Breast cancer is one of the major causes of
cancer-related morbidity and mortality among women worldwide
(1). Breast cancers originate from
the epithelial cells of the normal mammary gland. The ducts are
lined with luminal epithelial cells, which give rise to the
majority of breast cancers (2). As
a heterogeneous disease, breast cancer encompasses a wide variety
of pathological entities and this heterogeneity is reflected by the
differences in cell type composition and proportions, the
differences in the proliferation ability between glandular and
myoepithelial cells, the proliferation of progenitor cells, the
therapeutic responses and patient outcomes (3,4).
Breast cancer patients with the same clinical diagnostic and
prognostic profiles may exhibit markedly different clinical overall
outcomes and treatment responses (5), which may be due to the current breast
cancer taxonomies based on the morphological groups, dividing the
disease into clinical classes (6).
Therefore, the clinical behavior of cancer is not solely dependent
on morphology and a molecular taxonomy based on ‘signature’
profiles may facilitate a more accurate prediction of response to
therapy and prognosis (7).
The current molecular classifications of breast
cancer molecular subtypes are generally based on the gene
expression profiles according to i) luminal cell-related markers,
such as cytokeratins (CKs); ii) hormone receptors, such as estrogen
receptor (ER), progesterone receptor (PR) and androgen receptor
(AR); iii) growth factor receptors, such as human epidermal growth
factor receptor (HER); iv) anti-apoptosis markers, such as Bcl-2
and p53; v) cell proliferation indicators, such as Ki-67 and
survivin; vi) cell invasion-related factors, such as matrix
metalloproteinases (MMPs) and integrins; vii) signal transduction
pathway members, such as the PI3K/AKT pathway members
phosphatidylinositol-3-kinase (PI3K) and AKT; viii) cell cycle
control members, such as cyclins and cyclin-dependent kinases
(CDKs); ix) epithelial-to-mesenchymal transition-indicating factors
and regulating factors, such as cadherins and zinc-finger
transcription factors Snail, Slug, Zeb1 and Twist; x) metastatic
control factors; and xi) blood vessel-forming control factors
(8–10). This spectrum also includes stem cell
markers, tumor cell and microenvironment interacting factors and
other small regulatory molecules, such as microRNAs or other
non-coding RNAs. The currently established molecular classification
of breast cancers distinguishes breast cancer molecular subtypes
into five intrinsic subtypes: i) luminal subtype A (ER+
and/or PR+, HER2− and CK8/18+);
ii) luminal subtype B (ER+ and/or PR+,
HER2+ and CK8/18+); iii) HER2-enriched
subtype (ER− and/or PR− and
HER2+); iv) basal-like subtype [ER− and/or
PR−, HER2−, CK5/6+,
CK14+, CK17+ and epithelial growth factor
receptor (EGFR)+]; and v) normal breast-like type
(ER− and/or PR−, HER2−,
CK5/6−, CK14−, CK17−,
EGFR−) (11–14) (Table
I). Another subtype, referred to as the claudin-low subtype,
was later described (15,16). Furthermore, a subpopulation of the
luminal A subtype with a Ki-67 proliferation index of >14% was
designated as the luminal B subtype (17). As such, the breast cancer molecular
subtypes were redefined as follows: luminal A (ER+
and/or PR+, HER2− and Ki-67 ≤14%); luminal B
(ER+ and/or PR+, HER2− and Ki-67
>14%); luminal B HER2/neu+ (ER+ and/or
PR+, HER2+ and any Ki-67); HER2/neu subtype
(ER− and PR−, HER2/neu+ and any
Ki-67); and triple-negative subtype (ER−,
PR−, HER2− and any Ki-67) (18–20).
The luminal type of breast cancer tends to be
morphologically well differentiated and exhibits a relatively good
prognosis, whereas the ER− tumors are poorly
differentiated and exhibit a poor prognosis. The designation of the
luminal type of breast cancer was derived from the finding that
this type of breast cancer exhibits mRNA and protein expression of
CKs 8/18 (14), which is typically
associated with luminal epithelial cells, as opposed to basal
cells, which express CKs 5/6. The luminal type of breast cancer was
further subdivided into A and B subtypes, with the luminal B
subtype exhibiting significant expression differences and worse
outcomes (11). Thus, the luminal A
and B subtypes are collectively referred to as the luminal type,
which accounts for 65–70% of breast cancers, whereas the
HER2-enriched subtype accounts for ~10% of breast cancers and the
basal-like subtype accounts for 10–15% (14) or, according to other sources, 19% of
breast cancers (2,5,6). Those
molecular classification studies significantly contributed to the
better understanding of the complex properties of different breast
cancer types, their response to systemic treatment and their
clinical outcomes, including those that respond better to endocrine
treatment.
The luminal A subtype of breast cancer is
characterized by the luminal-type conventional molecular signatures
(ER, PR, Bcl-2 and CK8/18) and the luminal A subtype-specific
signatures of ER+ and/or PR+,
HER2− and Ki-67 ≤14%, which distinguishes luminal A from
luminal B subtype. The recognized luminal A subtype breast cancer
molecular signatures include GATA binding protein 3 (GATA-3), X-box
binding protein 1 (XBP-1), forkhead box A1 (FOXA1) and ADH1B
(23–26). The 5-year survival rate of luminal
type A breast cancer is 95%, which is the highest among the five
types, with a p53 mutation rate of 13% (11).
Studies of the crosstalk between estrogen receptor α
(ERα), FOXA1 and GATA-3 revealed that, in addition to the ER and PR
status, FOXA1 and GATA-3 are also correlated with the luminal A
subtype (11,14,25,27).
The interaction of FOXA1 with the cis-regulatory regions of
heterochromatin enhances the binding of ERα to DNA (28) and is involved in controlling almost
50% of the estrogen receptor target genes (29,30).
The expression of FOXA1 is significantly positively correlated with
the markers of good prognosis or ER-positivity (31) and FOXA1 was recently shown to be
required for almost all the ER-binding events in breast cancer
cells (32). The transcription
factor GATA-3 was recently identified as a key factor involved in
luminal cell differentiation in the mammary gland (33). The majority of breast cancers arise
from the luminal epithelial cells; therefore, GATA-3 appears to
regulate a set of genes involved in the differentiation and
proliferation of breast cancer cells (33). Low GATA-3 expression is
significantly associated with a higher histological grade, poor
differentiation, positive lymph nodes, ER− and
PR− status and HER2/neu overexpression, which are all
indicators of poor prognosis (34).
The expression of GATA-3 is strongly associated with the expression
of ERα in breast cancer and there is increasing evidence that
GATA-3 may be used as a clinical molecular signature to determine
the response to hormonal therapy and to refine the prognosis of
breast cancer patients (33,35,36).
The prognosis of the luminal A subtype breast cancer is more
favorable compared to that of other subtypes (2,6), which
may be due to the fact that the luminal A subtype expresses high
levels of GATA-3 that confer a favorable prognosis. It was
previously reported that GATA-3 functions as a critical regulator
of commitment and maturation of cancer cells in the luminal A
epithelial lineage, i.e., the expression of GATA-3 regulates
luminal differentiation (37). The
anti-apoptotic marker Bcl-2 has been proven to be an independent
molecular signature, alone or in combination with Ki-67 as a marker
pair, in the luminal type of breast cancer (38–42).
Estrogen is a steroid hormone that is crucial for
growth, development and reproduction (48). Estrogens have been shown to play an
important role in human breast cancer development and ~1/3 of
breast cancers are stimulated by estradiol (49). Estrogens exert their effects through
the action of the estrogen receptors α and β (ERα and ERβ), which
belong to the steroid hormone superfamily of nuclear receptors
(NRs) (50,51). ERs are members of the large NR
family of transcription factors that are typically activated upon
binding to small lipophilic molecules (52). The activities of steroid receptors,
particularly ERs, have been associated with the regulation of
breast epithelial cell cycle transition from the G0 into the S
phase (53,54). Since ~75% of breast cancers express
ERα, the significance of ERα in breast cancer is well-established
(55,56).
Based on the molecular signatures of luminal A
subtype breast cancer, the luminal A subtype signature genes, such
as GATA-3 and FOXA1, also appear to be promising therapeutic
targets. Among these, the expression of FOXA1 was positively
correlated with ER-positivity, particularly luminal A type
ER-positivity, and negatively correlated with tumor size, tumor
grade, nodal status, the expression of Ki-67 and HER2 and
basal-like subtype of breast cancer (31). A previous study reported that the
expression of FOXA1 was positively correlated with ER+
and PR+ status, but inversely correlated with nuclear
grade and the Ki-67 index, suggesting the therapeutic potentials of
FOXA1 targeting (73). Moreover,
the forkhead box O3a (FOXO3a) transcription factor was identified
as an intracellular mediator of ERα expression and an important
downstream target of the PI3K/AKT pathway, thus representing a
potential therapeutic target in ER+ breast cancer
(74).
An alternative way for the therapeutic
considerations in luminal A subtype of breast cancer is targeting
other members that are coexpressed with ER in the superfamily of
steroid receptors, including estrogen-related receptors, PRs
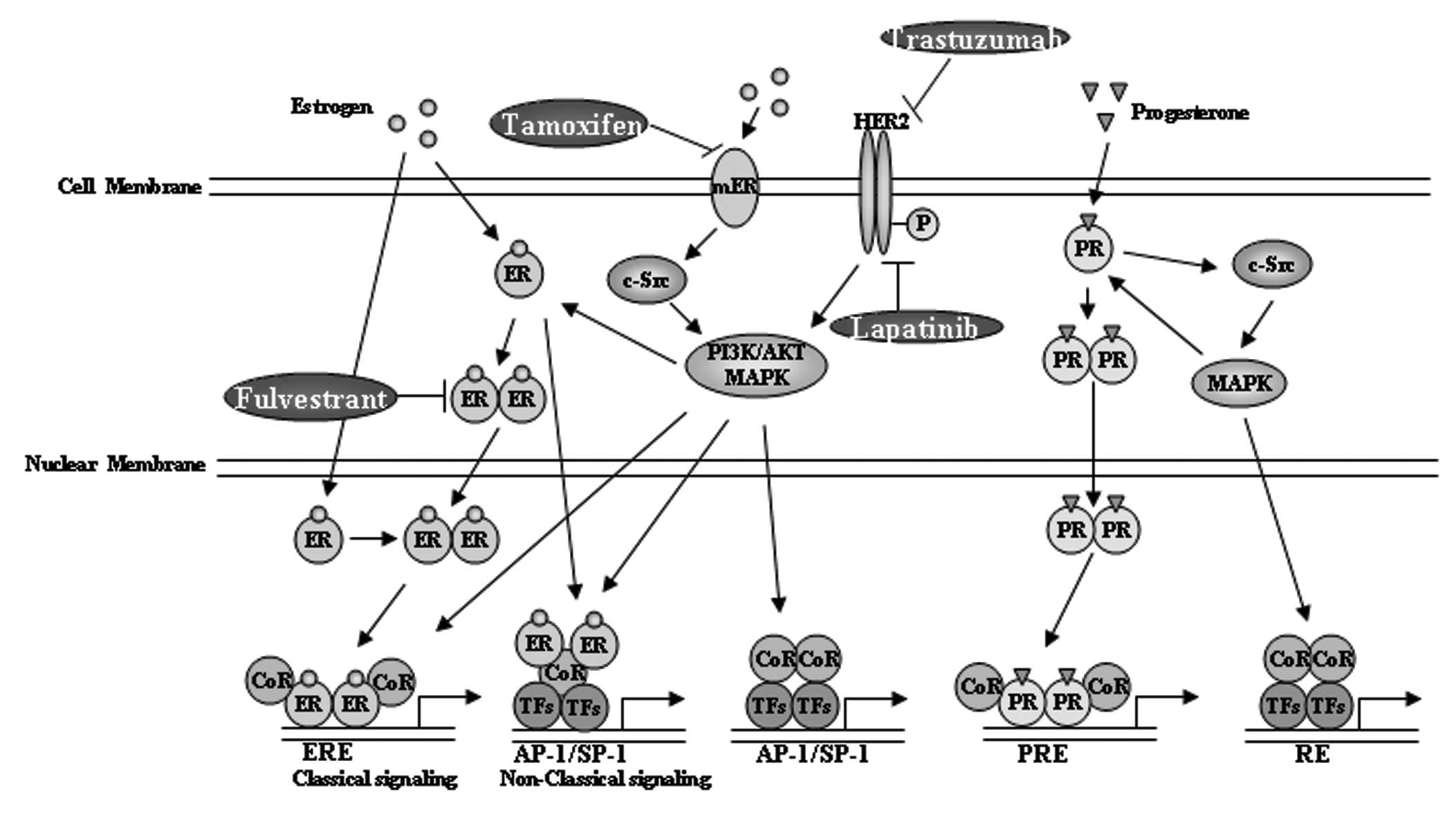
(Fig. 1), ARs, glucocorticoid and
mineralocorticoid receptors. The PR A and B isoforms were
demonstrated to play different roles in breast cancer cell growth
and have thus been considered as therapeutic targets of
antiprogestin (75). PR has been
clinically used for evaluating ER activity (64) and the loss of PR in ER+
tumors is considered to be predictive of the lack of response to
hormone therapy (76). However, the
estrogen response element (ERE) transcriptional activity remains a
better readout of ER function, as PR is just one of the numerous ER
target genes and is regulated by several other transcription
factors, such as Sp1 or AP-1 (77,78).
High ERE-activity is correlated with the luminal A type of breast
cancer and low ERE-activity is correlated with the malignancy
biomarker Ki-67 (79).
The major molecular distinctions between luminal
type A and B tumors are that luminal type A tumors exhibit a higher
expression of ER-related genes and luminal type B tumors exhibit a
higher expression of proliferation-related genes, such as CCNB1,
MKI67 and myeloblastosis oncogene-like 2 (MYBL2) (2,11,23,57,80).
In contrast to the luminal A subtype, the 5-year survival rate of
luminal B breast cancer is 50%, with a p53 mutation rate of 40%
(11), indicating the similarities
between luminal B subtype and p53-mutated tumors. Badve et
al(24) suggested that the
better prognosis of luminal A compared to that of luminal B breast
cancer may be due to the different function of ER in luminal A and
B cancers and the effect of additional factors, such as
coactivators, corepressors and transcription factors that modulate
ERα activity.
In addition to sharing similar signatures with
luminal subtype A, such as ER and Bcl-2, luminal subtype B tumors
also share similar signatures with the basal-like subtype tumors,
including the proliferation markers Ki-67, survivin and CCNB1, as
well as similar signatures with the HER2 subtype, such as the
overexpression of HER2 (4). The
Ki-67 proliferation index was previously used as a potential
unidimensional proliferation marker to distinguish luminal B from
luminal A tumors (17). Thus, the
Ki-67 index is the most useful signature that distinguishes
high-risk luminal B from low-risk luminal A tumors (81). In addition to the differences in
ER-related and proliferation-related gene expression, emerging
evidence demonstrated the amplification of growth receptor
signaling genes in luminal type B tumors, such as the
overexpression of fibroblast growth factor receptor 1 (FGFR1) in
luminal type B cancer patients (82), which may contribute to the poor
prognosis of luminal B compared to luminal A cancer patients
(2,11), despite their clinical ER+
status (83). Thus, the typical
signature genes in luminal subtype B tumors include FGFR1, HER1,
cyclin E1 and Ki-67 (2,11,12,17,82).
Using the luminal A subtype as a reference, according to the
multivariate analysis of untreated early-stage breast cancer, the
relapse-free survival of luminal B breast cancer exhibited a hazard
ratio of 2.43 (P<0.0001), similar to ErbB2/HER2 amplified tumors
with a hazard ratio of 2.53 (P=0.00012) (23,84).
The immunohistochemical analysis further demonstrated that ~20% of
luminal B cancers were HER2+ and ~30% of HER2-expressing
tumors were of the luminal B subtype (85). Clinically, the luminal (A and B)
type breast cancers are often grade I; however, the luminal B
breast cancers are often of the HER2+ genotype and are
more likely to be high-grade compared to luminal A cancers
(6,11,57).
More comprehensive gene signature profiles of the luminal type
breast cancers have been provided by assays based on the platforms
of the 21-gene signature assay Oncotype DX (Genomic Health, Redwood
City, CA, USA) and the 70-gene signature-based MammaPrint assay
(Agendia, Amsterdam, The Netherlands), as well as others (8,86).
A major characteristic of luminal type B breast
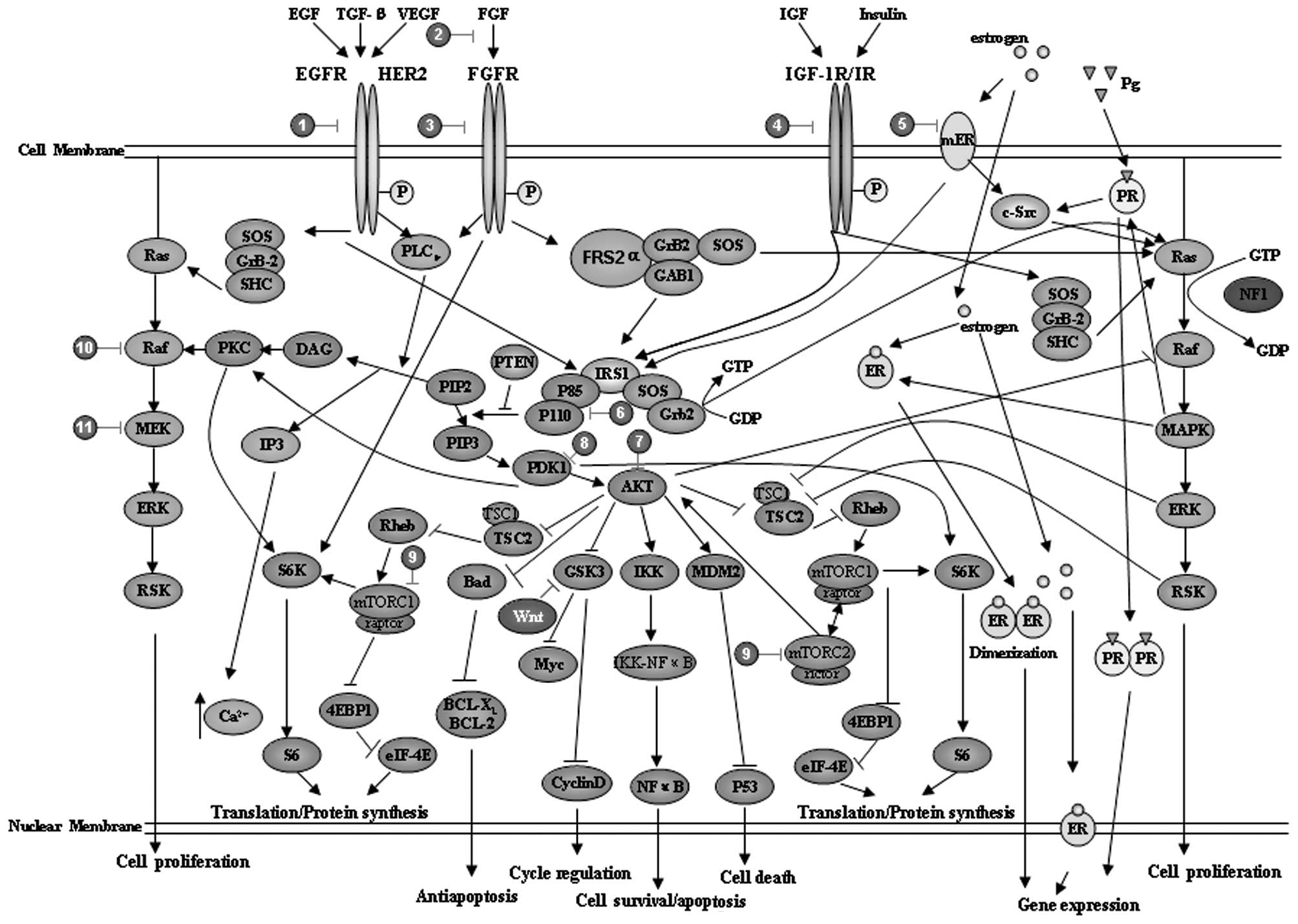
cancer cells is the expression of the HER2 gene (4,88). The
immunohistochemical results demonstrated that 30% of
HER2+ breast cancer cells are luminal type B and in this
type of cell the PI3K/AKT, Ras/mitogen-activated protein kinase
(MAPK) and phospholipase Cγ (PLCγ)/protein kinase C (PKC) signaling
pathways are also involved (84)
(Fig. 2 and Table II). Thus, recent clinical trials
considered targeting of alternative pathways in luminal B cancer,
such as using the drug gefitinib to target EGFR (89) and everolimus to target PI3K/AKT/mTOR
(90). Numerous small-molecule
inhibitors or antibodies targeting these signaling pathway elements
have been designed and investigated, such as BMS-754807,
cixutumumab, MK-0646, dalotuzumab, OSI-906 and CP-758171. Other
examples include targeting the IGF-1R pathway with TKI-258 and
AZD-4547 and targeting the PI3K/AKT pathway with MK-2206, XL-147
and XL-765 (84).
Another characteristic of luminal B tumors is the
high expression levels of Ki-67 combined with HER2 expression,
exhibiting high scores in the Oncotype DX gene expression profile.
Thus, for patients with HER2+ and ER+ tumors,
combination treatment with endocrine and anti-HER2 therapy may
achieve therapeutic benefits (91).
In addition, the Ki-67 and p53 signatured subtypes mainly belong to
the luminal B subtype (92,93), preconditioning for endocrine
resistance (94). Moreover,
previous studies demonstrated that, following neoadjuvant endocrine
treatment, changes in the expression of Ki-67 may predict long-term
outcome (95,96). Among the prognostic factors, ER
<10%, Ki-67 >14% and HER2 overexpression are considered as
risk factors (97).
In luminal B-type tumors, the high expression levels
of Ki-67 combined with HER2 expression exhibit high scores in the
Oncotype DX gene expression profile; thus, for patients with
HER2+ and ER+ tumors, the combination of
endocrine and anti-HER2 therapy may achieve therapeutic benefits
(91). However, in
ER+/PR− luminal breast tumors, aggressive
behavior and tamoxifen resistance are characteristic, despite the
ER+ status (98). This
subtype of luminal type breast cancers was classified as luminal B
tumors, with greater genomic instability and a higher proliferation
rate, as well as elevated growth factor signaling and membranous ER
activity (98). Luminal subtype B
cancer patients also exhibit a higher expression of HER1 and HER2
and active gowth factor signaling mediated by the PI3K/AKT/mTOR
pathway (98) (Fig. 2 and Table II). Thus, the optimal treatment
approach for this subset of patients may be the combination of
aromatase inhibitors, fulvestrant and chemotherapy. It was also
demonstrated that PR− luminal B tumors that were treated
with neoadjuvant chemotherapy (adriamycin/cyclophosphamide)
exhibited a significantly improved response compared to other types
of tumors (99).
This study was funded by the ‘Financial support for
selected researchers back from abroad (2011)’ of Liaoning
Province.
|
1
|
Pedraza V, Gomez-Capilla JA, Escaramis G,
et al: Gene expression signatures in breast cancer distinguish
phenotype characteristics, histologic subtypes, and tumor
invasiveness. Cancer. 116:486–496. 2010. View Article : Google Scholar
|
|
2
|
Sorlie T, Tibshirani R, Parker J, et al:
Repeated observation of breast tumor subtypes in independent gene
expression data sets. Proc Natl Acad Sci USA. 100:8418–8423. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bánkfalv A, Ludwig A, De-Hesselle B,
Buerger H, Buchwalow IB and Boecker W: Different proliferative
activity of the glandular and myoepithelial lineages in benign
proliferative and early malignant breast diseases. Mod Pathol.
17:1051–1061. 2004.PubMed/NCBI
|
|
4
|
Prat A and Perou CM: Deconstructing the
molecular portraits of breast cancer. Mol Oncol. 5:5–23. 2011.
View Article : Google Scholar
|
|
5
|
van’t Veer LJ, Dai H, van de Vijver MJ, et
al: Gene expression profiling predicts clinical outcome of breast
cancer. Nature. 415:530–536. 2002.PubMed/NCBI
|
|
6
|
Sotiriou C, Neo SY, McShane LM, et al:
Breast cancer classification and prognosis based on gene expression
profiles from a population-based study. Proc Natl Acad Sci USA.
100:10393–10398. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chung CH, Bernard PS and Perou CM:
Molecular portraits and the family tree of cancer. Nat Genet.
32:533–540. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paik S, Shak S, Tang G, et al: A multigene
assay to predict recurrence of tamoxifen-treated, node-negative
breast cancer. N Engl J Med. 351:2817–2826. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peinado H, Olmeda D and Cano A: Snail, Zeb
and bHLH factors in tumour progression: an alliance against the
epithelial phenotype? Nat Rev Cancer. 7:415–428. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu N, Yu Q, Liu TJ, et al: P-cadherin
expression and basal-like subtype in breast cancers. Med Oncol.
29:2606–2612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sørlie T, Perou CM, Tibshirani R, et al:
Gene expression patterns of breast carcinomas distinguish tumor
subclasses with clinical implications. Proc Natl Acad Sci USA.
98:10869–10874. 2001.PubMed/NCBI
|
|
12
|
Carey LA, Perou CM, Livasy CA, et al:
Race, breast cancer subtypes, and survival in the Carolina Breast
Cancer Study. JAMA. 295:2492–2502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Onitilo AA, Engel JM, Greenlee RT and
Mukesh BN: Breast cancer subtypes based on ER/PR and Her2
expression: comparison of clinicopathologic features and survival.
Clin Med Res. 7:4–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Perou CM, Sørlie T, Eisen MB, et al:
Molecular portraits of human breast tumours. Nature. 406:747–752.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herschkowitz JI, Simin K, Weigman VJ, et
al: Identification of conserved gene expression features between
murine mammary carcinoma models and human breast tumors. Genome
Biol. 8:R762007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Creighton CJ, Li X, Landis M, et al:
Residual breast cancers after conventional therapy display
mesenchymal as well as tumor-initiating features. Proc Natl Acad
Sci USA. 106:13820–13825. 2009. View Article : Google Scholar
|
|
17
|
Cheang MC, Chia SK, Voduc D, et al: Ki67
index, HER2 status, and prognosis of patients with luminal B breast
cancer. J Natl Cancer Inst. 101:736–750. 2009. View Article : Google Scholar
|
|
18
|
Fountzilas G, Dafni U, Bobos M, et al:
Differential response of immunohistochemically defined breast
cancer subtypes to anthracycline-based adjuvant chemotherapy with
or without paclitaxel. PLoS One. 7:e379462012. View Article : Google Scholar
|
|
19
|
Hannemann J, Kristel P, van Tinteren H, et
al: Molecular subtypes of breast cancer and amplification of
topoisomerase II alpha: predictive role in dose intensive adjuvant
chemotherapy. Br J Cancer. 95:1334–1341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goldhirsch A, Wood WC, Coates AS, Gelber
RD, Thurlimann B and Senn HJ: Strategies for subtypes - dealing
with the diversity of breast cancer: highlights of the St. Gallen
International Expert Consensus on the Primary Therapy of Early
Breast Cancer 2011. Ann Oncol. 22:1736–1747. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hoch RV, Thompson DA, Baker RJ and Weigel
RJ: GATA-3 is expressed in association with estrogen receptor in
breast cancer. Int J Cancer. 84:122–128. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jordan VC, Wolf MF, Mirecki DM, Whitford
DA and Welshons WV: Hormone receptor assays: clinical usefulness in
the management of carcinoma of the breast. Crit Rev Clin Lab Sci.
26:97–152. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hu Z, Fan C, Oh DS, et al: The molecular
portraits of breast tumors are conserved across microarray
platforms. BMC Genomics. 7:962006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Badve S, Turbin D, Thorat MA, et al: FOXA1
expression in breast cancer - correlation with luminal subtype A
and survival. Clin Cancer Res. 13:4415–4421. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oh DS, Troester MA, Usary J, et al:
Estrogen-regulated genes predict survival in hormone
receptor-positive breast cancers. J Clin Oncol. 24:1656–1664. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Smid M, Wang Y, Zhang Y, et al: Subtypes
of breast cancer show preferential site of relapse. Cancer Res.
68:3108–3114. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
van de Vijver MJ, He YD, van’t Veer LJ, et
al: A gene-expression signature as a predictor of survival in
breast cancer. N Engl J Med. 347:1999–2009. 2002.
|
|
28
|
Lacroix M and Leclercq G: About GATA3,
HNF3A, and XBP1, three genes co-expressed with the oestrogen
receptor-alpha gene (ESR1) in breast cancer. Mol Cell Endocrinol.
219:1–7. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Carroll JS and Brown M: Estrogen receptor
target gene: an evolving concept. Mol Endocrinol. 20:1707–1714.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Carroll JS, Liu XS, Brodsky AS, et al:
Chromosome-wide mapping of estrogen receptor binding reveals
long-range regulation requiring the forkhead protein FoxA1. Cell.
122:33–43. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mehta RJ, Jain RK, Leung S, et al: FOXA1
is an independent prognostic marker for ER-positive breast cancer.
Breast Cancer Res Treat. 131:881–890. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hurtado A, Holmes KA, Ross-Innes CS,
Schmidt D and Carroll JS: FOXA1 is a key determinant of estrogen
receptor function and endocrine response. Nat Genet. 43:27–33.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang SH, Chen Y and Weigel RJ: GATA-3 as a
marker of hormone response in breast cancer. J Surg Res.
157:290–295. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chou J, Provot S and Werb Z: GATA3 in
development and cancer differentiation: cells GATA have it! J Cell
Physiol. 222:42–49. 2010.
|
|
35
|
Mehra R, Varambally S, Ding L, et al:
Identification of GATA3 as a breast cancer prognostic marker by
global gene expression meta-analysis. Cancer Res. 65:11259–11264.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Dolled-Filhart M, Rydén L, Cregger M, et
al: Classification of breast cancer using genetic algorithms and
tissue microarrays. Clin Cancer Res. 12:6459–6468. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Asselin-Labat ML, Sutherland KD, Barker H,
et al: Gata-3 is an essential regulator of mammary-gland
morphogenesis and luminal-cell differentiation. Nat Cell Biol.
9:201–209. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dawson SJ, Makretsov N, Blows FM, et al:
BCL2 in breast cancer: a favourable prognostic marker across
molecular subtypes and independent of adjuvant therapy received. Br
J Cancer. 103:668–675. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ali HR, Dawson SJ, Blows FM, et al: A
Ki67/BCL2 index based on immunohistochemistry is highly prognostic
in ER-positive breast cancer. J Pathol. 226:97–107. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Abdel-Fatah TM, Powe DG, Ball G, et al:
Proposal for a modified grading system based on mitotic index and
Bcl2 provides objective determination of clinical outcome for
patients with breast cancer. J Pathol. 222:388–399. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Callagy GM, Webber MJ, Pharoah PD and
Caldas C: Meta-analysis confirms BCL2 is an independent prognostic
marker in breast cancer. BMC Cancer. 8:1532008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kallel-Bayoudh I, Hassen HB, Khabir A, et
al: Bcl-2 expression and triple negative profile in breast
carcinoma. Med Oncol. 28:S55–S61. 2010. View Article : Google Scholar
|
|
43
|
Saghatchian M, Mook S, Pruneri G, et al:
Additional prognostic value of the 70-gene signature
(MammaPrint®) among breast cancer patients with 4–9
positive lymph nodes. Breast. Jan 21–2013.(Epub ahead of
print).
|
|
44
|
Yokoyama J, Kobayashi T, Nakamura T and
Nakajima Y: A case of male breast cancer in which oncotype DX was
used to determine the therapeutic strategy. Gan To Kagaku Ryoho.
39:2057–2059. 2012.(In Japanese).
|
|
45
|
Morris SR and Carey LA: Molecular
profiling in breast cancer. Rev Endocr Metab Disord. 8:185–198.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Endo Y, Toyama T, Takahashi S, et al:
miR-1290 and its potential targets are associated with
characteristics of estrogen receptor α-positive breast cancer.
Endocr Relat Cancer. 20:91–102. 2013.
|
|
47
|
Prat A, Parker JS, Fan C, et al:
Concordance among gene expression-based predictors for ER-positive
breast cancer treated with adjuvant tamoxifen. Ann Oncol.
23:2866–2873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Swedenborg E, Power KA, Cai W, Pongratz I
and Rüegg J: Regulation of estrogen receptor beta activity and
implications in health and disease. Cell Mol Life Sci.
66:3873–3894. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
El-Tanani MK and Green CD: Interaction
between estradiol and growth factors in the regulation of specific
gene expression in MCF-7 human breast cancer cells. J Steroid
Biochem Mol Biol. 60:269–276. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Osz J, Brelivet Y, Peluso-Iltis C, et al:
Structural basis for a molecular allosteric control mechanism of
cofactor binding to nuclear receptors. Proc Natl Acad Sci USA.
109:E588–E594. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Choi KC and Jeung EB: The biomarker and
endocrine disruptors in mammals. J Reprod Dev. 49:337–345. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Chawla A, Repa JJ, Evans RM and
Mangelsdorf DJ: Nuclear receptors and lipid physiology: opening the
X-files. Science. 294:1866–1870. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Loi S, Sotiriou C, Haibe-Kains B, et al:
Gene expression profiling identifies activated growth factor
signaling in poor prognosis (luminal-B) estrogen receptor positive
breast cancer. BMC Med Genomics. 2:372009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Lee HR, Hwang KA, Park MA, Yi BR, Jeung EB
and Choi KC: Treatment with bisphenol A and methoxychlor results in
the growth of human breast cancer cells and alteration of the
expression of cell cycle-related genes, cyclin D1 and p21, via an
estrogen receptor-dependent signaling pathway. Int J Mol Med.
29:883–890. 2012.
|
|
55
|
Anderson E: The role of oestrogen and
progesterone receptors in human mammary development and
tumorigenesis. Breast Cancer Res. 4:197–201. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hartman J, Strom A and Gustafsson JA:
Estrogen receptor beta in breast cancer - diagnostic and
therapeutic implications. Steroids. 74:635–641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Brenton JD, Carey LA, Ahmed AA and Caldas
C: Molecular classification and molecular forecasting of breast
cancer: ready for clinical application? J Clin Oncol. 23:7350–7360.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Mauri D, Pavlidis N, Polyzos NP and
Ioannidis JP: Survival with aromatase inhibitors and inactivators
versus standard hormonal therapy in advanced breast cancer:
meta-analysis. J Natl Cancer Inst. 98:1285–1291. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Howell A, Cuzick J, Baum M, et al: Results
of the ATAC (Arimidex, Tamoxifen, Alone or in Combination) trial
after completion of 5 years’ adjuvant treatment for breast cancer.
Lancet. 365:60–62. 2005.
|
|
60
|
Fox EM, Arteaga CL and Miller TW:
Abrogating endocrine resistance by targeting ERα and PI3K in breast
cancer. Front Oncol. 2:1452012.
|
|
61
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
No authors listed. Tamoxifen for early
breast cancer: an overview of the randomised trials. Early Breast
Cancer Trialists’ Collaborative Group. Lancet. 351:1451–1467. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Harris TJ and McCormick F: The molecular
pathology of cancer. Nat Rev Clin Oncol. 7:251–265. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Hammond ME, Hayes DF, Dowsett M, et al:
American Society of Clinical Oncology/College Of American
Pathologists guideline recommendations for immunohistochemical
testing of estrogen and progesterone receptors in breast cancer. J
Clin Oncol. 28:2784–2795. 2010. View Article : Google Scholar
|
|
65
|
Pujol P, Daures JP, Thezenas S, Guilleux
F, Rouanet P and Grenier J: Changing estrogen and progesterone
receptor patterns in breast carcinoma during the menstrual cycle
and menopause. Cancer. 83:698–705. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shou J, Massarweh S, Osborne CK, et al:
Mechanisms of tamoxifen resistance: increased estrogen
receptor-HER2/neu cross-talk in ER/HER2-positive breast cancer. J
Natl Cancer Inst. 96:926–935. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Rutanen EM, Pekonen F, Nyman T and
Wahlström T: Insulin-like growth factors and their binding proteins
in benign and malignant uterine diseases. Growth Regul. 3:74–77.
1993.PubMed/NCBI
|
|
68
|
O’Toole SA, Dunn E, Sheppard BL, et al:
Oestrogen regulated gene expression in normal and malignant
endometrial tissue. Maturitas. 51:187–198. 2005.PubMed/NCBI
|
|
69
|
Millar EK, Graham PH, O’Toole SA, et al:
Prediction of local recurrence, distant metastases, and death after
breast-conserving therapy in early-stage invasive breast cancer
using a five-biomarker panel. J Clin Oncol. 27:4701–4708. 2009.
View Article : Google Scholar
|
|
70
|
Miller KD, Burstein HJ, Elias AD, et al:
Phase II study of SU11248, a multitargeted receptor tyrosine kinase
inhibitor (TKI), in patients (pts) with previously treated
metastatic breast cancer (MBC). J Clin Oncol. 23:5632005.
|
|
71
|
Coxon A, Bush T, Saffran D, et al: Broad
antitumor activity in breast cancer xenografts by motesanib, a
highly selective, oral inhibitor of vascular endothelial growth
factor, platelet-derived growth factor, and Kit receptors. Clin
Cancer Res. 15:110–118. 2009. View Article : Google Scholar
|
|
72
|
Ma CX, Crowder RJ and Ellis MJ: Importance
of PI3-kinase pathway in response/resistance to aromatase
inhibitors. Steroids. 76:750–752. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Hisamatsu Y, Tokunaga E, Yamashita N, et
al: Impact of FOXA1 expression on the prognosis of patients with
hormone receptor-positive breast cancer. Ann Surg Oncol.
19:1145–1152. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Habashy HO, Rakha EA, Aleskandarany M, et
al: FOXO3a nuclear localisation is associated with good prognosis
in luminal-like breast cancer. Breast Cancer Res Treat. 129:11–21.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Lanari C, Wargon V, Rojas P and Molinolo
AA: Antiprogestins in breast cancer treatment: are we ready? Endocr
Relat Cancer. 19:R35–R50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Cui X, Schiff R, Arpino G, Osborne CK and
Lee AV: Biology of progesterone receptor loss in breast cancer and
its implications for endocrine therapy. J Clin Oncol. 23:7721–7735.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Safe S and Kim K: Nuclear
receptor-mediated transactivation through interaction with Sp
proteins. Prog Nucleic Acid Res Mol Biol. 77:1–36. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Hewitt SC and Korach KS: Estrogen
receptors: structure, mechanisms and function. Rev Endocr Metab
Disord. 3:193–200. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Gohno T, Seino Y, Hanamura T, et al:
Individual transcriptional activity of estrogen receptors in
primary breast cancer and its clinical significance. Cancer Med.
1:328–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Perou CM, Jeffrey SS, van de Rijn M, et
al: Distinctive gene expression patterns in human mammary
epithelial cells and breast cancers. Proc Natl Acad Sci USA.
96:9212–9217. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Loi S, Haibe-Kains B, Desmedt C, et al:
Definition of clinically distinct molecular subtypes in estrogen
receptor-positive breast carcinomas through genomic grade. J Clin
Oncol. 25:1239–1246. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Turner N, Pearson A, Sharpe R, et al:
FGFR1 amplification drives endocrine therapy resistance and is a
therapeutic target in breast cancer. Cancer Res. 70:2085–2094.
2010. View Article : Google Scholar
|
|
83
|
de Azambuja E, Cardoso F, de Castro GJ Jr,
et al: Ki-67 as prognostic marker in early breast cancer: a
meta-analysis of published studies involving 12,155 patients. Br J
Cancer. 96:1504–1513. 2007.PubMed/NCBI
|
|
84
|
Tran B and Bedard PL: Luminal-B breast
cancer and novel therapeutic targets. Breast Cancer Res.
13:2212011. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Wirapati P, Sotiriou C, Kunkel S, et al:
Meta-analysis of gene expression profiles in breast cancer: toward
a unified understanding of breast cancer subtyping and prognosis
signatures. Breast Cancer Res. 10:R652008. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Prat A, Ellis MJ and Perou CM: Practical
implications of gene-expression-based assays for breast
oncologists. Nat Rev Clin Oncol. 9:48–57. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Nielsen TO, Parker JS, Leung S, et al: A
comparison of PAM50 intrinsic subtyping with immunohistochemistry
and clinical prognostic factors in tamoxifen-treated estrogen
receptor-positive breast cancer. Clin Cancer Res. 16:5222–5232.
2010. View Article : Google Scholar
|
|
88
|
Rakha EA, El-Sayed ME, Reis-Filho JS and
Ellis IO: Expression profiling technology: its contribution to our
understanding of breast cancer. Histopathology. 52:67–81. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Osborne CK, Neven P, Dirix LY, et al:
Gefitinib or placebo in combination with tamoxifen in patients with
hormone receptor-positive metastatic breast cancer: a randomized
phase II study. Clin Cancer Res. 17:1147–1159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Baselga J, Campone M, Piccart M, et al:
Everolimus in postmenopausal hormone-receptor-positive advanced
breast cancer. N Engl J Med. 366:520–529. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lønning PE: Poor-prognosis estrogen
receptor- positive disease: present and future clinical solutions.
Ther Adv Med Oncol. 4:127–137. 2012.PubMed/NCBI
|
|
92
|
Millar EK, Graham PH, McNeil CM, et al:
Prediction of outcome of early ER+breast cancer is
improved using a biomarker panel, which includes Ki-67 and p53. Br
J Cancer. 105:272–280. 2011.PubMed/NCBI
|
|
93
|
Jacquemier J, Charafe-Jauffret E, Monville
F, et al: Association of GATA3, P53, Ki67 status and vascular
peritumoral invasion are strongly prognostic in luminal breast
cancer. Breast Cancer Res. 11:R232009. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yamashita H, Toyama T, Nishio M, et al:
p53 protein accumulation predicts resistance to endocrine therapy
and decreased post-relapse survival in metastatic breast cancer.
Breast Cancer Res. 8:R482006. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Ellis MJ, Coop A, Singh B, et al:
Letrozole inhibits tumor proliferation more effectively than
tamoxifen independent of HER1/2 expression status. Cancer Res.
63:6523–6531. 2003.PubMed/NCBI
|
|
96
|
Dowsett M, Smith IE, Ebbs SR, et al:
Prognostic value of Ki67 expression after short-term presurgical
endocrine therapy for primary breast cancer. J Natl Cancer Inst.
99:167–170. 2007. View Article : Google Scholar
|
|
97
|
Cortesi L, De Matteis E, Cirilli C,
Marcheselli L, Proietto M and Federico M: Outcome evaluation in
pre-trastuzumab era between different breast cancer phenotypes: a
population-based study on Italian women. Tumori. 98:743–750.
2012.PubMed/NCBI
|
|
98
|
Wertheimer E, Gutierrez-Uzquiza A,
Rosemblit C, Lopez-Haber C, Sosa MS and Kazanietz MG: Rac signaling
in breast cancer: a tale of GEFs and GAPs. Cell Signal. 24:353–362.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Lips EH, Mulder L, Ronde JJ, et al:
Neoadjuvant chemotherapy in ER+HER2−breast
cancer: response prediction based on immunohistochemical and
molecular characteristics. Breast Cancer Res Treat. 131:827–836.
2012.PubMed/NCBI
|
|
100
|
Creighton CJ, Fu X, Hennessy BT, et al:
Proteomic and transcriptomic profiling reveals a link between the
PI3K pathway and lower estrogen-receptor (ER) levels and activity
in ER+breast cancer. Breast Cancer Res. 12:R402010.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Osborne CK, Shou J, Massarweh S and Schiff
R: Crosstalk between estrogen receptor and growth factor receptor
pathways as a cause for endocrine therapy resistance in breast
cancer. Clin Cancer Res. 11:865s–870s. 2005.PubMed/NCBI
|
|
102
|
Zhang Y, Su H, Rahimi M, Tochihara R and
Tang C: EGFRvIII-induced estrogen-independence,
tamoxifen-resistance phenotype correlates with PgR expression and
modulation of apoptotic molecules in breast cancer. Int J Cancer.
125:2021–2028. 2009. View Article : Google Scholar
|
|
103
|
Thakkar JP and Mehta DG: A review of an
unfavorable subset of breast cancer: estrogen receptor positive
progesterone receptor negative. Oncologist. 16:276–285. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Charafe-Jauffret E, Ginestier C, Monville
F, et al: Gene expression profiling of breast cell lines identifies
potential new basal markers. Oncogene. 25:2273–2284. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Katoh M: Genetic alterations of FGF
receptors: an emerging field in clinical cancer diagnostics and
therapeutics. Expert Rev Anticancer Ther. 10:1375–1379. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Sircoulomb F, Nicolas N, Ferrari A, et al:
ZNF703 gene amplification at 8p12 specifies luminal B breast
cancer. EMBO Mol Med. 3:153–166. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Karn T, Ruckhäberle E, Hanker L, et al:
Gene expression profiling of luminal B breast cancers reveals
NHERF1 as a new marker of endocrine resistance. Breast Cancer Res
Treat. 130:409–420. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Bergamaschi A, Christensen BL and
Katzenellenbogen BS: Reversal of endocrine resistance in breast
cancer: interrelationships among 14-3-3ζ, FOXM1, and a gene
signature associated with mitosis. Breast Cancer Res.
13:R702011.PubMed/NCBI
|
|
109
|
Glynn RW, Miller N, Mahon S and Kerin MJ:
Expression levels of HER2/neu and those of collocated genes at
17q12-21, in breast cancer. Oncol Rep. 28:365–369. 2012.PubMed/NCBI
|
|
110
|
Sotiriou C and Pusztai L: Gene-expression
signatures in breast cancer. N Engl J Med. 360:790–800. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Campbell RA, Bhat-Nakshatri P, Patel NM,
Constantinidou D, Ali S and Nakshatri H: Phosphatidylinositol
3-kinase/AKT- mediated activation of estrogen receptor α: a new
model for anti-estrogen resistance. J Biol Chem. 276:9817–9824.
2001.
|
|
112
|
Law JH, Habibi G, Hu K, et al:
Phosphorylated insulin-like growth factor-i/insulin receptor is
present in all breast cancer subtypes and is related to poor
survival. Cancer Res. 68:10238–10246. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Browne BC, O’Brien N, Duffy MJ, Crown J
and O’Donovan N: HER-2 signaling and inhibition in breast cancer.
Curr Cancer Drug Targets. 9:419–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Rodríguez-Pinilla SM, Sarrió D, Honrado E,
et al: Vimentin and laminin expression is associated with
basal-like phenotype in both sporadic and BRCA1-associated breast
carcinomas. J Clin Pathol. 60:1006–1012. 2007.PubMed/NCBI
|
|
116
|
Matos I, Dufloth R, Alvarenga M, Zeferino
LC and Schmitt F: p63, cytokeratin 5, and P-cadherin: three
molecular markers to distinguish basal phenotype in breast
carcinomas. Virchows Arch. 447:688–694. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG and
Weiss SJ: Canonical Wnt signaling regulates Slug activity and links
epithelial-mesenchymal transition with epigenetic breast cancer 1,
early onset (BRCA1) repression. Proc Natl Acad Sci USA.
109:16654–16659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Agus DB, Akita RW, Fox WD, et al:
Targeting ligand-activated ErbB2 signaling inhibits breast and
prostate tumor growth. Cancer Cell. 2:127–137. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Kurokawa H, Lenferink AE, Simpson JF, et
al: Inhibition of HER2/neu (erbB-2) and mitogen-activated protein
kinases enhances tamoxifen action against HER2-overexpressing,
tamoxifen-resistant breast cancer cells. Cancer Res. 60:5887–5894.
2000.
|
|
120
|
Moulder SL, Baetz T, Borges V, et al:
ARRY-380, a selective HER2 inhibitor: from drug design to clinical
evaluation. Mol Cancer Ther. 10:abs. A143. 2011. View Article : Google Scholar
|
|
121
|
Nelson JM and Fry DW: Akt, MAPK (Erk1/2),
and p38 act in concert to promote apoptosis in response to ErbB
receptor family inhibition. J Biol Chem. 276:14842–14847. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Rabindran SK, Discafani CM, Rosfjord EC,
et al: Antitumor activity of HKI-272, an orally active,
irreversible inhibitor of the HER-2 tyrosine kinase. Cancer Res.
64:3958–3965. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Lai CJ, Bao R, Tao X, et al: CUDC-101, a
multitargeted inhibitor of histone deacetylase, epidermal growth
factor receptor, and human epidermal growth factor receptor 2,
exerts potent anticancer activity. Cancer Res. 70:3647–3656. 2010.
View Article : Google Scholar
|
|
124
|
Hickinson DM, Klinowska T, Speake G, et
al: AZD8931, an equipotent, reversible inhibitor of signaling by
epidermal growth factor receptor, ERBB2 (HER2), and ERBB3: a unique
agent for simultaneous ERBB receptor blockade in cancer. Clin
Cancer Res. 16:1159–1169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Haluska P, Carboni JM, TenEyck C, et al:
HER receptor signaling confers resistance to the insulin-like
growth factor-I receptor inhibitor, BMS-536924. Mol Cancer Ther.
7:2589–2598. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Miknis G, Wallace E, Lyssikatos J, et al:
ARRY-334543, a potent, orally active small molecule inhibitor of
EGFR and ErbB-2. Proc Amer Assoc Cancer Res. 24:abs. 3399.
2005.
|
|
127
|
Kalous O, Conklin D, Desai AJ, et al:
Dacomitinib (PF-00299804), an irreversible Pan-HER inhibitor,
inhibits proliferation of HER2-amplified breast cancer cell lines
resistant to trastuzumab and lapatinib. Mol Cancer Ther.
11:1978–1987. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Ishikawa T, Seto M, Banno H, et al: Design
and synthesis of novel human epidermal growth factor receptor 2
(HER2)/epidermal growth factor receptor (EGFR) dual inhibitors
bearing a pyrrolo[3,2-d]pyrimidine scaffold. J Med Chem.
54:8030–8050. 2011.
|
|
129
|
Powis G, Ihle N and Kirkpatrick DL:
Practicalities of drugging the phosphatidylinositol-3-kinase/Akt
cell survival signaling pathway. Clin Cancer Res. 12:2964–2966.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Bos M, Mendelsohn J, Kim YM, Albanell J,
Fry DW and Baselga J: PD153035, a tyrosine kinase inhibitor,
prevents epidermal growth factor receptor activation and inhibits
growth of cancer cells in a receptor number-dependent manner. Clin
Cancer Res. 3:2099–2106. 1997.
|
|
131
|
Wilhelm SM, Dumas J, Adnane L, et al:
Regorafenib (BAY 73–4506): A new oral multikinase inhibitor of
angiogenic, stromal and oncogenic receptor tyrosine kinases with
potent preclinical antitumor activity. Int J Cancer. 129:245–255.
2011.
|
|
132
|
Wilhelm SM, Carter C, Tang L, et al: BAY
43-9006 exhibits broad spectrum oral antitumor activity and targets
the RAF/MEK/ERK pathway and receptor tyrosine kinases involved in
tumor progression and angiogenesis. Cancer Res. 64:7099–7109. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Falcon BL, Barr S, Gokhale PC, et al:
Reduced VEGF production, angiogenesis, and vascular regrowth
contribute to the antitumor properties of dual mTORC1/mTORC2
inhibitors. Cancer Res. 71:1573–1583. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Mendel DB, Laird AD, Xin X, et al: In vivo
antitumor activity of SU11248, a novel tyrosine kinase inhibitor
targeting vascular endothelial growth factor and platelet-derived
growth factor receptors: determination of a
pharmacokinetic/pharmacodynamic relationship. Clin Cancer Res.
9:327–337. 2003.
|
|
135
|
Hu-Lowe DD, Zou HY, Grazzini ML, et al:
Nonclinical antiangiogenesis and antitumor activities of axitinib
(AG-013736), an oral, potent, and selective inhibitor of vascular
endothelial growth factor receptor tyrosine kinases 1, 2, 3. Clin
Cancer Res. 14:7272–7283. 2008. View Article : Google Scholar
|
|
136
|
Wedge SR, Kendrew J, Hennequin LF, et al:
AZD2171: a highly potent, orally bioavailable, vascular endothelial
growth factor receptor-2 tyrosine kinase inhibitor for the
treatment of cancer. Cancer Res. 65:4389–4400. 2005. View Article : Google Scholar
|
|
137
|
Wedge SR, Ogilvie DJ, Dukes M, et al:
ZD6474 inhibits vascular endothelial growth factor signaling,
angiogenesis, and tumor growth following oral administration.
Cancer Res. 62:4645–4655. 2002.PubMed/NCBI
|
|
138
|
Matsui J, Funahashi Y, Uenaka T, Watanabe
T, Tsuruoka A and Asada M: Multi-kinase inhibitor E7080 suppresses
lymph node and lung metastases of human mammary breast tumor
MDA-MB-231 via inhibition of vascular endothelial growth
factor-receptor (VEGF-R) 2 and VEGF-R3 kinase. Clin Cancer Res.
14:5459–5465. 2008. View Article : Google Scholar
|
|
139
|
Fletcher GC, Brokx RD, Denny TA, et al:
ENMD-2076 is an orally active kinase inhibitor with antiangiogenic
and antiproliferative mechanisms of action. Mol Cancer Ther.
10:126–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Strumberg D, Schultheis B, Adamietz IA, et
al: Phase I dose escalation study of telatinib (BAY 57-9352) in
patients with advanced solid tumours. Br J Cancer. 99:1579–1585.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Albert DH, Tapang P, Magoc TJ, et al:
Preclinical activity of ABT-869, a multitargeted receptor tyrosine
kinase inhibitor. Mol Cancer Ther. 5:995–1006. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Mi YJ, Liang YJ, Huang HB, et al: Apatinib
(YN968D1) reverses multidrug resistance by inhibiting the efflux
function of multiple ATP-binding cassette transporters. Cancer Res.
70:7981–7991. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Yakes FM, Chen J, Tan J, et al:
Cabozantinib (XL184), a novel MET and VEGFR2 inhibitor,
simultaneously suppresses metastasis, angiogenesis, and tumor
growth. Mol Cancer Ther. 10:2298–2308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Mordant P, Loriot Y, Leteur C, et al:
Dependence on phosphoinositide 3-kinase and RAS-RAF pathways drive
the activity of RAF265, a novel RAF/VEGFR2 inhibitor, and RAD001
(Everolimus) in combination. Mol Cancer Ther. 9:358–368. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Gendreau SB, Ventura R, Keast P, et al:
Inhibition of the T790M gatekeeper mutant of the epidermal growth
factor receptor by EXEL-7647. Clin Cancer Res. 13:3713–3723. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Turner N and Grose R: Fibroblast growth
factor signalling: from development to cancer. Nat Rev Cancer.
10:116–129. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Gavine PR, Mooney L, Kilgour E, et al:
AZD4547: an orally bioavailable, potent, and selective inhibitor of
the fibroblast growth factor receptor tyrosine kinase family.
Cancer Res. 72:2045–2056. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
García-Echeverría C, Pearson MA, Marti A,
et al: In vivo antitumor activity of NVP-AEW541-A novel, potent,
and selective inhibitor of the IGF-IR kinase. Cancer Cell.
5:231–239. 2004.PubMed/NCBI
|
|
149
|
Sabbatini P, Rowand JL, Groy A, et al:
Antitumor activity of GSK1904529A, a small-molecule inhibitor of
the insulin-like growth factor-I receptor tyrosine kinase. Clin
Cancer Res. 15:3058–3067. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Mulvihill MJ, Cooke A, Rosenfeld-Franklin
M, et al: Discovery of OSI-906: a selective and orally efficacious
dual inhibitor of the IGF-1 receptor and insulin receptor. Future
Med Chem. 1:1153–1171. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wen B, Deutsch E, Marangoni E, et al:
Tyrphostin AG 1024 modulates radiosensitivity in human breast
cancer cells. Br J Cancer. 85:2017–2021. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Sabbatini P, Korenchuk S, Rowand JL, et
al: GSK1838705A inhibits the insulin-like growth factor-1 receptor
and anaplastic lymphoma kinase and shows antitumor activity in
experimental models of human cancers. Mol Cancer Ther. 8:2811–2820.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Simoncini T, Hafezi-Moghadam A, Brazil DP,
Ley K, Chin WW and Liao JK: Interaction of oestrogen receptor with
the regulatory subunit of phosphatidylinositol-3-OH kinase. Nature.
407:538–541. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Migliaccio A, Castoria G, Di Domenico M,
et al: Steroid-induced androgen receptor-oestradiol receptor β-Src
complex triggers prostate cancer cell proliferation. EMBO J.
19:5406–5417. 2000.PubMed/NCBI
|
|
155
|
Junttila TT, Akita RW, Parsons K, et al:
Ligand-independent HER2/HER3/PI3K complex is disrupted by
trastuzumab and is effectively inhibited by the PI3K inhibitor
GDC-0941. Cancer Cell. 15:429–440. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Yu K, Toral-Barza L, Shi C, Zhang WG and
Zask A: Response and determinants of cancer cell susceptibility to
PI3K inhibitors: combined targeting of PI3K and Mek1 as an
effective anticancer strategy. Cancer Biol Ther. 7:307–315.
2008.PubMed/NCBI
|
|
157
|
Smirnova T, Zhou ZN, Flinn RJ, et al:
Phosphoinositide 3-kinase signaling is critical for ErbB3-driven
breast cancer cell motility and metastasis. Oncogene. 31:706–715.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Knight SD, Adams ND, Burgess JL, et al:
Discovery of GSK2126458, a highly potent inhibitor of PI3K and the
mammalian target of rapamycin. ACS Med Chem Lett. 1:39–43. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Mallon R, Hollander I, Feldberg L, et al:
Antitumor efficacy profile of PKI-402, a dual phosphatidylinositol
3-kinase/mammalian target of rapamycin inhibitor. Mol Cancer Ther.
9:976–984. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Sutherlin DP, Bao L, Berry M, et al:
Discovery of a potent, selective, and orally available class I
phosphatidylinositol 3-kinase (PI3K)/mammalian target of rapamycin
(mTOR) kinase inhibitor (GDC-0980) for the treatment of cancer. J
Med Chem. 54:7579–7587. 2011. View Article : Google Scholar
|
|
161
|
Venkatesan AM, Dehnhardt CM, Delos Santos
E, et al: Bis(morpholino-1,3,5-triazine) derivatives: potent
adenosine 5′-triphosphate competitive
phosphatidylinositol-3-kinase/mammalian target of rapamycin
inhibitors: discovery of compound 26 (PKI-587), a highly
efficacious dual inhibitor. J Med Chem. 53:2636–2645.
2010.PubMed/NCBI
|
|
162
|
Rhodes N, Heerding DA, Duckett DR, et al:
Characterization of an Akt kinase inhibitor with potent
pharmacodynamic and antitumor activity. Cancer Res. 68:2366–2374.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Grimshaw KM, Hunter LJ, Yap TA, et al:
AT7867 is a potent and oral inhibitor of AKT and p70 S6 kinase that
induces pharmacodynamic changes and inhibits human tumor xenograft
growth. Mol Cancer Ther. 9:1100–1110. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Meuillet EJ, Zuohe S, Lemos R, et al:
Molecular pharmacology and antitumor activity of PHT-427, a novel
Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin
homology domain inhibitor. Mol Cancer Ther. 9:706–717. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Lu CH, Wyszomierski SL, Tseng LM, et al:
Preclinical testing of clinically applicable strategies for
overcoming trastuzumab resistance caused by PTEN deficiency. Clin
Cancer Res. 13:5883–5888. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Feldman RI, Wu JM, Polokoff MA, et al:
Novel small molecule inhibitors of 3-phosphoinositide-dependent
kinase-1. J Biol Chem. 280:19867–19874. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Cully M, You H, Levine AJ and Mak TW:
Beyond PTEN mutations: the PI3K pathway as an integrator of
multiple inputs during tumorigenesis. Nat Rev Cancer. 6:184–192.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Shor B, Zhang WG, Toral-Barza L, et al: A
new pharmacologic action of CCI-779 involves FKBP12-independent
inhibition of mTOR kinase activity and profound repression of
global protein synthesis. Cancer Res. 68:2934–2943. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Rivera VM, Squillace RM, Miller D, et al:
Ridaforolimus (AP23573; MK-8669), a potent mTOR inhibitor, has
broad antitumor activity and can be optimally administered using
intermittent dosing regimens. Mol Cancer Ther. 10:1059–1071. 2011.
View Article : Google Scholar
|
|
170
|
Yuan J, Mehta PP, Yin MJ, et al:
PF-04691502, a potent and selective oral inhibitor of PI3K and mTOR
kinases with antitumor activity. Mol Cancer Ther. 10:2189–2199.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Chresta CM, Davies BR, Hickson I, et al:
AZD8055 is a potent, selective, and orally bioavailable
ATP-competitive mammalian target of rapamycin kinase inhibitor with
in vitro and in vivo antitumor activity. Cancer Res. 70:288–298.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Bhagwat SV, Gokhale PC, Crew AP, et al:
Preclinical characterization of OSI-027, a potent and selective
inhibitor of mTORC1 and mTORC2: distinct from rapamycin. Mol Cancer
Ther. 10:1394–1406. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Wallin JJ, Edgar KA, Guan J, et al:
GDC-0980 is a novel class I PI3K/mTOR kinase inhibitor with robust
activity in cancer models driven by the PI3K pathway. Mol Cancer
Ther. 10:2426–2436. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Yu K, Toral-Barza L, Shi C, et al:
Biochemical, cellular, and in vivo activity of novel
ATP-competitive and selective inhibitors of the mammalian target of
rapamycin. Cancer Res. 69:6232–6240. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Guichard SM, Howard Z, Heathcote D, et al:
AZD2014, a dual mTORC1 and mTORC2 inhibitor is differentiated from
allosteric inhibitors of mTORC1 in ER+breast cancer.
Cancer Res. 72:abs. 917. 2012. View Article : Google Scholar
|
|
176
|
Liu Q, Thoreen C, Wang J, Sabatini D and
Gray NS: mTOR mediated anti-cancer drug discovery. Drug Discov
Today Ther Strateg. 6:47–55. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
177
|
Hawkins W, Mitchell C, McKinstry R, et al:
Transient exposure of mammary tumors to PD184352 and UCN-01 causes
tumor cell death in vivo and prolonged suppression of tumor
regrowth. Cancer Biol Ther. 4:1275–1284. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Ohren JF, Chen H, Pavlovsky A, et al:
Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe
novel noncompetitive kinase inhibition. Nat Struct Mol Biol.
11:1192–1197. 2004. View Article : Google Scholar : PubMed/NCBI
|