol. Three CpG island
regions (as shown in Fig. 2)
overlapping the ABCB1 promoter and transcription start site were
identified using MethPrimer software (33). Quantitative DNA methylation analysis
was performed by pyrosequencing of bisulfite-treated DNA from
pre-treatment (n=66) and post-treatment (n=105) breast carcinoma
specimens, as previously described (22,27,34).
ABCB1 methylation analysis by HRM
In the present study, HRM analysis covering the
entire region of the ABCB1 promoter was estimated using
pyrosequencing and was established in genomic DNA samples (100–500
ng) from pre-treatment breast tumor tissues (n=59). A total of 10
ng bisulfite-converted DNA sample was used for each HRM methylation
analysis. PCR amplification and subsequent HRM analysis was
performed on a RG6000 system (Corbett Life Science; Qiagen GmbH)
using the EpiTect HRM kit (Qiagen GmbH) according to the
recommendations of the manufacturer. The
reverse-transcription-quantitative PCR (RT-qPCR) cycling conditions
and the sequences of the primers for HRM analysis of all the
examined ABCB1 regions are summarized in Table I. A standard curve including
bisulfite-converted human control DNA (Qiagen GmbH) as the fully
methylated control and dilutions (0, 5, 10, 20, 30, 40, 50, 60, 70,
80, 90 and 100%) with unmethylated control DNA (Qiagen GmbH) was
included in each replicate. The collected HRM data were analyzed
using Rotor-Gene software version 6.0 (Corbett Life Science; Qiagen
GmbH). Subsequently, the optimal conditions for HRM analysis were
used for ABCB1 promoter methylation status in ovarian tumor
tissue samples (n=61), as described in Table I.
 | Table I.Primer sequences, amplicon sizes and
conditions for real-time PCR and the following HRM analysis of the
examined ABCB1 promoter. |
Table I.
Primer sequences, amplicon sizes and
conditions for real-time PCR and the following HRM analysis of the
examined ABCB1 promoter.
| Target
ABCB1 | Primers
(5′-3′) | Amplicon size
(bp) |
Annealinga |
|---|
| Region 1 | Forward:
TTAGAGAGGTGTAATGGAAGTTAGAATATTTT | 140 | 30 sec/59°C |
|
| Reverse:
CACTATTCCTACCCAACCAATCAA |
|
|
| Region 2 | Forward:
GTTGATTGGTTGGGTAGGAAT | 132 | 30 sec/55°C |
|
| Reverse:
CAAACAACAACCTCTACTTCTTTAAA |
|
|
| Region 3 | Forward:
TTTTTTAGGTTTTTTTATTAAAGT | 124 | 30 sec/50°C |
|
| Reverse:
CTTAAAAACTATCCCATAATAACTC |
|
|
| Region 4 | Forward:
AGATTTAGGAGTTTTTGGAGTAG | 101 | 30 sec/50°C |
|
| Reverse:
CTCAAAAAACAAATCCCC |
|
|
| Region 5 | Forward:
TTGTGGAGATGTTGGAGATT | 132 | 30 sec/61°C |
|
| Reverse:
ACACAAAATCTCCAACATCTCCA |
|
|
| Region 6 | Forward:
TTGTGGAGATGTTGGAGATTT | 116 | 60 sec/58°C |
|
| Reverse:
CCATCAAAACCAAAAAACAAT |
|
|
| Region 7 | Forward:
TGATGGGGGATTAGAGGTTAGTT | 136 | 30 sec/57°C |
|
| Reverse:
AAAATTCTTCTTCTTTACTCCTCCATTA |
|
|
Quantification of ABCB1 gene
expression
Total RNA was isolated from frozen tissue using
TRIzol reagent (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
according to the procedure provided by the manufacturer. The
quality and quantity of the extracted RNA were assessed by
spectrophotometry and agarose gel electrophoresis (28S/18S
ribosomal ratio). Complementary DNA (cDNA) was synthesized using
0.5 µg total RNA and random hexamer primers with the RevertAid
First Strand cDNA Synthesis kit (Fermentas; Thermo Fisher
Scientific, Inc). Contamination with genomic DNA was assessed by
PCR amplification of ubiquitin C (UBC) fragment capable of
discriminating between products amplified from cDNA (190 bp) and
from genomic DNA (1009 bp) as previously described (35).
Subsequently, absolute quantification of
ABCB1 transcript levels in breast carcinoma samples (n=34
pre-treatment and n=55 post-treatment) was performed by RT-qPCR as
previously described (8). Human
peptidylprolyl isomerase A (PPIA) was used as a reference gene for
normalization of the ABCB1 transcript levels. Standards for
the construction of the calibration curve were prepared using
Gateway Cloning Technology (Thermo Fisher Scientific, Inc.) by
cloning ABCB1 and PPIA gene fragments into the
pDONR201 vector (Thermo Fisher Scientific, Inc.) and propagation of
vectors in Escherichia coli DH5α Maximum Efficiency Cells
(Thermo Fisher Scientific, Inc.) as previously described (8). In ovarian carcinoma samples (n=61), our
recently published method of relative quantification of
ABCB1 expression with normalization to 3 reference genes,
namely peptidylprolyl isomerase A (PPIA), ubiquitin C (UBC) and
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation
protein ζ (YWHAZ, 32) was used. Amplification efficiencies for each
reference and target gene were calculated applying the formula
Efficiency=10−1/slope−1. The qPCR study design adhered
to the Minimum Information for Publication of Quantitative
Real-time PCR Experiments Guidelines (36).
Statistical analyses
Due to the deviation of the data from the normal
distribution, gene methylation and expression levels were analyzed
with non-parametric statistical tests. The correlation between
transcript and methylation levels was assessed using Spearman's
rank correlation. Mann-Whitney test, Kruskal-Wallis test and
Spearman's rank correlation were used for analysis of associations
of transcript and methylation levels with clinical data. In
general, the evaluated clinical and pathological variables in
breast carcinomas were as follows: Stage (I/II vs. III/IV), grade
(1 or 2 vs. 3), and histological type (invasive ductal vs. other
invasive carcinoma). Pathological lymph node categorization (pN)
was performed as follows; pN0 indicates that regional lymph node
metastasis was not found, pN1-3 indicates that micrometastases or
metastases in axillary lymph nodes were identified in 1–3 nodes
(pN1), 4–9 nodes (pN2) or ≥10 nodes (pN3); pNx indicates that
regional lymph nodes cannot be assessed. Pathological tumor size
(pT) was described as follows: pT1, the tumor is ≥2 centimeters
(cm) large; pT2, the tumor size is >2 but ≤5 cm; pT3, the tumor
is ≥5 cm large; pT4, a tumor of any size with direct extension to
chest wall and/or to the skin; pTx, a tumor size cannot be assessed
(37). Expression of estrogen and
progesterone receptors and p53 protein was estimated as positive
vs. negative. Expression of p53 protein, as a potential marker of
aggressive type of breast cancer, was available in a limited number
of samples because unlike ER and PR, main prognostic factors for
breast cancer, p53 is not routinely assessed in clinical practice.
In ovarian carcinomas, the evaluated clinical data were: Stage
(I/II vs. III/IV), grade (1 or 2 vs. 3), tumor size (pT1 vs pT2-4),
histological type (high grade serous vs. others) and expression of
the proliferation marker Ki67 (expressed as a percentage). Survival
functions were plotted by Kaplan-Meier curves, and the statistical
significance was evaluated by the log-rank test. Progression-free
survival (PFS), which was defined as the time elapsed between
surgical treatment and disease progression or mortality from any
cause, was used for survival analysis. Two-sided P<0.05 was
considered to indicate a statistically significant difference. The
Bonferroni's test was used for adjustment of P-values due to
multiple comparisons. All statistical analyses were performed using
SPSS v16.0 (SPSS Inc., Chicago, IL, USA). Due to the heterogeneity
between the pre-treatment and post-treatment sets, the data were
analyzed separately.
Results
Study population characteristics
Breast carcinoma samples from pre-treatment and
post-treatment patients (n=83 and 112, respectively) were collected
in the present study. The clinical data from all patients with
breast carcinomas are described in Table
II. Both sets of patients significantly differed in stage
distribution and estrogen and progesterone receptors expression
(P<0.001; according to the results of χ2 test). As an
additional type of solid tumor, samples from patients with ovarian
carcinoma were collected in the present study (n=61). Of these, 50
samples were collected during surgery prior to any treatment while
11 samples were collected following neoadjuvant chemotherapy. All
the post-treatment samples were tumors of high-grade serous
carcinoma subtype with advanced stages (III or IV) and grade 3. The
clinical data of the examined patients with ovarian carcinoma are
described in Table III.
 | Table II.Clinical characteristics of the
patients with breast carcinoma involved in the present study. |
Table II.
Clinical characteristics of the
patients with breast carcinoma involved in the present study.
|
Characteristics | Pre-treatment set
(n=83) n (%)a | Post-treatment set
(n=112) n (%)a |
|---|
| Stageb |
|
Ic | 31 (37.3) | 0 (0) |
| II | 35 (42.1) | 0 (0) |
|
III | 7 (8.4) | 87 (77.7) |
| IV | 2 (2.4) | 20 (17.9) |
|
N/A | 8 (9.6) | 5 (4.5) |
| Lymph node
metastasis |
| Present
(pN1-3) | 44 (53.0) | 71 (63.4) |
| Absent
(pN0) | 32 (38.6) | 36 (32.1) |
|
pNx | 7 (8.4) | 5 (4.5) |
| Tumor
sizeb |
|
pT1 | 45 (54.2) | 0 (0) |
|
pT2 | 30 (36.1) | 5 (4.5) |
|
pT3 | 1
(1.2) | 63
(56.3) |
|
pT4 | 3
(3.6) | 39
(34.8) |
|
pTx | 4 (4.8) | 5 (4.5) |
| Histological
type |
|
Invasive ductal carcinoma | 67 (80.7) | 93 (83.0) |
| Other
types of carcinomac | 16 (19.3) | 19 (17.0) |
|
N/A | 0 (0) | 0 (0) |
| Expression of
ERb |
|
Positive | 48 (57.8) | 89 (79.5) |
|
Negative | 31 (37.3) | 23 (20.5) |
|
N/A | 4 (4.8) | 0 (0) |
| Expression of
PRb |
|
Positive | 46 (55.4) | 76 (67.9) |
|
Negative | 33 (39.8) | 36 (32.1) |
|
N/A | 4 (4.8) | 0 (0) |
| Expression of
p53 |
|
Positive | 21 (25.3) | 35 (31.3) |
|
Negative | 49 (59.0) | 67 (59.8) |
|
N/A | 13 (15.5) | 10 (8.9) |
 | Table III.Clinical characteristics of the
patients with ovarian carcinoma involved in the present study. |
Table III.
Clinical characteristics of the
patients with ovarian carcinoma involved in the present study.
|
Characteristics | Pre-treatment set
(n=50) n (%)a | Post-treatment set
(n=11) n (%)a |
|---|
| Stage |
| I | 2 (4.0) | 0 (0) |
| II | 2 (4.0) | 0 (0) |
|
III | 37 (74.0) | 10 (90.9) |
| IV | 4 (8.0) | 1 (9.1) |
|
N/A | 5 (10.0) | 0 (0) |
| EOC type |
| High
grade serous | 40 (80.0) | 11 (100.0) |
| Other
type | 5 (10.0) | 0 (0) |
|
N/A | 5 (10.0) | 0 (0) |
| Histological
grade |
| 1 | 1 (2.0) | 0 (0) |
| 2 | 9 (18.0) | 0 (0) |
| 3 | 35 (70.0) | 11 (100) |
|
N/A | 5 (10.0) | 0 (0) |
| Ki-67 protein
expression |
| Median
± SD (%) | 39.0±24.0 | 31.8±17.4 |
|
N/A | 15 (30.0) | 0 (0) |
Development of HRM analysis for
estimation of ABCB1 promoter methylation levels
For rapid, affordable and simple screening of the
ABCB1 promoter methylation status, a HRM methylation
analysis was developed. First, the ABCB1 methylation levels
were estimated in a subset of the aforementioned pre-treatment
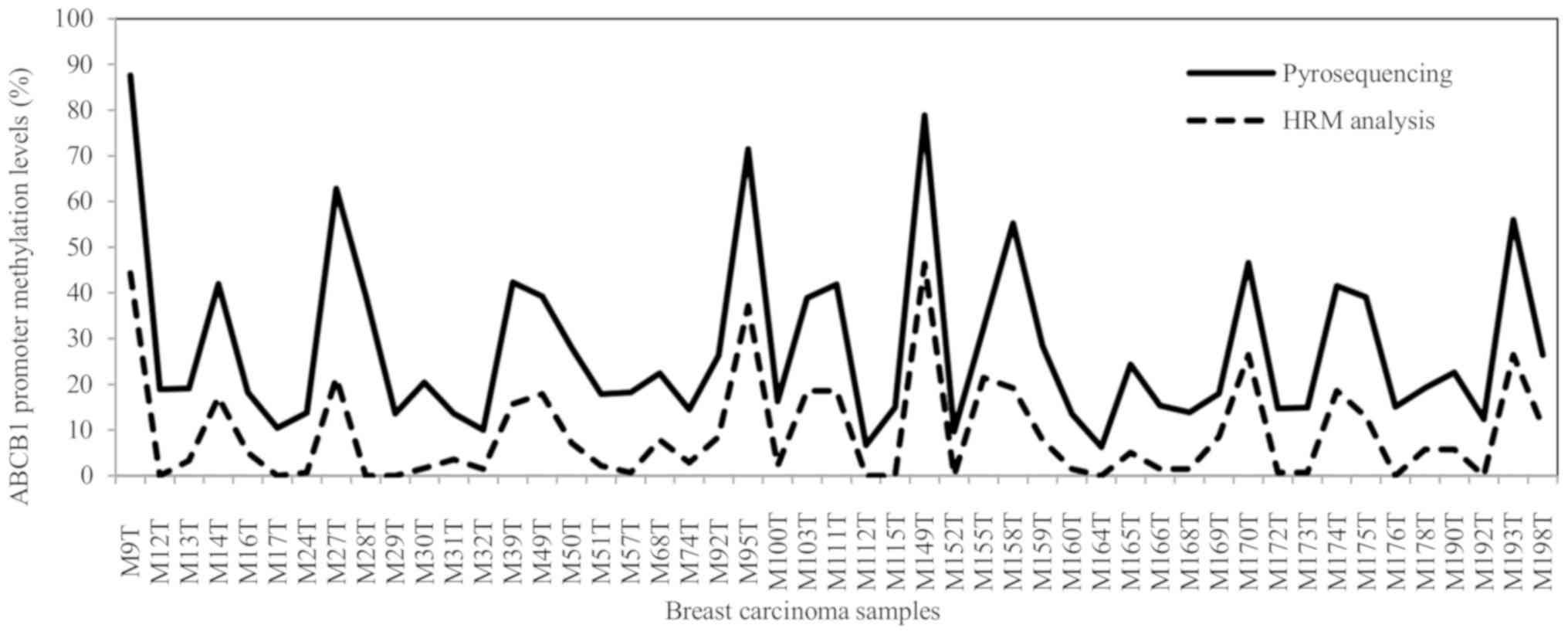
breast carcinoma samples (n=59) (Fig.
1). DNA methylation levels were then compared with the DNA
methylation levels of the ABCB1 promoter that were assessed
previously in tumor samples from 66 pre-treatment and 105
post-treatment patients with breast carcinoma by pyrosequencing
(22,27,34). The
methylation levels estimated using HRM analysis were closely
associated with the pyrosequencing methylation levels in the breast
carcinoma samples as estimated by Spearman's rho correlation
(P=0.001, ρ=0.699, n=59; significant after the Bonferroni's test)
(Fig. 3).
The ABCB1 sequence covered by pyrosequencing and HRM
analysis was the same and overlapped the transcriptional start site
of the ABCB1 gene. Neverthless, for accurate and optimal PCR
amplicon lengths in HRM analysis, this estimated sequence of the
ABCB1 gene was divided into 7 regions (Fig. 2). Typical results of methylation
analysis using HRM are presented in Fig.
4A (normalized graph of distribution of differentially
methylated samples along calibration curves ranging from 0 to 100%
methylation). Fig. 4B shows the
distribution of ABCB1 methylation using a baseline of 50%
methylation. Subsequently, this newly developed HRM method was used
for ABCB1 methylation analysis and evaluation of the
clinical consequences of ABCB1 methylation status in ovarian
carcinoma samples (n=61).
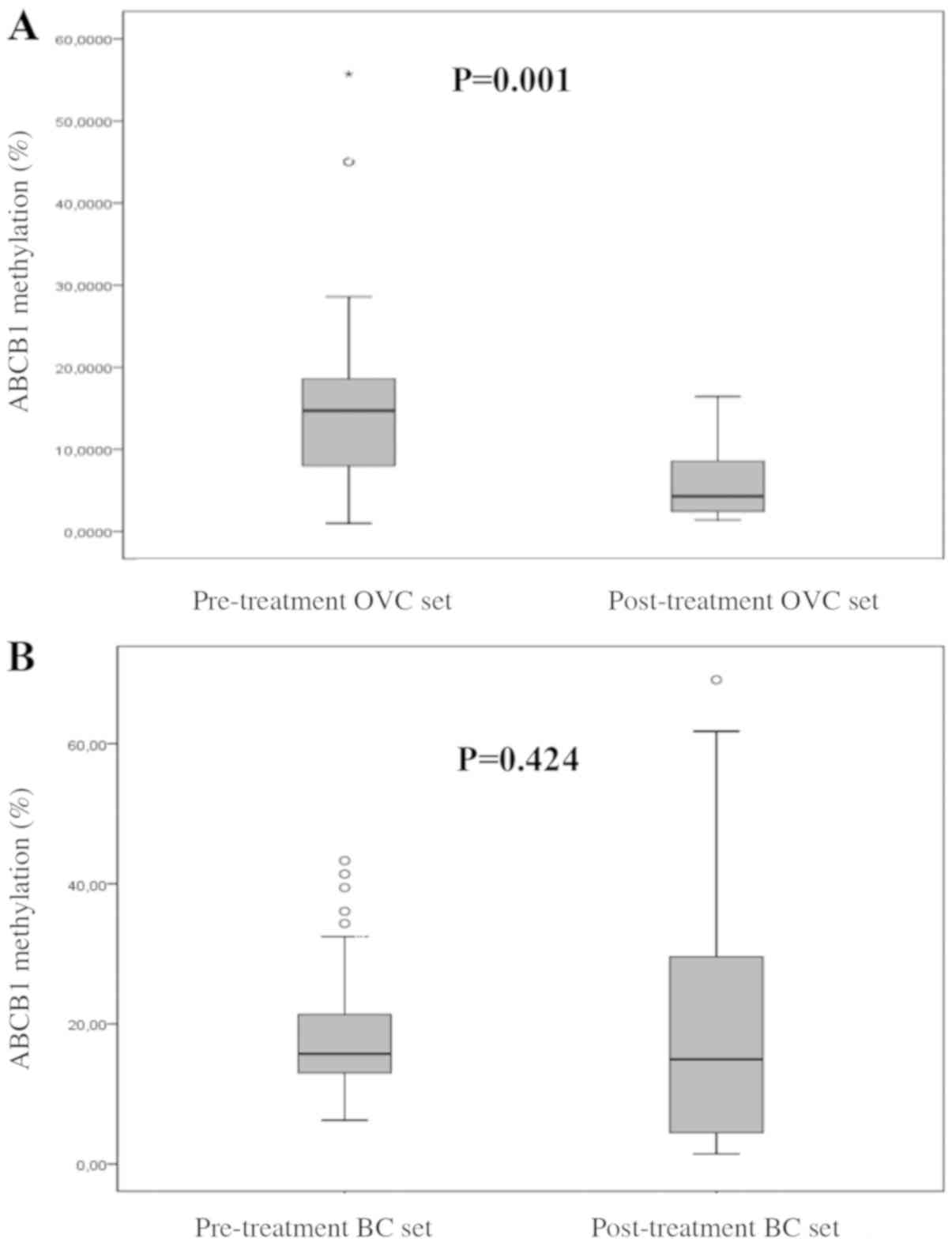
Variability of ABCB1 promoter
methylation
Extensive variability was observed in the
ABCB1 promoter methylation status ranging from 0 to 80%,
with mean methylation levels of 19 and 14% in breast and ovarian
carcinoma samples, respectively. Comparison of pre-treatment and
post-treatment levels of ABCB1 methylation revealed
significantly higher methylation of ABCB1 prior to
chemotherapy treatment in ovarian carcinoma samples as estimated by
the Mann-Whitney test (n=61; P=0.001; significant after the
Bonferroni's test; Fig. 5A), in which
paclitaxel and platinum derivatives were normally used. By
contrast, the difference in ABCB1 methylation between
pre-treatment and post-treatment breast carcinoma samples was not
statistically significant, as shown in Fig. 5B (Mann-Whitney test, n=171;
P=0.424).
When comparison between tumor and control tissue
samples were performed, no significant changes in ABCB1
methylation were observed for patients with breast carcinoma. In
ovarian carcinoma samples, the comparison of all (pre-treatment and
post-treatment) ovarian tumor tissues with control ovarian tissues
(n=11) revealed significant hypermethylation in 85.2% of tumor
samples as estimated by non-parametric the Kruskal-Wallis test
(n=61; P<0.001, Table IV). This
hypermethylation was confirmed by the multiple comparison post-hoc
Bonferroni's test; (n=50; P=0.002) in pretreatment ovarian tumor
samples when compared with control ovarian tissues.
 | Table IV.Significant changes in methylation
status of the ABCB1 promoter in the set of pre-treatment and
post-treatment ovarian carcinomas in comparison with control
ovarian tissue samples. |
Table IV.
Significant changes in methylation
status of the ABCB1 promoter in the set of pre-treatment and
post-treatment ovarian carcinomas in comparison with control
ovarian tissue samples.
| Gene | ABCB1
methylation status | Ovarian control
tissue samples (n=11) n (%)a | Ovarian carcinoma
tissue samples (n=61) n (%)a |
|---|
| ABCB1 | Hypomethylated | 0 (0) | 0 (0) |
|
| Normal-like | 11 (100) | 9 (14.8) |
|
|
Hypermethylated | 0 (0) | 52
(85.2)b |
Functional analysis of ABCB1
methylation levels
To assess a potential functional meaning of the
observed promoter methylation status, ABCB1 transcript
levels were determined by RT-qPCR, and the effect of methylation on
ABCB1 transcript levels was analyzed. In breast carcinoma
samples, correlations of ABCB1 methylation and transcript
levels were evaluated separately in samples from pre- and
post-treatment tumors (n=34 and 55, respectively), in which both
total RNA and DNA from tumor tissues were available. In the
pre-treatment set, ABCB1 DNA methylation levels negatively
correlated with ABCB1 transcript levels, as estimated by
Spearman's rho correlation (P=0.025, ρ=0.397; P=0.05 after the
Bonferroni's test), while the results were not statistically
significant in the post-treatment set (P=0.308). Similar findings
were observed in ovarian carcinoma samples, in which high
ABCB1 DNA methylation levels significantly correlated with
lower transcript levels (Spearman's rho correlation test; P=0.001,
ρ=0.470; P=0.002 after the Bonferroni's test) in the pre-treatment
set of ovarian carcinoma samples, but not in the post-treatment set
(P=0.069).
Clinical associations of ABCB1
promoter methylation and expression with prognosis of patients
Comparison of ABCB1 promoter methylation
levels and clinical features revealed various significant
associations. In the post-treatment breast carcinoma samples,
significantly higher ABCB1 intratumoral methylation levels
were observed in patients with negative p53 expression
(Mann-Whitney test; n=70, P=0.039; Table
V). In the pre-treatment ovarian carcinoma samples, higher
ABCB1 methylation levels were observed in tumors at stage I
in comparison with a cohort of advanced stages II–IV cases
(Mann-Whitney test; n=45, P=0.028; Table
V).
 | Table V.Evaluation of associations between
ABCB1 promoter methylation levels in breast and ovarian
carcinoma tissues and clinicopathological data of patients. |
Table V.
Evaluation of associations between
ABCB1 promoter methylation levels in breast and ovarian
carcinoma tissues and clinicopathological data of patients.
| Patient sets | Associated clinical
characteristics (n)a | ABCB1 methylation
(%) | P-value |
|---|
| Post-treatment
breast carcinoma set | p53
expression |
|
|
|
|
Positive (21) | 21.2±2.0 | 0.039b |
|
|
Negative (49) | 15.9±2.8 |
|
| Pre-treatment
ovarian carcinoma set | Stage |
|
|
|
| I
(2) | 31.1±13.9 | 0.028b |
|
|
Advanced stages II–IV
(43) | 18.6±2.1 |
|
Subsequently, analyses of ABCB1 transcript
levels and clinical features were performed. Intratumoral
ABCB1 transcript levels in the post-treatment breast or
ovarian carcinoma samples did not exhibit any association. In the
pre-treatment carcinoma samples from breast patients with grade-1
or 2 tumors, significantly higher ABCB1 transcript levels
were observed compared with patients exhibiting grade-3 tumors
(Mann-Whitney test; n=70, P=0.011). However, the trend test did not
show a statistically significant difference (P>0.05), and thus,
this association was not further discussed. Conversely,
significantly higher levels of ABCB1 in high-grade (grade 3)
tumors compared with low-grade (grades 1 or 2) tumors were observed
in the pre-treatment set of ovarian carcinoma samples (Mann-Whitney
test; n=45, P=0.021; Table VI).
 | Table VI.Evaluation of associations between
ABCB1 transcript levels in breast and ovarian carcinoma tissues and
clinicopathological data of the patients. |
Table VI.
Evaluation of associations between
ABCB1 transcript levels in breast and ovarian carcinoma tissues and
clinicopathological data of the patients.
| Patient set | Associated clinical
characteristics (n)a | ABCB1 expression
normalized to control genesb | P-value |
|---|
| Pre-treatment
ovarian carcinoma set | Grade |
|
|
|
| Low
grade 1/2 (10) | 1.74±0.03 | 0.021c |
|
| High
grade 3 (35) | 1.59±0.03 |
|
In the survival analysis, ABCB1 promoter
methylation or ABCB1 transcript expression was not
significantly associated with PFS of patients with breast or
ovarian carcinoma in the post-treatment set. In the pre-treatment
sets, low expression levels of ABCB1 were associated with
longer PFS estimated by the Kaplan-Meier method in patients with
breast carcinoma. Differencies between groups were compared using
the log-rank test (n=68; P=0.001; significant after the
Bonferroni's test; P=0.004; Fig. 6A).
The same trend, although non-significant, was also found in ovarian
carcinoma (n=50; P=0.05; P=0.2 after the Bonferroni's test;
Fig. 6B).
Discussion
In the present study, HRM methylation analysis of
the most frequently studied ABC drug-efflux transporter (namely
ABCB1, which is associated with MDR in cancer cells), was
developed. This method was compared with ABCB1 promoter
methylation status obtained by pyrosequencing and a strong
correlation of both methods was found.
DNA methylation and histone modifications are
important reversible mechanisms of epigenetic gene expression
regulation and serve a role in cancer development. Numerous recent
studies have suggested a direct role for epigenetic inactivation of
genes in determining tumor chemosensitivity (38). Methylation-specific PCR technology,
which has previously been used for ABCB1 gene analysis
mainly encompassing the SP-1 site in the 3′-region of ABCB1,
was applied (23–25). In the present study, the three CpG
islands identified by MethPrimer software in a promoter region
encompassing the start site were analyzed. This type of analysis
allows an exact estimation of a realistic methylation pattern of
the complete promoter region in contrast to only using small
regions of the ABCB1 gene separately. Furthermore, HRM
analysis allows a simple, rapid and reasonably economic (and
therefore, suitable for routine clinical practice) method for
detection of the percentage of methylated CpG islands of genes with
potential clinical importance.
The present study reports the first set of results
of ABCB1 promoter methylation assessments in ovarian
carcinoma. In breast carcinoma, ABCB1 promoter methylation
was estimated for the first time in 2010 (20). The patients with breast carcinoma in
that study displayed widespread aberrant CpG island methylation
(mean, 39.3% in 28 invasive breast tumors and 40.7% in 27 ductal
carcinoma in situ). Furthermore, comparison of tumor tissues
with normal breast tissues revealed significant ABCB1
hypermethylation in breast tumors in comparison with normal breast
tissue (20). Dejeux et al
(21) analyzed the methylation
patterns in the promoter regions of 14 genes including ABCB1
in 75 well-described pre-treatment samples from patients with
locally advanced breast carcinoma in comparison with 6 normal
breast tissues. Absence of methylation was observed in all normal
breast tissue samples, while 3 amplification products of the
ABCB1 gene were observed to be methylated in 70, 64 and 81%
of tumor samples. Klajic et al (22) estimated ABCB1 DNA methylation
by pyrosequencing in a series of 238 breast carcinoma tissue
samples (ranging from ductal carcinoma in situ to invasive
tumors of stages IV). ABCB1 was the most frequently
hypermethylated gene in all invasive and ductal carcinoma in
situ samples (mean methylation, 16.2%) in comparison with
normal breast tissue samples (mean methylation, 2.6%).
Collectively, it was demonstrated by previous studies and confirmed
in the present study that breast carcinoma exhibits frequent
ABCB1 promoter hypermethylation. The present study extends
the knowledge on this topic by the additional observation of such
hypermethylation in ovarian carcinomas.
The negative significant correlation between
ABCB1 methylation and gene expression levels observed in
samples prior to chemotherapy treatment in the two types of tumors
evaluated in the present study demonstrates a clear functional
effect of ABCB1 methylation status. It confirms the results
of Sharma et al (24), who
reported ABCB1 hypomethylation in tumor samples (n=41) with
high P-glycoprotein levels (as estimated using
immunohistochemistry) of patients with breast carcinoma prior to
chemotherapy or radiation treatment. The lack of correlation
between methylation status and expression levels in samples
following neoadjuvant therapy that was observed in the present
study is in the agreement with the results of Dejeux et al
(21), who reported a lack of such
correlation in post-treatment sets of samples. In ovarian
carcinoma, the correlation of ABCB1 promoter methylation
with the transcript levels of this gene that was observed in the
present study is the novelty of the present report.
Regarding clinical consequences, the present study
observed associations between ABCB1 expression and patient
survival. High ABCB1 expression levels significantly
predicted poor progressive-free survival (PFS) of patients with
breast and ovarian carcinoma prior to treatment. These observations
indirectly suggest that silencing of ABCB1 expression may
affect patient prognosis. Although a few significant associations
have been suggested between ABCB1 methylation levels and
prognostic factors of patients with breast or ovarian carcinomas,
none of these associations were confirmed by the post-hoc
tests.
Nevertheless, the present observation that
ABCB1 methylation does not appear to have a direct
prognostic role was unexpected, suggesting that the connection
between ABCB1 and drug resistance may be a complex
phenomenon. By contrast, it has been previously shown that patients
with breast carcinoma and hypermethylated ABCB1 promoter had
significantly longer median overall survival (OS) compared with
patients exhibiting a hypomethylated ABCB1 promoter
(21). Similarly, ABCB1
promoter hypermethylation in circulating DNA was significantly
associated with longer OS (23). By
contrast, Klajic et al (22)
did not observe a significant correlation between ABCB1
methylation and survival. Taken together, the correlation of
ABCB1 methylation with gene expression reported in the
present study may be of interest as a potential chemoresistance
biomarker although its biological relevance for drug transport
remains to be evaluated in vitro and confirmed in
vivo.
A modest sample size and a relatively low number of
samples corresponding to patients with ovarian carcinoma upon
receiving treatment may be considered as major limitations of the
present study. Our study primarily focused on the establishment and
validation of a novel method for assessment of epigenetic
regulation of an important gene implicated in chemoresistance.
Thus, all the present clinical findings should be interpreted with
caution and replicated by independent validation studies. Protein
levels of ABCB1 gene product called P-glycoprotein (P-gp)
are usually estimated in different tumor cells and they are
associated with multidrug resistance of tumor cells. In our
previous studies, we successfully identified P-gp by western blot
assay, but only in highly taxane-resistant SK-BR-3/PacR, MCF-7/PacR
and NCI/ADR-RES carcinoma cell lines (39,40). We
tried to estimate P-gp protein levels in breast and ovarian tumor
tissues with high and low level of methylation in the frame of the
present study. However, we did not succeed to unambiguously detect
the levels of P-gp in any of these tumor tissue samples by
immunoblotting analysis and thus it was not possible to connect
differences reflecting methylation status of the ABCB1 gene
with its protein product levels. Finally, but equally important,
methylation belongs to epigenetic factors controlling the rate of
transcription and may or may not contribute also to the translation
rate. This is probably a gene- and protein-specific process and
also presumably individually different due to variation in RNA
processing and protein translation and degradation machineries.
Nevertheless, we perceive this fact as a limitation of the study
and we are planing to collect more samples and estimate P-gp levels
by immunohistochemistry in the near future.
In conclusion, the present study reports the
successful development of a cost-effective HRM methylation analysis
method of the ABCB1 promoter region. Hypermethylation of the
analyzed ABCB1 promoter region significantly correlated with
downregulation of its transcript levels in tumors from
pre-treatment subsets of patients with breast and ovarian
carcinoma. The observed association of low ABCB1 transcript
levels with longer survival suggesting good prognosis in the
pre-treatment subsets of patients with breast and ovarian carcinoma
opens the potential use of ABCB1 as a prognostic biomarker.
Its clinical utility should be further evaluated in larger
independent cohorts of samples.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Ministry of Health of the Czech Republic (project no. 17-28470A) by
the project of the Czech Science Foundation (no. P303/12/G163), by
the project Progres Q28 ONCOLOGY from Charles University and by the
National Sustainability Program I (NPU I) (grant no. LO1503)
provided by the Ministry of Education Youth and Sports of the Czech
Republic.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
RV, VNK and PaS conceived and planned the study. RV,
VB, KE and GIGA performed the experiments. JK, RV and JT analyzed
the data and carried out statistical analyses. LR, RK, PeS and MM
prepared and characterized the tumor samples and collected the
clinical data. RV, VNK and PaS wrote the manuscript. All authors
read and approved the final manuscript, and agree to be accountable
for all aspects of the research in ensuring that the accuracy or
integrity of any part of the work are appropriately investigated
and resolved.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Commission of the National Institute of Public Health in Prague
(ethic codes: IGA no. 9799-4 of 30 January 2008, 14055-3 of 2 July
2012, and 14056-3 of 2 July 2012) and Institutional Review Board of
Norwegian Radiumhospital (Oslo, Norway) in the frame of the
Norwegian Cancer Society (D-03067) and the Norwegian Research
Council (163027/V409) projects. The methods were carried out in
accordance with guidelines approved by the above Ethics
Commissions. The study was conducted in accordance with the ethical
standards of the Declaration of Helsinki of 1975 and its most
recent version. All subjects were informed about the study topic
and provided their written consent to participate in the study.
Patient consent to publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gottesman MM and Pastan I: Biochemistry of
multidrug resistance mediated by the multidrug transporter. Annu
Rev Biochem. 62:385–427. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Leonessa F and Clarke R: ATP binding
cassette transporters and drug resistance in breast cancer. Endocr
Relat Cancer. 10:43–73. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ling V: Multidrug resistance: Molecular
mechanisms and clinical relevance. Cancer Chemother Pharmacol. 40
(Suppl):S3–S8. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sakaeda T, Nakamura T and Okumura K:
Pharmacogenetics of drug transporters and its impact on the
pharmacotherapy. Curr Top Med Chem. 4:1385–1398. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Arnal M, Franco N, Fargeot P, Riedinger
JM, Brunet-Lecomte P and Lizard-Nacol S: Enhancement of mdr1 gene
expression in normal tissue adjacent to advanced breast cancer.
Breast Cancer Res Treat. 61:13–20. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mairinger S, Erker T, Muller M and Langer
O: PET and SPECT radiotracers to assess function and expression of
ABC transporters in vivo. Curr Drug Metab. 12:774–792. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Charpin C, Vielh P, Duffaud F, Devictor B,
Andrac L, Lavaut MN, Allasia C, Horschowski N and Piana L:
Quantitative immunocytochemical assays of P-glycoprotein in breast
carcinomas: Correlation to messenger RNA expression and to
immunohistochemical prognostic indicators. J Natl Cancer Inst.
86:1539–1545. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vaclavikova R, Nordgard SH, Alnaes GI,
Hubackova M, Kubala E, Kodet R, Mrhalova M, Novotny J, Gut I,
Kristensen VN and Soucek P: Single nucleotide polymorphisms in the
multidrug resistance gene 1 (ABCB1): Effects on its expression and
clinicopathological characteristics in breast cancer patients.
Pharmacogenet Genomics. 18:263–273. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ehrlichova M, Mohelnikova-Duchonova B,
Hrdy J, Brynychova V, Mrhalova M, Kodet R, Rob L, Pluta M, Gut I,
Soucek P and Vaclavikova R: The association of taxane resistance
genes with the clinical course of ovarian carcinoma. Genomics.
102:96–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Johnatty SE, Beesley J, Gao B, Chen X, Lu
Y, Law MH, Henderson MJ, Russell AJ, Hedditch EL, Emmanuel C, et
al: ABCB1 (MDR1) polymorphisms and ovarian cancer progression and
survival: A comprehensive analysis from the ovarian cancer
association consortium and the cancer genome atlas. Gynecol Oncol.
131:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lili X and Xiaoyu T: Expression of PKCα,
PKCε, and P-gp in epithelial ovarian carcinoma and the clinical
significance. Eur J Gynaecol Oncol. 36:181–185. 2015.PubMed/NCBI
|
|
12
|
Burger H, Foekens JA, Look MP, Meijer-van
Gelder ME, Klijn JG, Wiemer EA, Stoter G and Nooter K: RNA
expression of breast cancer resistance protein, lung
resistance-related protein, multidrug resistance-associated
proteins 1 and 2, and multidrug resistance gene 1 in breast cancer:
Correlation with chemotherapeutic response. Clin Cancer Res.
9:827–836. 2003.PubMed/NCBI
|
|
13
|
Chintamani, Singh JP, Mittal MK, Saxena S,
Bansal A, Bhatia A and Kulshreshtha P: Role of p-glycoprotein
expression in predicting response to neoadjuvant chemotherapy in
breast cancer-a prospective clinical study. World J Surg Oncol.
3:612005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Surowiak P, Materna V, Matkowski R,
Szczuraszek K, Kornafel J, Wojnar A, Pudelko M, Dietel M, Denkert
C, Zabel M and Lage H: Relationship between the expression of
cyclooxygenase 2 and MDR1/P-glycoprotein in invasive breast cancers
and their prognostic significance. Breast Cancer Res. 7:R862–R870.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu L, Katsaros D, Wiley A, Rigault de la
Longrais IA, Puopolo M and Yu H: Expression of MDR1 in epithelial
ovarian cancer and its association with disease progression. Oncol
Res. 16:395–403. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Egger G, Liang G, Aparicio A and Jones PA:
Epigenetics in human disease and prospects for epigenetic therapy.
Nature. 429:457–463. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tabolacci E and Chiurazzi P: Epigenetics,
fragile X syndrome and transcriptional therapy. Am J Med Genet A
161A. 2797–2808. 2013. View Article : Google Scholar
|
|
18
|
O'Hagan HM, Mohammad HP and Baylin SB:
Double strand breaks can initiate gene silencing and
SIRT1-dependent onset of DNA methylation in an exogenous promoter
CpG island. PLoS Genet. 4:e10001552008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tabish AM, Poels K, Hoet P and Godderis L:
Epigenetic factors in cancer risk: Effect of chemical carcinogens
on global DNA methylation patternin human TK6 cells. PLoS One.
7:e346742012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Muggerud AA, Rønneberg JA, Wärnberg F,
Botling J, Busato F, Jovanovic J, Solvang H, Bukholm I,
Børresen-Dale AL, Kristensen VN, et al: Frequent aberrant DNA
methylation of ABCB1, FOXC1, PPP2R2B and PTEN in ductal carcinoma
in situ and early invasive breast cancer. Breast Cancer Res.
12:R32010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dejeux E, Rønneberg JA, Solvang H, Bukholm
I, Geisler S, Aas T, Gut IG, Børresen-Dale AL, Lønning PE,
Kristensen VN and Tost J: DNA methylation profiling in doxorubicin
treated primary locally advanced breast tumours identifies novel
genes associated with survival and treatment response. Mol Cancer.
9:682010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Klajic J, Fleischer T, Dejeux E, Edvardsen
H, Warnberg F, Bukholm I, Lønning PE, Solvang H, Børresen-Dale AL,
Tost J and Kristensen VN: Quantitative DNA methylation analyses
reveal stage dependent DNA methylation and association to
clinico-pathological factors in breast tumors. BMC Cancer.
13:4562013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sharma G, Mirza S, Parshad R, Srivastava
A, Datta Gupta S, Pandya P and Ralhan R: CpG hypomethylation of
MDR1 gene in tumor and serum of invasive ductal breast carcinoma
patients. Clin Biochem. 43:373–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sharma G, Mirza S, Parshad R, Gupta SD and
Ralhan R: DNA methylation of circulating DNA: A marker for
monitoring efficacy of neoadjuvant chemotherapy in breast cancer
patients. Tumour Biol. 33:1837–1843. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Enokida H, Shiina H, Igawa M, Ogishima T,
Kawakami T, Bassett WW, Anast JW, Li LC, Urakami S, Terashima M, et
al: CpG hypermethylation of MDR1 gene contributes to the
pathogenesis and progression of human prostate cancer. Cancer Res.
64:5956–5962. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Vaclavikova R, Ehrlichova M, Hlavata I,
Pecha V, Kozevnikovova R, Trnkova M, Adamek J, Edvardsen H,
Kristensen VN, Gut I and Soucek P: Detection of frequent ABCB1
polymorphisms by high-resolution melting curve analysis and their
effect on breast carcinoma prognosis. Clin Chem Lab Med.
50:1999–2007. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Klajic J, Busato F, Edvardsen H, Touleimat
N, Fleischer T, Bukholm I, Børresen-Dale AL, Lønning PE, Tost J and
Kristensen VN: DNA methylation status of key cell-cycle regulators
such as CDKNA2/p16 and CCNA1 correlates with treatment response to
doxorubicin and 5-fluorouracil in locally advanced breast tumors.
Clin Cancer Res. 20:6357–6366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Aas T, Børresen AL, Geisler S,
Smith-Sørensen B, Johnsen H, Varhaug JE, Akslen LA and Lønning PE:
Specific P53 mutations are associated with de novo resistance to
doxorubicin in breast cancer patients. Nat Med. 2:811–814. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Geisler S, Lønning PE, Aas T, Johnsen H,
Fluge O, Haugen DF, Lillehaug JR, Akslen LA and Børresen-Dale AL:
Influence of TP53 gene alterations and c-erbB-2 expression on the
response to treatment with doxorubicin in locally advanced breast
cancer. Cancer Res. 61:2505–2512. 2001.PubMed/NCBI
|
|
30
|
Geisler S, Børresen-Dale AL, Johnsen H,
Aas T, Geisler J, Akslen LA, Anker G and Lønning PE: TP53 gene
mutations predict the response to neoadjuvant treatment with
5-fluorouracil and mitomycin in locally advanced breast cancer.
Clin Cancer Res. 9:5582–5588. 2003.PubMed/NCBI
|
|
31
|
Brynychová V, Hlaváč V, Ehrlichová M,
Václavíková R, Pecha V, Trnková M, Wald M, Mrhalová M, Kubáčková K,
Pikus T, et al: Importance of transcript levels of caspase-2
isoforms S and L for breast carcinoma progression. Future Oncol.
9:427–438. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Elsnerova K, Mohelnikova-Duchonova B,
Cerovska E, Ehrlichova M, Gut I, Rob L, Skapa P, Hruda M, Bartakova
A, Bouda J, et al: Gene expression of membrane transporters:
Importance for prognosis and progression of ovarian carcinoma.
Oncol Rep. 35:2159–2170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tost J and Gut IG: DNA methylation
analysis by pyrosequencing. Nat Protoc. 2:2265–2275. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Soucek P, Anzenbacher P, Skoumalová I and
Dvorák M: Expression of cytochrome P450 genes in CD34+
hematopoietic stem and progenitor cells. Stem Cells. 23:1417–1422.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bustin SA, Benes V, Garson JA, Hellemans
J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL,
et al: The MIQE guidelines: Minimum information for publication of
quantitative real-time PCR experiments. Clin Chem. 55:611–622.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Amin MB, Edge SB, Greene FL, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton CC, Hess KR,
Sullivan DC, et al: AJCC cancer staging manual. 8th. Springer;
Cham, Switzerland: 2017, View Article : Google Scholar
|
|
38
|
Chen Z, Shi T, Zhang L, Zhu P, Deng M,
Huang C, Hu T, Jiang L and Li J: Mammalian drug efflux transporters
of the ATP binding cassette (ABC) family in multidrug resistance: A
review of the past decade. Cancer Lett. 370:153–164. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Václavíková R, Boumendjel A, Ehrlichová M,
Kovár J and Gut I: Modulation of paclitaxel transport by flavonoid
derivatives in human breast cancer cells. Is there a correlation
between binding affinity to NBD of P-gp and modulation of
transport? Bioorg Med Chem. 14:4519–4525. 2006.
|
|
40
|
Němcová-Fürstová V, Kopperová D,
Balušíková K, Ehrlichová M, Brynychová V, Václavíková R, Daniel P,
Souček P and Kovář J: Characterization of acquired paclitaxel
resistance of breast cancer cells and involvement of ABC
transporters. Toxicol Appl Pharmacol. 310:215–228. 2016. View Article : Google Scholar : PubMed/NCBI
|