Introduction
Febrile rash illness (FRI) is highly contagious and
clinically manifests as fever (temperature ≥37.5°C) and a systemic,
local skin or mucous membrane rash (1). FRI may be accompanied by other clinical
symptoms, such as upper respiratory inflammation, conjunctivitis
and systemic mucosal plaques. Viruses associated with FRI include
measles (2), rubella (3), enterovirus, varicella-zoster and
parvovirus B19 (4). In 1978, China
implemented a policy of planned immunization in order to control
the incidence of measles. As a result, the incidence of measles has
reduced significantly. However, enteroviruses can also cause a
similar rash and fever syndrome normally associated with atypical
measles. The main enteroviruses include Coxsackie virus A16 and
EV71, which cause hand, foot and mouth disease in children. Ju
et al (5) reported that the
main viruses causing FRI in Guangdong, China between 2010 and 2012
were EV71 and CVA16, accounting for <82% of all cases. Coxsackie
B3 virus (CVB3) is the main pathogen associated with human viral
myocarditis and dilated cardiomyopathy (6). In addition, studies have reported that
CVB3 is associated with aseptic meningitis and encephalitis
(7) and has also been isolated from
patients suffering from hand, foot and mouth disease (8). However, to the best of our knowledge,
there are few studies regarding the association between the
enterovirus infections, particularly CVB3 infection and FRI. To
improve the understanding of the genetic characteristics of
enterovirus and its association with RFI in China, a study was
conducted of the etiology of fever-rash illnesses in Jilin, China
in 2013.
Materials and methods
Sample processing
Twenty throat swab samples were obtained from the
Center for Disease Control and Prevention (CDC) of Jilin, China.
Each sample was resuspended in 3 ml of Dulbecco's modified Eagle's
medium (DMEM) supplemented with 2% fetal bovine serum. The samples
were subsequently stored at −80°C until further use. Vero cells in
DMEM containing 2% fetal bovine serum were inoculated with 200 µl
of resuspended sample and incubated at 37°C in a 5% CO2
atmosphere. The cells were observed daily for cytopathic effects
(CPE). At least two passages were performed before the culture was
regarded as negative for CPE. Following complete CPE, the cells
were freeze/thawed three times and subsequently centrifuged at
8,000 × g at 4°C for 15 min. The supernatant was collected and
stored at −80°C.
Viral titer determination
Ten-fold serial dilutions (10−1 to
10−10) were made for each virus. Each dilution of virus
was inoculated into 8 wells of a 96-well plate containing Vero
cells. The plates were incubated at 37°C for 7 days. Viral titers
were calculated using the Reed-Muench method (9).
RNA extraction, reverse
transcription-polymerase chain reaction (RT-PCR) and
sequencing
Total RNA was extracted from the infected cell
suspension using the Qiagen Viral RNA Mini kit (Qiagen, Hilden,
Germany) following the manufacturer's instructions. The extracted
RNA was used as a template for RT-PCR amplification using Takara
RT-PCR kit version 3.0. The enterovirus universal primers and
specific primers of the VP1 genes for CVB3,
CVA16 and EV71 are listed in Table I. RT was performed at 42°C for 30 min
followed by a denaturation step at 95°C for 5 min. The cycling
conditions for enterovirus specific PCR were as follows: Initial
denaturation at 94°C for 1 min, 40 cycles at 94°C for 1 min, 50°C
for 1 min and 72°C for 90 sec, and a final extension at 72°C for 10
min. The thermocycling conditions for CVB3 were as follows:
Initial denaturation at 94°C for 2 min, 30 cycles at 94°C for 30
sec, 55°C for 45 sec and 72°C for 45 sec, and a final elongation
step at 72°C for 8 min. RT-PCR products were gel purified and sent
for sequencing at the Sangon Biotech Institute Co., Ltd. (Shanghai,
China).
 | Table I.RT-PCR primers. |
Table I.
RT-PCR primers.
| Primer | Sequence | Product, bp |
|---|
| Enterovirus universal
primer | F:
CAAGCACTTCTGTTTCCCCGG | 450 |
|
| R:
ATTGTCACCATAAGCAGCCA |
|
| CVB3 specific
primer | F:
AGGAATTCATGGAAGACGCGATAAC | 772 |
|
| R:
TGTCTAGATGCTTTGCCTAGTAGTG |
|
| CVA16 specific
primer | F:
TTGCAGACATGATTGACCAG | 211 |
|
| R:
GAGTGATGGTTCAACACACA |
|
| EV71 specific
primer | F:
GCAGCCCAAAAGAACTTCAC | 226 |
|
| R:
ATTTCAGGAGCTTGGAGTGC |
|
Data analysis
The GenBank database was searched for similar
sequences using the BLAST program. Nucleotide and deduced amino
acid sequences of the viruses were aligned using the BioEdit
7.0.5.3 software (10) and
subsequently compared with each other and with the reference
sequences of homologous CVB3 retrieved from the GenBank
database. Phylogenetic trees were generated using Mega 6.0
(11) and the maximum likelihood
method and Kimura two-parameter model. Bootstrapping was performed
with 1,000 replicates.
Results
Isolation and virus
identification
Twenty throat swabs were collected from patients who
presented with symptoms of FRI (temperature ≥37.5°C and a rash on
the face, neck or trunk) between April and June 2013 in three
different areas of Jilin, China. All the samples were provided by
the Jilin Provincial CDC. The 20 samples were all positive for
enterovirus by RT-PCR. When specific primers for CVB3,
CVA16 and EV71 were used, only five samples were
positive for CVB3. The other 15 samples were negative for
these three viruses. Specific details of the five positive samples
(JL-01, JL-02, JL-03, JL-04 and JL-05) are shown in Table II.
 | Table II.CVB3 sample data. |
Table II.
CVB3 sample data.
| No. | Gender | Age | Area | Specimen type | Rash and fever |
|---|
| JL-01 | Male | 2 y | Hunchun | Throat swab | Yes |
| JL-02 | Male | 39 y | Tumen | Throat swab | Yes |
| JL-03 | Female | 27 y | Tumen | Throat swab | Yes |
| JL-04 | Male | 4 m | Dunhua | Throat swab | Yes |
| JL-05 | Male | 1 y | Hunchun | Throat swab | Yes |
Virus titers
The titers of JL-01, JL-02, JL-03, JL-04 and JL-05
were 106.5, 106.4, 107.5,
106.5 and 106.5 tissue culture infectious
dose50/100 µl, respectively, indicating a strong ability
to infect Vero cells.
Sequence analysis of VP1
The amplified VP1 fragments of CVB3 were 772
base pairs. Comparisons of the VP1 gene fragments indicated
that the five CVB3 viruses were between 99.99–100% identical at the
nucleotide and amino acid levels. In addition, all the isolates
were 99.99% identical at the nucleotide and amino acid levels to
the CVB3/MKP and CVB3/Macocy viruses, which have been reported to
induce myocarditis and central nervous system disease (12,13),
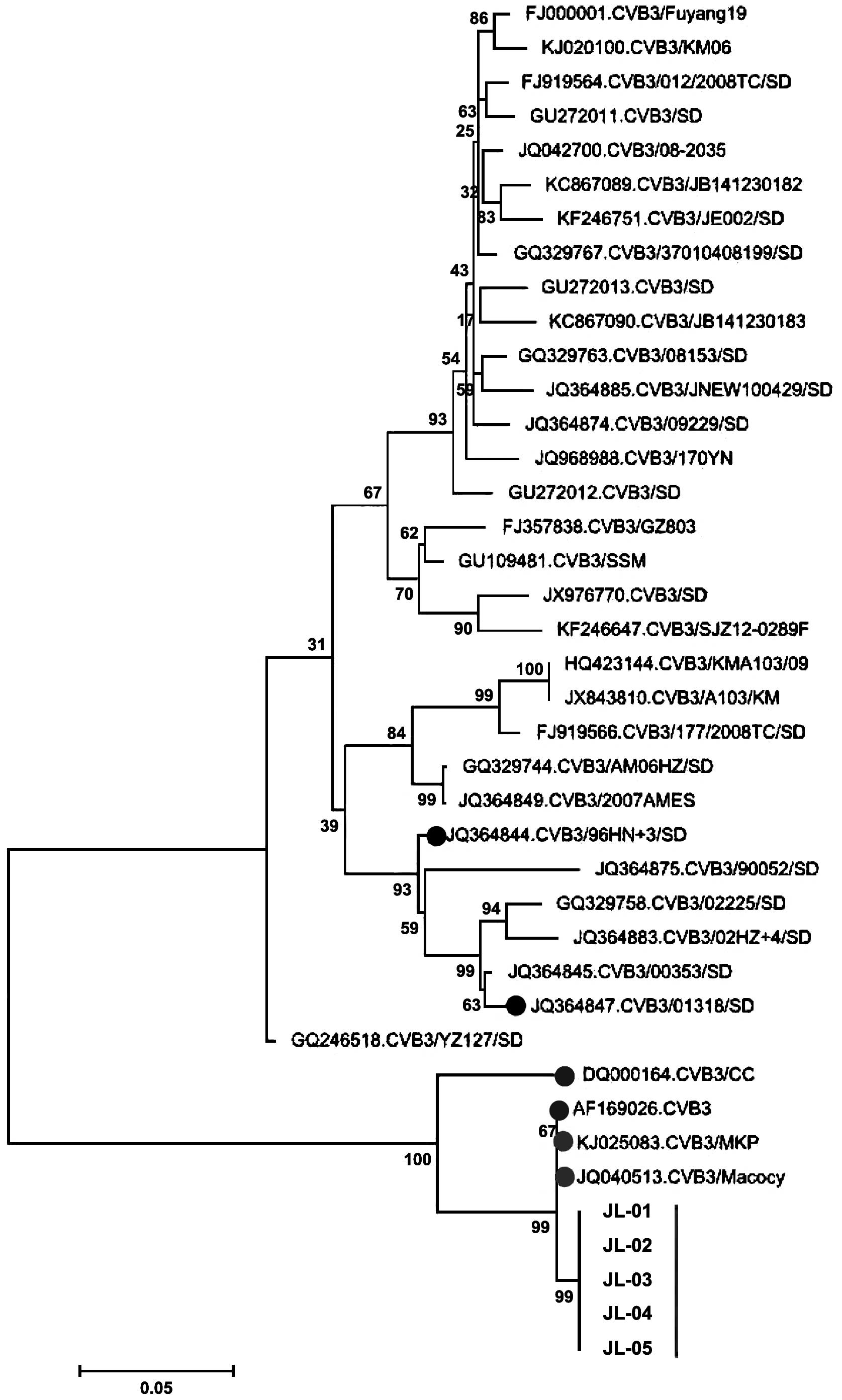
respectively. The phylogenetic analysis using the VP1
sequences (Fig. 1) showed that the
five CVB3 isolates were genetically distinct from the
CVB3/96HN.3/SD (JQ364844) and CVB3/01318/SD (JQ364847) viruses,
which have been isolated from Shandong and are known to cause
aseptic meningitis (14). DQ000164 and
AF169026, which were isolated from Harbin and Beijing in 2005 and
1999, are located on the same branch with the five CVB3 isolates in
the present study.
Discussion
Rash and fever can be caused by infections,
allergies and connective tissue diseases. Currently, the primary
viruses known to cause rash and fever are measles, varicella,
parvovirus B19 and enterovirus. Measles is a highly contagious
disease and is often accompanied by serious complications. Since
the global implementation of the measles immunization program, the
incidence of measles has decreased significantly, while parvovirus
B19 infection rates have been increasing. The enteroviruses most
frequently reported are EV71 and CVA16, which cause hand, foot and
mouth disease in children. CVB3 is reported less often, however, it
is most often associated with viral myocarditis. With the
development of molecular biological techniques, the biology of CVB3
has become clearer (15). However, due
to variability in the CVB3 genome, the same serotypes can
exhibit differences in virulence and cell tropism (16). The susceptibility and clinical
manifestations of different populations to CVB3 can vary. In
addition, CVB3 infections are predominantly asymptomatic, so they
can be missed or misdiagnosed.
In the present study, 20 samples from individuals
suffering from RFI tested negative for measles and positive for
enterovirus. Specifically, CVB3 was isolated from five of the
samples. The isolates were able to infect Vero cells, indicating
pathogenicity (12). Further studies
are required to identify the enterovirus(es) present in the
remaining 15 samples.
The VP1 region is particularly important in the
molecular characterization of enteroviruses. VP1 possesses
neutralizing epitopes that directly affect the antigenicity of the
virus. By analyzing the CVB3 VP1 sequence, five CVB3 viruses
isolated in the present study were similar to each other and other
local Chinese isolates. The homology of nucleotides and amino acids
were 99.73–99.99% and 99.93–99.99%, respectively, suggesting that
the VP1 region of Chinese CVB3 isolates is highly conservative.
These results indicate that the CVB3/MKP and CVB3/Macocy viruses,
which induce myocarditis and central nervous system disease,
respectively, are located in the same clade as the five isolates
identified in the present study, suggesting that they may have
tropism characteristics for the heart and the central nervous
system. However, further studies are required to confirm this
observation.
In conclusion, CVB3 may be responsible for RFI. In
addition, sequence data was generated for the VP1 gene from
CVB3 isolates. The present study provides a basis for future
research on these viruses in Jilin, China.
Acknowledgements
The authors would like to thank Dr Jiang Bian from
the CDC of Jilin, China, for collecting the samples used in the
present study. The study was supported by the National Natural
Science Foundation of China (grant nos. 81271897 and 81472662),
Foundation of Jilin Provincial Health Department (grant no.
2012Z083) and the Basic Scientific Research Program of Jilin
University.
References
|
1
|
de Moraes JC, Toscano CM, de Barros EN,
Kemp B, Lievano F, Jacobson S, Afonso AMS, Strebel PM and Cairns
KL: VigiFex Group: Etiologies of rash and fever illnesses in
Campinas, Brazil. J Infect Dis. 204:(Suppl 2). S627–S636. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization: Manual for the
laboratory diagnosis of measles and rubella virus infection2nd.
Geneva, Switzerland: 2007
|
|
3
|
Andrus JK and Periago MR: Elimination of
rubella and congenital rubella syndrome in the Americas: Another
opportunity to address inequities in health. Rev Panam Salud
Publica. 15:145–146. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ramsay M, Reacher M, O'Flynn C, Buttery R,
Hadden F, Cohen B, Knowles W, Wreghitt T and Brown D: Causes of
morbilliform rash in a highly immunised English population. Arch
Dis Child. 87:202–206. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ju XF, Xu AG, Fang QY and Huang JM:
Etiology study on febrile rash illness in Guangdong 2010–2012. Chin
J Dis Control Prev ISTIC. 17:670–673. 2013.
|
|
6
|
Fairweather D, Stafford KA and Sung YK:
Update on coxsackievirus B3 myocarditis. Curr Opin Rheumatol.
24:401–407. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tao Z, Wang H, Li Y, Liu G, Xu A, Lin X,
Song L, Ji F, Wang S, Cui N, et al: Molecular epidemiology of human
enterovirus associated with aseptic meningitis in Shandong
Province, China, 2006–2012. PLoS One. 9:e897662014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wong AH, Lau CS, Cheng PK, Ng AY and Lim
WW: Coxsackievirus B3-associated aseptic meningitis: An emerging
infection in Hong Kong. J Med Virol. 83:483–489. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reed LJ and Muench H: A simple method of
estimating fifty per cent endpoints. Am J Epidemiol. 27:493–497.
1938.
|
|
10
|
Hall TA: BioEdit: A user-friendly
biological sequence alignment editor and analysis program for
Windows 95/98/NT. Nucl Acids Symp Ser. 41:95–98. 1999.
|
|
11
|
Tamura K, Stecher G, Peterson D, Filipski
A and Kumar S: MEGA6: Molecular Evolutionary Genetics Analysis
version 6.0. Mol Biol Evol. 30:2725–2729. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu B, Li Z, Xiang F, Li F, Zheng Y and
Wang G: The whole genome sequence of coxsackievirus B3 MKP strain
leading to myocarditis and its molecular phylogenetic analysis.
Virol J. 11:332014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Dong C, Chen D-E and Song Z:
Coxsackievirus-induced acute neonatal central nervous system
disease model. Int J Clin Exp Pathol. 7:858–869. 2014.PubMed/NCBI
|
|
14
|
Tao Z, Song Y, Li Y, Liu Y, Jiang P, Lin
X, Liu G, Song L, Wang H and Xu A: Coxsackievirus B3, Shandong
Province, China, 1990–2010. Emerg Infect Dis. 18:1865–1867. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chapman NM1, Tu Z, Tracy S and Gauntt CJ:
An infectious cDNA copy of the genome of a non-cardiovirulent
coxsackievirus B3 strain: its complete sequence analysis and
comparison to the genomes of cardiovirulent coxsackieviruses. Arch
Virol. 135:115–130. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tu Z, Chapman NM, Hufnagel G, Tracy S,
Romero JR, Barry WH, Zhao L, Currey K and Shapiro B: The
cardiovirulent phenotype of coxsackievirus B3 is determined at a
single site in the genomic 5′ nontranslated region. J Virol.
69:4607–4618. 1995.PubMed/NCBI
|