Introduction
Non-small cell lung cancer (NSCLC) accounts for ~85%
of all cases of lung cancer, and can be further classified as lung
adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC) and
large cell carcinoma (1). However,
the majority of lung cancer cases are diagnosed at a late stage
after tumour cell metastasis has usually occurred, which leads to a
poor prognosis and high mortality rates worldwide (2). Therefore, identifying biomarkers for
the early diagnosis of lung cancer and therapeutic targets are
essential in order to enhance clinical outcomes.
The netrin (Ntn) family consists of Ntn-1, −2, −3,
−4, -G1 and G2, which all belong to the superfamily of laminins.
These proteins were initially described as chemoattractive or
chemorepulsive cues to guide axonal migration and neuronal growth
in the developing central nervous system (3,4). Over
the course of the past few decades, Ntns have been shown to be
involved in diverse biological processes other than neuronal
development, including processes such as organogenesis,
angiogenesis, tumorigenesis and inflammation (5,6). Ntn-4
was cloned in 2000 and has been found to be involved in several
different types of cancer. Ntn-4 was shown to inhibit angiogenesis
and the progression of colorectal cancer by binding to the
cell-surface receptor, neogenin, or uncoordinated receptor 5 (Unc5)
(7–10); by contrast, other studies have
reported that Ntn-4 promotes the proliferation and invasion of
gastric cancer cells (11), and it
has also been shown to increase the survival and migration rates of
neuroblastoma cells (12),
suggesting that the role of Ntn-4 in tumour progression remains
controversial. Previously, Reuten et al (13) demonstrated that Ntn-4 plays a key
role in regulating basement membrane (BM) composition and stiffness
by interacting with the γ1 chain of laminin, rather than binding to
any of the Ntn receptors. It has also been demonstrated that the
presence of Ntn-4 maintains a softer BM, and a decrease in the
level of Ntn-4 leads to an increase in BM stiffness and the
alteration of the tumour microenvironment, which consequently
promotes the migration and metastasis of breast cancer cells
(14,15). The level of Ntn-4 in breast cancer
tumours has been found to be associated with a longer disease-free
survival and overall survival rates, and it has been shown to be an
independent prognostic factor in patients with early-stage breast
cancer (16,17). Data from a public NSCLC dataset from
the Netherlands Cancer Institute (NKI) revealed that there was a
1.5-fold decrease in the expression level of Ntn-4 in patients with
NSCLC; however, the biological effects of Ntn-4 on NSCLC have yet
to be fully elucidated. Moreover, the precise reason why Ntn-4 is
downregulated in NSCLC needs to be further investigated.
Owing to alternative splicing, the RNA-binding
protein quaking (Qki) gene is translated into three main protein
isoforms, namely Qki-5, Qki-6 and Qki-7, which differ in terms of
their C-terminal amino acid sequences (18). Qki regulates the
post-transcriptional level of target genes through selectively
interacting with the Qki response element (19). Increasing evidence has revealed that
Qki fulfils an important role in lung cancer as a tumour suppressor
by alternatively splicing cancer-associated genes. Notably, a low
expression level of Qki in NSCLC is an independent prognostic
factor for disease-free survival (20). It has also been reported that a low
expression level of Qki-6 is positively associated with poor
overall survival rates in patients with NSCLC. The upregulation of
Qki-6 expression has been found to inhibit NSCLC
cell proliferation, migration and epithelial-mesenchymal transition
(EMT) (21). The overexpression of
Qki-5 has also been shown to suppress the progression and EMT of
NSCLC cells by inhibiting the level of β-catenin (22), the inhibition of Notch signalling
(23) and TGF-β/Smad signalling
(24), or by alternatively
repressing the splicing of the cytoskeletal gene adducin3 in lung
cancer cells (25).
Since low levels of Qki-5 and Ntn-4 have been shown
to contribute to the development and progression of NSCLC, the
present study aimed to investigate whether Qki-5 can affect NSCLC
progression by regulating the expression of Ntn-4. To address this
question, the correlation between Qki-5 and Ntn-4 was first
analysed according to the NKI data. As was anticipated, Qki-5 and
Ntn-4 were found to be strongly correlated in NSCLC. Subsequently,
as detailed in the Materials and methods section, a series of
experiments were performed which confirmed that a decrease in the
level of Qki-5 promoted the proliferation, migration, invasion and
EMT of NSCLC by downregulating Ntn-4. Taken together, the findings
of the present study shed light onto the biological role of Ntn-4
in NSCLC progression, also helping to elucidate one of the
underlying mechanisms responsible for the downregulation of Ntn-4
in lung cancer.
Materials and methods
The Cancer Genome Atlas (TCGA) data
analysis
TCGA data analysis was performed using the publicly
accessible TCGA database (https://tcga-data.nci.nih.gov/tcga/). The mRNA values
of Ntn-4 and Qki were collected from the samples of 483 patients
with LUAD and 59 control samples, and from the samples of 486
patients with LUSC and 50 control samples, which were subsequently
quantified according to TCGA normalization protocol. The mRNA
levels of Qki and Ntn-4 that were associated with the survival data
for the patients with LUAD and LUSC were also analysed according to
TCGA datasets.
Clinical sample collection
For the Ntn-4 and Qki-5 mRNA expression level
analysis, a total of 30 patients diagnosed with LUAD, and 30
patients diagnosed with LUSC, at the Fourth Affiliated Hospital of
China Medical University were enrolled in the present study. The
Ethics Committee of the Fourth Hospital of China Medical University
approved the present study (approval no. EC-2018-HX-013), and all
the patients involved in this research signed informed consent
forms. Tumour tissues, and the corresponding adjacent normal
tissues were dissected and stored in liquid nitrogen for periods up
to 90 days prior to performing the reverse
transcription-quantitative PCR (RT-qPCR) experiments.
Immunohistochemistry (IHC)
The tumour samples from patients with LUAD or LUSC
were collected between July, 2018 and August, 2019. For the Ntn-4
and Qki-5 IHC staining of the human LUAD and LUSC tissues, slides
were prepared using an ultrathin semiautomatic microtome,
deparaffinized in 100% xylene and rehydrated in a graded ethanol
series (70, 80, 90, 95 and 100%, Beijing InnoChem Science &
Technology Co.,Ltd.). For antigen retrieval, the slides were
treated with sodium citrate solution (pH 6.0) (Beijing Solarbio
Science & Technology Co., Ltd.) for 6 min at 100°C. After
incubating the slides in 1% bovine serum albumin in
phosphate-buffered saline (PBS) for 30 min at room temperature, the
slides were incubated with anti-Ntn-4 (rabbit, 1:1,000, cat. no.
NBP191343, Novus Biologicals, LLC) and anti-Qki-5 (rabbit, 1:1,000,
cat. no. AB9904, MilliporeSigma) antibodies overnight at 4°C. After
washing the slides several times with PBS, they were incubated with
a goat anti-rabbit IgG secondary antibody conjugated to horseradish
peroxidase (HRP; 1:300, cat. no. BF03008, Biodragon Immunotech) for
30 min at room temperature. Staining was visualized using a DAB
Horseradish Peroxidase Color Development kit (p0203, Beyotime
Institute of Biotechnology). An appropriate amount of DAB dyeing
solution was then added, and the tissues were fully covered and
incubated at room temperature for 30 min away from light. The
nuclei were counterstained for 30 sec with haematoxylin at room
temperature. (C0107, Beyotime Institute of Biotechnology) For the
negative control experiments, sections were incubated with PBS
instead of the primary antibodies. Positive staining controls were
performed using para-carcinoma tissues of breast cancer and renal
cancer. The slides were finally covered with neutral balata and
cover glass slips. The qualitative scoring system used for
evaluation of the intensity of staining was as follows: Negative
(no staining, 0); weakly positive (+); moderately positive (++);
and strongly positive (+++). Images of the stained tissues were
obtained using a positive fluorescence microscope (BX53/DP73,
Olympus Corporation).
Cells, cell culture and treatment
The human LUAD cell lines, A549 (cat. no. TCHu150)
and H23 (cat. no. SCSP-5002) were purchased from The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China). The LUSC cell line, NCI-H2107 (cat. no.
CRL-5983_FL), and the human bronchial epithelial cell line, 16HBE
(cat. no. PCS-300-010) were purchased from Jennino Biological
Technology (Guangzhou, China). The cells were cultured in
Gibco® Roswell Park Memorial Institute (RPMI)-1640
medium (Thermo Fisher Scientific, Inc.) supplemented with 10% (v/v)
Gibco® foetal bovine serum (FBS; Thermo Fisher
Scientific, Inc.) and maintained in a humidified atmosphere with 5%
CO2 at 37°C.
Lentiviral production and
transfection
To generate the Ntn-4- and Qki-5-overexpressing cell
lines (oe-Ntn-4 and oe-Qki-5), the Ntn-4 or Qki-5 coding sequence,
respectively, was cloned into the GV492 (11.9 kb) lentiviral vector
(Shanghai Genechem Co., Ltd.) to form recombinant plasmids.
Subsequently, the recombinant GV492, Helper 1.0 (12.0 kb) and
Helper 2.0 (5.8 kb) plasmids were co-transfected into 293T cells
(Wuhan Procell Life Technology Co., Ltd.). After 48 or 72 h, the
culture supernatant was harvested, concentrated and purified
(82,700 g for 2 h at 4°C). The concentrations of lentivirus were
measured using Immunostaining Plaque Assay (26), and the data obtained revealed that
the titres of LV-Ntn-4 and LV-Qki-5 were 1.5E+09 and
1.2E+09 (TU/ml), respectively.
To produce the short hairpin RNAs (shRNAs) of human
Ntn-4 and Qki-5, the target genes for Ntn-4 (sh-Ntn-4) (sequence,
5′-CCGGGCGCTATTTGTACTTCTAAAT-3′), Qki-5 (sh-Qki-5) (sequence,
5′-CCGGCTATTAACCCACAGCATTTT-3′) and the negative control (NC)
scramble (sh-NC, the scramble sequences of shNtn-4 and shQki-5 are
TTCTCCGAACGTGTCACGT and TATTAACCACAATGCGCTTTGCCC, respectively)
were inserted into the GV118 vector (7.4 kb) (Shanghai Genechem
Co., Ltd.). The recombinant plasmids Helper 1.0 (12.0 kb) and
Helper 2.0 (5.8 kb) were again co-transfected into 293T cells. The
production and quantification of LV-Ntn-4-RNAi were performed
following the protocol described above (26). The titre of LV-Ntn-4-RNAi was
determined to be 2.00E+08 TU/ml. Lentiviral production
was performed by Shanghai Genechem Co., Ltd. Finally, the cells
(~1×105 cells/ml) were seeded in cell culture dishes and
subsequently transfected with the lentivirus, following the
manufacturer's instructions.
Cell viability analysis
The transfected A549 and H2107 cells were seeded
into 96-well plates at a density of 4×103 cells/well,
and allowed to grow for 2 days. Cell proliferation was determined
using a Cell Counting Kit-8 (CCK-8) (Beyotime Institute of
Biotechnology) assay. The absorbance at 450 nm was read using a
microplate reader (BioTek Instruments, Inc.).
RT-qPCR analysis
Total RNA was extracted using Invitrogen®
TRIzol™ reagent (Thermo Fisher Scientific, Inc.), and first-strand
cDNA was generated using an RT system in a 20 µl reaction mixture
[SYBR-Green Master Mix (2X) (No ROX) 10 µl, PCR forward primer (10
µM) 0.4 µl, PCR reverse primer (10 µM) 0.4 µl, DNA 2 µl and
ddH2O 7.2 µl] containing 1 µg total RNA. Aliquots (0.5
µl) of cDNA were amplified using Fast SYBR-Green PCR Master Mix
(Vazyme Biotech Co., Ltd.) in each 20 µl reaction. PCRs were run on
a Roche Light Cycler 480 II with the following primers: Qki-5
forward, 5′-TCCGAGGCAAAGGCTCAATGAG-3′ and reverse,
5′-GCTCTGTTCTGAGCATCTTCCAC-3′; Ntn-4 forward,
5′-GTACTTTGCGACTAACTGCTCC-3′ and reverse,
5′-TCCAGTGCATGGAAAAGGACT-3′; and GAPDH forward,
5′-GAAGGTGAAGGTCGGAGT-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
The PCR thermocycling conditions were pre-denaturation for 30 sec
at 95°C, denaturation for 20 sec at 95°C, annealing for 15 sec at
58°C, and extension for 15 sec at 72°C. The relative expression
values of Qki-5 and Ntn-4 were calculated and normalized to those
of GAPDH in each sample using the 2−ΔΔCq method
(27).
Western blot analysis
Following transfection of the cells for 48 h, the
cells were collected, washed and lysed with cell lysis buffer
(P0013B, Beyotime Institute of Biotechnology) containing protease
inhibitors phenylmethylsulfonyl fluoride (PMSF; ST506-2, Beyotime
Institute of Biotechnology), and the protein concentrations were
quantified using an enhanced BCA protein assay kit (Biosharp Life
Sciences). Protein electrophoresis was performed using 10% sodium
dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE;
kgb113k, Nanjing KeyGen Biotech Co., Ltd.), and western blotting
was performed on a nitrocellulose transfer membrane. After using 5%
skimmed milk powder closed 2 h at room temperature, the
nitrocellulose filter membranes (66485, Pall Life Sciences) were
incubated at 4°C overnight with the following primary antibodies:
Anti-β-actin (mouse, 1:5,000; cat. no. 66009-1-Ig, Proteintech
Group, Inc.), anti-Ntn-4 (rabbit, 1:1,000; cat. no. NBP191343,
Novus Biologicals, LLC), anti-Qki-5 (rabbit, 1:1,000; cat. no.
AB9904, MilliporeSigma), anti-E-cadherin (rabbit, 1:5,000; cat. no.
20874-1-AP), anti-N-cadherin (rabbit, 1:5,000; cat. no.
22018-1-AP), anti-vimentin (rabbit, 1:5,000; cat. no. 13066-1-AP)
and anti-Snail (rabbit, 1:500; 13099-1-AP) (all from Proteintech
Group, Inc.). After washing three times with Tris-buffered saline
containing Tween (TBST; cat. no. t917680; Beijing Innochem
Technology Co., Ltd), the membranes were incubated with
HRP-conjugated secondary antibody (Goat anti-Rabbit, 1:5000; cat.
no. A21020; Abbkine Scientific Co., Ltd.) at room temperature for 2
h. The protein bands were scanned using an Odyssey infrared imager
(LI-COR Biosciences). The relative grey values of the proteins of
interest were analysed using ImageJ software (Version 1.51j8,
National Institutes of Health) and normalized against those of
β-actin.
Wound healing assay
The transfected A549 and H2107 cells were grown
until they had reached ~90% confluency in a 12-well plate, and a
cell wound was created by scratching the cells with a sterile
200-µl pipette tip. After washing the cells with PBS twice, they
were cultured in RPMI-1640 medium supplemented with 0.5% FBS for 48
h. Cell images were captured under a microscope (IX73, Olympus
Corporation). The gap sizes were measured using ImageJ software
(version 1.51J8) and calculated as the percentages after 48 h
relative to the gap sizes at 0 h.
Cell migration assay
The cells were collected and resuspended in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.). Aliquots (200
µl) of cell suspension were placed in the upper chamber of a
Transwell insert (Corning, Inc.). Subsequently, the Transwell
chamber was inserted into a 24-well plate. Following incubation of
the cells in RPMI-1640 medium containing 10% (v/v) FBS for 24 h at
37°C, the cells on the upper membrane of the insert were gently
removed using a cotton swab, whereas those cells that had migrated
to the bottom surface were fixed with cold methanol (Wuhan
Servicebio Co.,Ltd) and stained with crystal violet (Wuhan
Servicebio Co., Ltd.) for 30 min at room temperature. Finally, the
cell numbers on the bottom surface were photographed and quantified
using an inverted microscope system (Olympus IX73; Olympus
Corporation).
RNA immunoprecipitation (RIP)
assay
RIP experiments were performed according to the
manufacturer's instructions using the BersinBio™ RIP kit (Guangzhou
Bersinbio Co., Ltd.). Briefly, the A549 cells were lysed using RIP
lysis buffer, and the cell extract was incubated with protein A/G
agarose beads (Guangzhou Bersinbio Co., Ltd.) conjugated with
either Qki-5 antibody (rabbit, 1:400; cat. no. AB9904,
MilliporeSigma) or control IgG (rabbit, 1:4000; cat. no. abs20035,
Absin) for 2 h. To purify RNA from the immunoprecipitation, the
beads were washed, and the proteins were removed using Proteinase K
(0.5 mg/ml). Subsequently, RT-qPCR experiments were performed to
analyse the mRNA expression level of Ntn-4.
Xenograft tumour growth
BALB/c (nu/nu) nude female mice (6 weeks old,
weighing 20–23 g, n=20 for one trial) were randomly divided into
four groups with 5 mice in each group as follows: The oe-NC,
oe-Qki-5 + sh-NC, oe-Qki-5 + sh-Ntn-4 and oe-Qki-5 + oe-Ntn-4. The
A549 cells (1×107 cells/ml, 0.2 ml) stably transfected
with the vectors overexpression (oe)-Qki-5, oe-Qki-5 and oe-Ntn-4,
oe-Qki-5 and sh-Ntn-4, or empty vector were subcutaneously injected
into the flanks of mice. During the procedure, the mice were
anaesthetised using 4% isoflurane (Shandong Keyuan Pharmaceutical
Co.; in oxygen), with the subsequent maintenance of anaesthesia
with 2.5% isoflurane. Tumour diameters were measured using
callipers, and volumes were calculated based on the formula
V= (a × b2)/2, where a and
b represent the longest and the shortest diameter of the
tumour, respectively. To minimize animal suffering, the mice were
housed under standard conditions and cared for (23°C; 50% humidity;
12:12 light/dark cycle; food and water were available ad
libitum) according to institutional guidelines. At the end of
the experiment, the animals were euthanized with 5% isoflurane
inhalation for 5–10 min; the vital signs were checked to ensure
their disappearance, and cervical dislocation was finally performed
to avoid the possibility of recovery. Finally, the tumours were
isolated, weighed and photographed. The animal experiments were
approved by the Animal Research Ethics Committee of the Animal
Centre of Shengjing Hospital affiliated with China Medical
University (No. KT201945).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism version 8.0.0 for Windows by observers blinded to the
experimental design. The data are presented as the mean ± standard
error of the mean (SEM). The data between two groups were compared
using an unpaired ttest, whereas oneway analysis of variance
(ANOVA) followed by the post-hoc Tukey's Student Range (HSD) test
was adopted for data comparisons among multiple groups. The
correlation between the mRNA expression levels of Qki-5 and Ntn-4
was analysed using Pearson's correlation (R2
correlation). The optical density values and tumour volume at
different time points were compared by twofactor ANOVA. A value of
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression of Ntn-4 is downregulated
in NSCLC tissues and cells
The NSCLCassociated expression dataset was retrieved
from TCGA database (https://portal.gdc.cancer.gov/). A total of 1,078
samples were obtained, including 483 LUAD samples and 59 control
samples, as well as 486 LUSC samples and 50 control samples. The
Ntn-4 expression data were subsequently obtained and analysed, and
the results confirmed that the mRNA expression level of Ntn-4 was
lower in both the LUAD and LUSC samples compared with the control
samples (Fig. 1A). No statistically
significant differences in the overall survival of the patients
with LUSC were observed when comparing between the high and low
Ntn-4 mRNA level groups (Fig. 1B);
however, the patients with LUAD with a low mRNA expression level of
Ntn-4 exhibited poorer overall survival rates compared with the
patients with high Ntn-4 mRNA expression levels (Fig. 1C). In addition, the results of
RT-qPCR revealed that the expression of Ntn-4 in NSCLC tissues was
lower compared with that in the control tissues (Fig. 1D). Moreover, IHC staining analysis
also revealed comparatively lower expression levels of Ntn-4 in
NSCLC tissues relative to the control tissues (Fig. 1E). With respect to the in
vitro experiments, the expression level of Ntn-4 was detected
in the human NSCLC cell lines, H23, A549 and H2170, and Ntn-4 was
found to be expressed at low levels in the human NSCLC cell lines
compared with the 16HBE control cell line (Fig. 1F). Moreover, the western blot
analysis revealed that, compared with that in the control cells,
the Ntn-4 protein expression levels in the NSCLC cells were lower
(Fig. 1G).
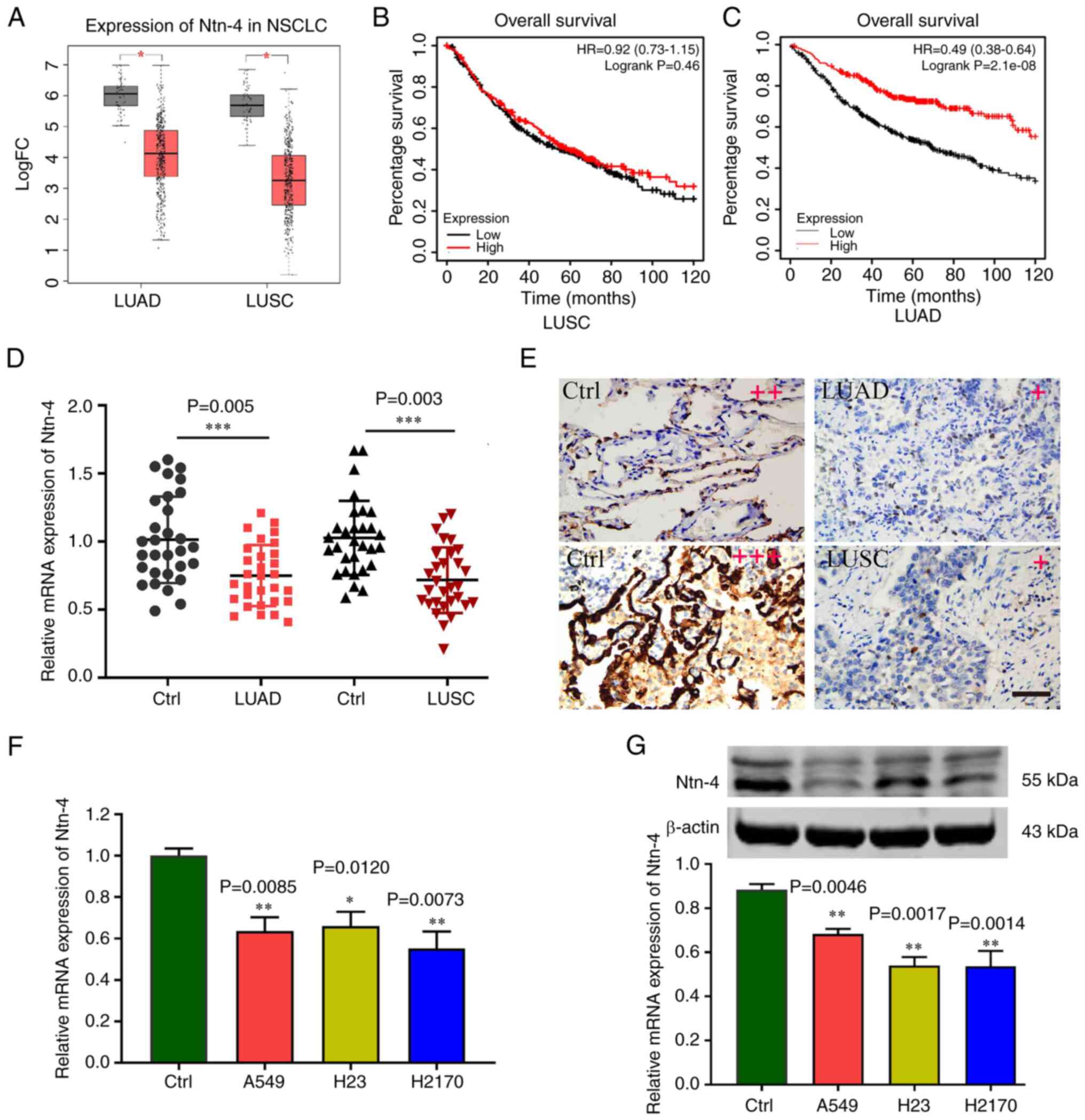 | Figure 1.Level of Ntn-4 is decreased in NSCLC
tissues and cells. (A) Box-plot of Ntn-4 expression in the LUAD and
LUSC datasets; the red boxes portray the cancer groups and the grey
boxes represent the control groups. *P<0.05 vs. the control. (B
and C) The differences in the overall survival rates between the
high and low Ntn-4 mRNA expression level groups in patients with
NSCLC are shown. (D) Expression of Ntn-4 mRNA in LUAD and LUSC
tissues and adjacent normal tissues (Ctrl) detected using RT-qPCR
(n=30). ***P<0.001 vs. Ctrl. (E) Representative images of Ntn-4
immunohistochemical staining in LUAD, LUSC and Ctrl tissues are
shown. Scale bar, 100 µm. The qualitative score for staining was as
follows: Negative (0: no staining), weakly positive (+), moderate
positive (++), and strongly positive (+++). (F) The expression of
Ntn-4 mRNA in LUAD cells (NCI-H23 and A549 cell lines), LUSC cells
(H2170 cell line) and normal human bronchial epithelial cells
(16HBE cell line) was measured using RT-qPCR. Data are presented as
the mean ± SEM of three independent experiments (n=3). *P<0.05
and **P<0.01 vs. 16HBE cells (Ctrl). (G) Levels of Ntn-4 protein
in LUAD, LUSC and Ctrl cells were detected using western blot
analysis, and were quantified using ImageJ software and normalized
against β-actin. LUAD, lung adenocarcinoma; LUSC, lung squamous
cell carcinoma; NSCLC, non-small cell lung cancer; Ntn, netrin;
Ctrl, control; RT-qPCR, reverse transcription-quantitative PCR. |
Ntn-4 inhibits the proliferation,
migration and invasion of NSCLC cells
To further examine the effects of Ntn-4 on NSCLC
cells, the A549 and H2170 cells were transfected with Ntn-4 shRNA
or Ntn-4 overexpression lentivirus. The results from the ensuing
RT-qPCR and western blot analysis experiments confirmed that the
levels of Ntn-4 were increased in the cells with the
lentivirus-mediated overexpression of Ntn-4 compared with the
control cells (Fig. 2A and B),
whereas the levels were decreased in both the A549 and H2170 cells
with Ntn-4 shRNA lentivirus transfection (Fig. 2C and D). The OD values of both A549
(Fig. 2E) and H2170 (Fig. 2F) cells were found to be
significantly increased in the sh-Ntn-4 group compared with the
sh-NC group, although they were decreased in the cells transfected
with the oe-Ntn-4 lentivirus compared with the cells transfected
with oe-NC lentivirus, indicating that Ntn-4 may exert its effect
through the inhibition of NSCLC cell proliferation. Cell migration
and invasion were subsequently assessed using wound healing and
Transwell assays. The quantification of the resultant data revealed
that the efficiency of scratch wound healing was lower in the cells
transfected with oe-Ntn-4 compared with the cells transfected with
oe-NC, whereas Ntn-4 silencing led to the opposite findings in
terms of the scratch wound healing ability of the cells (Fig. 2G and H). Moreover, the numbers of
invading cells were also reduced upon transfection with the
oe-Ntn-4 vector compared with sh-NC transfection (Fig. 2I). Collectively, these data
suggested that Ntn-4 can reduce the migratory and invasive
capabilities of the NSCLC cells.
 | Figure 2.Ntn-4 inhibits the proliferation,
migration and invasion of NSCLC cells. (A and B) The mRNA and
protein levels of Ntn-4 in NSCLC cells transfected with oe-Ntn-4
lentivirus were detected using RT-qPCR and western blot analysis
(n=3). (C and D) The mRNA and protein levels of Ntn-4 in NSCLC
cells transfected with Ntn-4 shRNA lentivirus were measured using
RT-qPCR and western blot analysis (n=3). (E and F) The
proliferation of A549 and H2170 cells was analysed using a CCK-8
assay (n=6). (G and H) A scratch wound assay was used to examine
the cell migration capability (n=3). Scale bar, 100 µm. (I)
Representative images of Transwell-based cell invasion of NSCLC
cells are shown, in addition to the quantification of the number of
invaded cells (n=3). Scale bar, 100 µm. Data are presented as the
mean ± SEM of three independent experiments. *P<0.05,
**P<0.01 and ***P<0.001, vs. shRNA negative control (sh-NC);
#P<0.05, ##P<0.01 and
###P<0.001, vs. overexpression negative control
(oe-NC); NSCLC, non-small cell lung cancer; Ntn, netrin; RT-qPCR,
reverse transcription-quantitative PCR. |
Qki-5 increases the expression of
Ntn-4 in NSCLC cells
According to the LUAD and LUSC datasets, the mRNA
expression level of Qki was lower in the LUAD and LUSC tissue
samples compared with the control samples (Fig. 3A). No statistically significant
differences in the overall survival of patients with LUSC were
observed comparing between the high and low Qki mRNA level groups;
however, patients with LUAD with a low mRNA expression level of Qki
exhibited poorer overall survival rates compared with those with
high Qki mRNA expression levels (Fig.
S1). RT-qPCR experiments were subsequently performed to further
confirm that the mRNA expression level of Qki-5 was lower in NSCLC
tissues compared with that in the adjacent tissues (Fig. 3B). Furthermore, the IHC staining
experiments also revealed a low level of Qki-5 in NSCLC tissues
compared with the adjacent normal tissues (Fig. 3C). Relative to the control cells,
the mRNA expression level of Qki-5 was also decreased in NSCLC cell
lines (Fig. 3D). To clarify the
association between Qki-5 and Ntn-4, the NSCLC cells were
subsequently transfected with oe-Qki-5 lentivirus. The mRNA and
protein levels of Qki-5 were found to be markedly upregulated in
the A549 and H2170 cells transfected with oe-Qki-5 compared with
the cells that were transfected with oe-NC lentivirus (Fig. 3E and F). As was anticipated, the
expression of Ntn-4 was upregulated in the oe-Qki-5 cells (Fig. 3G and H). Furthermore, the degree of
correlation between Qki-5 and Ntn-4 in the LUAD and LUSC datasets
was analysed, and the level of Qki-5 was found to be positively
correlated with the level of Ntn-4 (Fig. 3I and J). Pearson's correlation
analysis also confirmed that the level of Qki-5 positively
correlated with that of Ntn-4 in clinical samples of NSCLC
(Fig. 3K and L). To provide further
evidence that Qki-5 may be responsible for regulating Ntn-4, Qki-5
was silenced in 16HBE cells, where both Qki-5 and Ntn-4 had been
overexpressed. In these experiments, the Ntn-4 protein levels were
found to be downregulated in the cells in which Qki-5 was knocked
down, even though Qki-5 and Ntn-4 had been overexpressed (Fig. 3M and N). Taken together, these
findings indicated that Qki-5 is able to upregulate the expression
level of Ntn-4 in NSCLC cells.
 | Figure 3.Qki-5 increases the expression of
Ntn-4 in NSCLC cells. (A) Box-plot of Qki-5 expression in the LUAD
and LUSC datasets; the red boxes represent the cancer groups and
the grey boxes represent the control groups. *P<0.05 vs. control
(Ctrl). (B) The expression of Qki-5 mRNA in LUAD and LUSC tissues
and adjacent normal tissues was measured using RT-qPCR (n=30).
**P<0.01 vs. Ctrl. (C) Representative images of
immunohistochemical staining for Qki-5 in LUAD and LUSC tissues and
adjacent normal tissues are shown. Scale bar, 100 µm. (D) mRNA
Expression of Qki-5 in NSCLC cells. Data are presented as the mean
± SEM of three independent experiments (n=3). **P<0.01 vs. Ctrl.
(E and F) The mRNA and protein expression levels of Qki-5 were
detected using RT-qPCR and western blot analysis, respectively.
Data are presented as the mean ± SEM of three independent
experiments (n=3). **P<0.01 and ***P <0.001 vs. the oe-NC
group. (G and H) The levels of Ntn-4 mRNA and protein were detected
using RT-qPCR and western blot analysis. Data are presented as the
mean ± SEM of three independent experiments (n=3). **P<0.01 and
***P<0.001 vs. the oe-NC group. (I and J) Expression correlation
diagram of Qki-5 and Ntn-4 in the LUAD and LUSC datasets. (K and L)
Correlation analysis of Qki-5 and Ntn-4 in the LUAD and LUSC
tissues, as determined using Pearson's correlation analysis. (M and
N) Protein levels of Qki-5 and Ntn-4 were detected in 16HBE cells
using western blot analysis. Data are presented as the mean ± SEM
of three independent experiments (n=3). *P<0.05, ***P<0.001
and ****P<0.0001. LUAD, lung adenocarcinoma; LUSC, lung squamous
cell carcinoma; NSCLC, non-small cell lung cancer; Ntn, netrin;
Qki, quaking; Ctrl, control; RT-qPCR, reverse
transcription-quantitative PCR. |
Qki-5 inhibits the proliferation,
migration and invasion of NSCLC cells via the upregulation of
Ntn-4
To investigate whether Qki-5 inhibits NSCLC
progression by upregulating the expression of Ntn-4, Qki-5 was
first overexpressed in A549 cells. The level of Ntn-4 was found to
be increased in the cells following Qki-5 and/or Ntn-4
overexpression, whereas the level was decreased in cells following
the silencing of Ntn-4 (Fig. 4A and
B). Moreover, the OD values were found to be significantly
decreased in the oe-Qki-5 + sh-NC group compared with the oe-NC
group, although, by contrast, they were increased in the cells
co-transfected with oe-Qki-5 and sh-Ntn-4. In addition, the OD
values of the cells co-transfected with oe-Qki-5 and oe-Ntn-4 were
found to be lower compared with those of the oe-Qki-5 + sh-NC group
(Fig. 4C). The results from the
scratch wound healing and Transwell assays revealed that Ntn-4
silencing was able to reverse the inhibitory effects of Qki-5 on
cell migration and invasion, and both the efficiency of wound
healing and the numbers of invaded cells were increased in the
cells co-transfected with oe-Qki-5 and sh-Ntn-4. By contrast, the
migratory and invasive capabilities of the cells were further
decreased in the cells co-transfected with oe-Qki-5 and oe-Ntn-4
compared with those co-transfected with oe-Qki-5 and sh-NC
(Fig. 4D and E). To further
determine the role of Qki-5 and Ntn-4 in EMT, the levels of EMT
marker molecules were detected, and it was found that the
overexpression of Qki-5 and/or Ntn-4 led to an increase in the
level of E-cadherin (Fig. 4F and
G), whereas the levels of N-cadherin, vimentin and the EMT
transcription factor, Snail, in A549 cells were decreased compared
with the control group (Fig. 4F-J).
However, by overexpressing Qki-5 and inhibiting the expression of
Ntn-4 in the A549 cells, the suppressive effects of Qki-5 on EMT
were found to be partly reversed (Fig.
4F-J). Subsequently, RIP assay was performed to determine
whether Qki-5 is able to bind to Ntn-4 mRNA. The data obtained
revealed that Ntn-4 mRNA was highly enriched in the Qki-5
antibody-precipitated RNA fraction (Fig. 4K), suggesting that Qki-5 could
inhibit NSCLC progression partly by upregulating the expression of
Ntn-4.
 | Figure 4.Qki-5 inhibits the progression of
NSCLC cells by upregulating Ntn-4. (A) A549 cells were transfected
with oe-Qki-5, either alone or combined with oe-Ntn-4 or sh-Ntn-4
lentivirus. mRNA levels of Qki-5 and Ntn-4 were detected using
reverse transcription-quantitative PCR (n=3). (B) The protein
levels of Qki-5 and Ntn-4 in A549 cells were measured using western
blot analysis (n=3). (C) Cell proliferation was detected using a
CCK8 assay (n=6). (D) The cell migratory capability of the cells
was tested using a scratch wound healing assay (n=3). Scale bar,
100 µm. (E) Transwell assays were used to analyse the cell invasive
capability (n=3). Scale bar, 100 µm. (F-J) The EMT marker molecules
E-cadherin, N-cadherin, vimentin and Snail were detected by western
blotting, and subsequently quantified (n=3). (K) RIP assay was used
to confirm the interaction between Qki-5 and Ntn-4. Data are
presented as the mean ± SEM of three independent experiments.
**P<0.01, ***P<0.001, ****P<0.0001, vs. overexpression
negative control (oe-NC); #P<0.05,
##P<0.01 and ###P<0.001, vs.
overexpression Qki-5 and shRNA negative control (oe-Qki-5+sh-NC);
&&P<0.01,
&&&P<0.001 and
&&&&P<0.0001, vs. overexpression
Qki-5 and shRNA Ntn-4 (oe-Qki-5+ sh-Ntn-4); NSCLC, non-small cell
lung cancer; Ntn, netrin; Qki, quaking; EMT, epithelial-mesenchymal
transition; RIP, radioimmunoprecipitation. |
Qki-5 inhibits the tumorigenesis of
NSCLC by upregulating Ntn-4 expression in vivo
To confirm the effects of Ntn-4 on tumour growth
inhibition in vivo, the A549 cells were initially
transfected with the oe-NC, oe-Qki-5 + sh-NC, oe-Qki-5 + sh-Ntn-4
or oe-Qki-5 + oe-Ntn-4 lentiviruses, and subsequently the cells
were collected and subcutaneously injected into nude mice. The
average volumes of the growing tumours were measured each week, and
5 weeks later, the tumours were separated and weighed. Decreases in
both the volume and weight of tumours were noted in the mice
injected with oe-Qki-5 + sh-NC cells compared with those in mice
injected with oe-NC cells. However, the volume and weight of the
tumours were increased in the mice injected with oe-Qki-5 +
sh-Ntn-4 cells, whereas the cells transfected with oe-Ntn-4 and
oe-Qki-5 together led to the most prominent repressive effects on
tumour growth (Fig. 5).
Discussion
Over the past decade, a large number of published
studies have demonstrated that alternative splicing is involved in
the origin and progression of lung cancer. Alterations in
particular splicing regulators result in changes in both the
expression and function of their target genes in lung cancer; for
example, Qki has been shown to contribute to lung cancer by
regulating the splicing of the ESYT2, NFIB, ENAH, SPAG9,
NUMB and FN1 genes (20,28).
The present study identified that the levels of Ntn-4 and Qki-5
were downregulated in lung cancer tissue and cell lines, and a high
correlation existed between Ntn-4 and Qki-5. The low expression of
Qki-5 suggests that it may fulfil a role in the progression of
NSCLC, and that there could be an association with the poorer
overall survival of patients with NSCLC. The effects of Ntn-4 on
the proliferation, migration, invasion and EMT of lung cancer cells
were further investigated, and it was demonstrated that the
expression of Ntn-4 was regulated by Qki-5. The findings of the
present study thus suggest that Ntn-4 is a potential therapeutic
target for the progression of lung cancer.
It has been reported that the balance of Qki-5 and
Qki-6 influences the conditions of lung cancer (29). Although both the Qki-5 and Qki-6
isoforms are detectable in lung cancer, Qki-5 is mainly expressed
in tumour tissue, whereas Qki-6 is predominantly expressed in
matched normal tissue in patients with NSCLC (20). Recently, several studies have shed
light upon both the function and the underlying mechanisms of Qki-5
with respect to the inhibition of NSCLC progression (22–25).
Circ-MTO1 serves as a ‘sponge’ of miR-17 to enhance the level of
Qki-5, which has been found to further inhibit the Notch signalling
pathway, thereby suppressing the proliferation of LUAD cells
(23). At the transcriptional
level, the overexpression of KLF6 has been shown to inhibit
TGF-β/Smad signalling, which subsequently suppresses EMT and the
invasion of LUAD mediated through the upregulation of the
expression of Qki-5 (24). It has
also been reported that Qki-5 can suppress the aggressiveness of
NSCLC cells, either by inhibiting the β-catenin signalling pathway
(22) or by alternatively
suppressing the splicing of cytoskeletal gene adducing 3 (25). Consistent with the findings of the
aforementioned studies, the results of the present study revealed
that the mRNA and protein expression of Qki-5 was lower in lung
cancer tissues compared with that in corresponding adjacent
tissues. Moreover, the inhibitory effects of Qki-5 on cell
proliferation, migration, invasion and EMT were confirmed by
overexpressing Qki-5 in NSCLC cell lines.
As an axon guidance signal, Ntn-4 has been shown to
be extensively expressed in the brain, where it contributes to
multiple physiological functions in the development of the nervous
system (30). In addition, Ntn-4
plays a critical role in non-nervous systems; however, the exact
role(s) and mechanism(s) of Ntn-4 in different cells and tissues
have yet to be fully elucidated. A high concentration of Ntn-4 (5
µg/ml) has been found to inhibit endothelial cell proliferation and
promote endothelial cell apoptosis, whereas a low concentration of
Ntn-4 (100 ng/ml) exerts the opposite effect (31). The majority of previously published
studies have suggested that Ntn-4 binds to the receptor, Unc5B, to
promote both the proliferation and differentiation of endothelial
progenitor cells and the angiogenesis of ischaemic hind limbs in
mice (32), whereas the
proliferation and angiogenesis of human placental endothelial cells
and retinas is inhibited (10,33,34).
By contrast, it has been demonstrated that the role of Ntn-4 is
mediated by regulating the BM through its interaction with laminin,
rather than by binding to receptors of the Ntn family (13,15).
Previous studies have revealed the function of Ntn-4 in several
different types of tumours, including colorectal, gastric and
breast cancer (9,11,15–17).
In the present study, NKI data were analysed, and it was found that
the level of Ntn-4 was decreased in patients with NSCLC.
Furthermore, the inhibitory effects of Ntn-4 on NSCLC cell
proliferation, migration and invasion were confirmed by performing
gain-and-loss function experiments for Ntn-4. Of note, a study
confirmed that Qki silencing suppressed the expression of Ntn-4 by
directly binding to Ntn-4 mRNA in endothelial cells (35). In the present study, to clarify
whether Qki-5 is able to inhibit the aggressiveness of NSCLC by
regulating Ntn-4, a correlation analysis of Qki-5 and Ntn-4 was
performed, which subsequently detected the expression of Ntn-4 in
Qki-5-overexpressing NSCLC cells. As was anticipated, the Qki-5
mRNA level was found to be positively correlated with the level of
Ntn-4 mRNA in patients with NSCLC, and the expression of Ntn-4 was
upregulated in NSCLC cells overexpressing Qki-5. Furthermore, the
results of RIP assay illustrated that Qki-5 could directly bind to
Ntn-4 mRNA in NSCLC cells. In addition, the suppressive role of
Qki-5 in the progression of NSCLC was reversed by knocking down
Ntn-4 both in vitro and in vivo.
Taken together, the findings from the present study
demonstrated that the levels of both Qki-5 and Ntn-4 were
downregulated in patients with NSCLC and in NSCLC cell lines, and
that the overexpression of Qki-5 or Ntn-4 via gene manipulation was
able to suppress the proliferation, migration, invasion and EMT of
NSCLC cells and tumorigenesis in vivo. Moreover, the
inhibitory effects of Qki-5 on NSCLC progression were partly
mediated via the upregulation of the expression of Ntn-4,
suggesting that Ntn-4 is a target of Qki-5. In conclusion, Ntn-4
was shown to be negatively associated with NSCLC progression;
therefore, it may serve as a potential biomarker for patients with
NSCLC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the Natural Science
Foundation of Liaoning Province (grant no. 20180551268).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZW, SL and HJ were involved in the conception and
design of the study. ZW and SL performed the experiments, collected
the images, analysed the data and wrote the manuscript. SL and GP
conducted the histological examination of the lung tissue, qPCR
experiments and analysed the data. HJ provided funds, supervised
and revised the manuscript. All authors confirmed the authenticity
of all the raw data, and have read and approved the final
manuscript.
Ethics approval and consent to
participate
Patients diagnosed with LUAD and patients diagnosed
with LUSC at the Fourth Affiliated Hospital of China Medical
University were enrolled in the present study. The Ethics Committee
of the Fourth Hospital of China Medical University approved the
study (approval no. EC-2018-HX-013). The patients involved in this
research signed informed consent forms. The animal experiments were
approved by the Animal Research Ethics Committee of Animal Center
of Shengjing Hospital affiliated to China Medical University (no.
KT201945).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gazdar AF: Should we continue to use the
term non-small-cell lung cancer? Ann Oncol. 21 (Suppl
7):vii225–vii229. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ellis PM and Vandermeer R: Delays in the
diagnosis of lung cancer. J Thorac Dis. 3:183–188. 2011.PubMed/NCBI
|
|
3
|
Cirulli V and Yebra M: Netrins: Beyond the
brain. Nat Rev Mol Cell Biol. 8:296–306. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yurchenco PD and Wadsworth WG: Assembly
and tissue functions of early embryonic laminins and netrins. Curr
Opin Cell Biol. 16:572–579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bruikman CS, Zhang H, Kemper AM and van
Gils JM: Netrin family: Role for protein isoforms in cancer. J
Nucleic Acids. 2019:39471232019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ziegon L and Schlegel M: Netrin-1: A
modulator of macrophage driven acute and chronic inflammation. Int
J Mol Sci. 23:2752021. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nacht M, St Martin TB, Byrne A, Klinger
KW, Teicher BA, Madden SL and Jiang Y: Netrin-4 regulates
angiogenic responses and tumor cell growth. Exp Cell Res.
315:784–794. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Eveno C, Broqueres-You D, Feron JG,
Rampanou A, Tijeras-Raballand A, Ropert S, Leconte L, Levy BI and
Pocard M: Netrin-4 delays colorectal cancer carcinomatosis by
inhibiting tumor angiogenesis. Am J Pathol. 178:1861–1869. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eveno C, Contreres JO, Hainaud P, Nemeth
J, Dupuy E and Pocard M: Netrin-4 overexpression suppresses primary
and metastatic colorectal tumor progression. Oncol Rep. 29:73–78.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lejmi E, Leconte L, Pedron-Mazoyer S,
Ropert S, Raoul W, Lavalette S, Bouras I, Feron JG, Maitre-Boube M,
Assayag F, et al: Netrin-4 inhibits angiogenesis via binding to
neogenin and recruitment of Unc5B. Proc Natl Acad Sci USA.
105:12491–12496. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lv B, Song C, Wu L, Zhang Q, Hou D, Chen
P, Yu S, Wang Z, Chu Y, Zhang J, et al: Netrin-4 as a biomarker
promotes cell proliferation and invasion in gastric cancer.
Oncotarget. 6:9794–9806. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villanueva AA, Falcon P, Espinoza N, R LS,
Milla LA, Hernandez-SanMiguel E, Torres VA, Sanchez-Gomez P and
Palma V: The Netrin-4/Neogenin-1 axis promotes neuroblastoma cell
survival and migration. Oncotarget. 8:9767–9782. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Reuten R, Patel TR, McDougall M, Rama N,
Nikodemus D, Gibert B, Delcros JG, Prein C, Meier M, Metzger S, et
al: Structural decoding of netrin-4 reveals a regulatory function
towards mature basement membranes. Nat Commun. 7:135152016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mehlen P and Fattet L: Netrin-4 regulates
stiffness and metastasis. Nat Mater. 20:722–723. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Reuten R, Zendehroud S, Nicolau M,
Fleischhauer L, Laitala A, Kiderlen S, Nikodemus D, Wullkopf L,
Nielsen SR, McNeilly S, et al: Basement membrane stiffness
determines metastases formation. Nat Mater. 20:892–903. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esseghir S, Kennedy A, Seedhar P, Nerurkar
A, Poulsom R, Reis-Filho JS and Isacke CM: Identification of
NTN4, TRA1, and STC2 as prognostic
markers in breast cancer in a screen for signal sequence encoding
proteins. Clin Cancer Res. 13:3164–3173. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yi L, Lei Y, Yuan F, Tian C, Chai J and Gu
M: NTN4 as a prognostic marker and a hallmark for immune
infiltration in breast cancer. Sci Rep. 12:105672022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chenard CA and Richard S: New implications
for the QUAKING RNA binding protein in human disease. J Neurosci
Res. 86:233–242. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Biedermann B, Hotz HR and Ciosk R: The
Quaking family of RNA-binding proteins: Coordinators of the cell
cycle and differentiation. Cell Cycle. 9:1929–1933. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
de Miguel FJ, Pajares MJ, Martinez-Terroba
E, Ajona D, Morales X, Sharma RD, Pardo FJ, Rouzaut A, Rubio A,
Montuenga LM and Pio R: A large-scale analysis of alternative
splicing reveals a key role of QKI in lung cancer. Mol Oncol.
10:1437–1449. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang H, Li J, Tian F, Su X, Wang X, Tang
D, Zhang L, Zhang T and Ni Y: QKI-6 suppresses cell proliferation,
migration, and emt in non-small cell lung cancer. Front Oncol.
12:8975532022. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhou X, Li X, Sun C, Shi C, Hua D, Yu L,
Wen Y, Hua F, Wang Q, Zhou Q and Yu S: Quaking-5 suppresses
aggressiveness of lung cancer cells through inhibiting β-catenin
signaling pathway. Oncotarget. 8:82174–82184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang B, Chen M, Jiang N, Shi K and Qian
R: A regulatory circuit of circ-MTO1/miR-17/QKI-5 inhibits the
proliferation of lung adenocarcinoma. Cancer Biol Ther.
20:1127–1135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang S, Tong X, Li C, Jin E, Su Z, Sun Z,
Zhang W, Lei Z and Zhang HT: Quaking 5 suppresses TGF-β-induced EMT
and cell invasion in lung adenocarcinoma. EMBO Rep. 22:e520792021.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang JZ, Fu X, Fang Z, Liu H, Zong FY, Zhu
H, Yu YF, Zhang XY, Wang SF, Huang Y and Hui J: QKI-5 regulates the
alternative splicing of cytoskeletal gene ADD3 in lung cancer. J
Mol Cell Biol. 13:347–360. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li N, Deng CL, Zhang B and Ye HQ: Viral
titer quantification of west nile virus by immunostaining plaque
assay. Methods Mol Biol. 2585:15–21. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Coomer AO, Black F, Greystoke A, Munkley J
and Elliott DJ: Alternative splicing in lung cancer. Biochim
Biophys Acta Gene Regul Mech. 1862:1943882019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sebestyen E, Zawisza M and Eyras E:
Detection of recurrent alternative splicing switches in tumor
samples reveals novel signatures of cancer. Nucleic Acids Res.
43:1345–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Staquicini FI, Dias-Neto E, Li J, Snyder
EY, Sidman RL, Pasqualini R and Arap W: Discovery of a functional
protein complex of netrin-4, laminin gamma1 chain, and integrin
alpha6beta1 in mouse neural stem cells. Proc Natl Acad Sci USA.
106:2903–2908. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han Y, Shao Y, Liu T, Qu YL, Li W and Liu
Z: Therapeutic effects of topical netrin-4 inhibits corneal
neovascularization in alkali-burn rats. PLoS One. 10:e01229512015.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee NG, Jeung IC, Heo SC, Song J, Kim W,
Hwang B, Kwon MG, Kim YG, Lee J, Park JG, et al: Ischemia-induced
Netrin-4 promotes neovascularization through endothelial progenitor
cell activation via Unc-5 Netrin receptor B. FASEB J. 34:1231–1246.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dakouane-Giudicelli M, Brouillet S,
Traboulsi W, Torre A, Vallat G, Si Nacer S, Vallée M, Feige JJ,
Alfaidy N and de Mazancourt P: Inhibition of human placental
endothelial cell proliferation and angiogenesis by netrin-4.
Placenta. 36:1260–1265. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kociok N, Crespo-Garcia S, Liang Y, Klein
SV, Nurnberg C, Reichhart N, Skosyrski S, Moritz E, Maier AK,
Brunken WJ, et al: Lack of netrin-4 modulates pathologic
neovascularization in the eye. Sci Rep. 6:188282016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang H, Vreeken D, Leuning DG, Bruikman
CS, Junaid A, Stam W, de Bruin RG, Sol WMPJ, Rabelink TJ, van den
Berg BM, et al: Netrin-4 expression by human endothelial cells
inhibits endothelial inflammation and senescence. Int J Biochem
Cell Biol. 134:1059602021. View Article : Google Scholar : PubMed/NCBI
|



















