Introduction
The ERM family is composed of the proteins ezrin,
moesin and radixin, which are cell structure-related proteins
(1). Their sequence of the
amino-terminal halves (300 amino acids) is highly conserved and is
found in band 4.1, an erythrocyte membrane protein. Together the
proteins are called the band 4.1 superfamily and their common
domain is referred to as the FERM domain, in combination with
merlin/schwannomin, the NF2 tumour suppressor protein and Talin.
These highly homologous proteins constitute a family with
structural and viable relationships. Moreover, the proteins are
localized to the submembranous cytoskeleton and are traditionally
known as molecules responsible for maintaining cell integrity and
morphology. ERM proteins are also involved in the binding process
between the plasma membrane and the actin cytoskeleton, and are
involved in a variety of cellular functions, such as cell adhesion,
migration and the organization of cell surface structures.
It has been suggested that radixin is involved in
anchoring the ‘pointed’ ends of actin filaments to the membrane in
stereocilia (2). Using
antigen-activated T cells, ERM proteins regulate cytoskeleton
relaxation and promote T cell-APC (antigen-presenting cell)
conjugation. Ezrin-radixin-moesin (ERM) proteins are rapidly
inactivated through a VAV1-RAC1 pathway (3). The resulting disanchoring of the
cortical actin cytoskeleton from the plasma membrane decreased
cellular rigidity, resulting in a more efficient T cell-APC
conjugate formation. Thus, this pathway favours immunological
synapse formation and the development of an effective immune
response.
Ezrin was shown to act as an anchorage protein for
CD44, a cell adhesion molecule that is widely involved in
metastatic cells, as well as ICAM2 (4). We reported that ezrin interacts with
the cadherin complex and co-ordinates the cell-cell adhesion
mechanisms in epithelial cells (5). Thus, the above studies showed that
the disruption or deletion of ezrin and its family members
disrupted homotypic and heterotypic cell-cell adhesion. ERM protein
functions, and their link to CD44 and ICAM2, suggest a potential
role of the ERM family in cancer invasion and metastasis.
Despite the detection of the viable roles of ERM
family proteins, the impact of these molecules in cancer
pathogenesis has yet to be investigated. Evidence emerging from
clinical and translational studies showed that the ERM family is
linked to disease progression in clinical cancers. For example, in
patients with head and neck squamous cell carcinoma, high levels of
ezrin protein staining were associated with a shorter survival
(6). In prostate tissues, ezrin
immunoreactivity is associated with the Gleason score and seminal
vesicle invasion of the patients (7). In paediatric soft tissue sarcoma
(STS), high levels of ezrin immunoreactivity are associated with
disease-free and overall survival of the patients (8). Using a tissue microarray of >5000
tumours, Bruce et al (9)
have shown that a link exists between ezrin and clinical outcome in
patients with a variety of tumour types.
In the present study, we aimed to establish the
pattern of expression of the ERM proteins and deduce a possible
relationship between these molecules and clinical outcome in a
cohort of human breast cancers.
Materials and methods
Materials
RNA extraction and RT kits were obtained from AbGene
Ltd., Surrey, England, UK. PCR primers were designed using Beacon
Designer (CA, USA) and synthesised by Invitrogen Ltd. (Pasley,
Scotland, UK). Molecular biology grade agarose and DNA ladder were
purchased from Invitrogen (Pasley). Master mix for routine PCR and
customised master mix for quantitative PCR were from AbGene.
Polyclonal antibody to human moesin and radixin, and mouse
monoclonal antibody to human ezrin were purchased from Santa Cruz
Biotechnology Ltd. (Santa Cruz, CA, USA) and Affinity Antibodies,
Inc. (Exeter, England, UK), respectively. Anti-ER (543) and
anti-ER-β (8974) antibodies were purchased from Santa Cruz
Biotechnology, Inc. Peroxidase conjugated anti-rabbit and anti-goat
antibodies were from Sigma and a biotin universal staining kit was
from Vector Laboratories (Nottingham, England, UK).
Cells and tissue samples
Breast cancer tissues (n=120) and normal background
tissues (n=32) were collected immediately after surgery and stored
in the deep freezer until use. Details of histology and clinical
outcome were obtained from pathology reports as was previously
reported (Table I). Patients were
routinely followed after surgery. The median follow-up period was
120 months. The presence of tumour cells in the collected tissues
was verified by a consultant pathologist who examined
H&E-stained frozen sections (10,11).
 | Table I.Clinicopathological details of the
study cohort. |
Table I.
Clinicopathological details of the
study cohort.
| Clinical
information | No. |
|---|
| Nodal status | |
| Negative | 65 |
| Positive | 55 |
| Grade | |
| 1 | 23 |
| 2 | 41 |
| 3 | 56 |
| Histology | |
| Ductal | 94 |
| Lobular | 14 |
| Medullary | 2 |
| Tubular | 2 |
| Mucinous | 4 |
| Other | 4 |
| TNM staging | |
| TNM 1 | 69 |
| TNM 2 | 40 |
| TNM 3 | 7 |
| TNM 4 | 4 |
| Clinical outcome | |
| Disease-free | 81 |
| Metastasis | 7 |
| Local
recurrence | 5 |
| Succumbed to breast
cancer | 20 |
| Succumbed to
unrelated disease | 7 |
Tissue processing, RNA extraction, cDNA
synthesis and RT-PCR
Frozen sections of tissues were cut at 5–10 μm and
were kept for immunohistochemistry and routine histology (12). A further 15–20 sections were mixed
and homogenised using a hand-held homogeniser, in ice-cold RNA
extraction solution. RNA concentration was determined using a UV
spectrophotometer. Reverse transcription was carried out using a RT
kit with an anchored oligo(dt) primer supplied by AbGene, and 1 pg
total RNA in a 96-well plate. The quality of cDNA was verified
using β-actin primers.
Quantitative analysis of ezrin, radixin
and moesin transcripts
The level of ERM transcripts from the cDNA prepared
as above was determined using a real-time quantitative PCR, based
on the Amplifluor™ technology as previously reported (13,14),
which has been modified from a method previously described
(13). Briefly, PCR primer pairs
were designed using the Beacon Designer software (version 2, CA,
USA) (sequence given in Table II).
An additional sequence, known as the Z sequence
(5′actgaacctgaccgtaca’3), which is complementary to the universal Z
probe (13) (Intergen Inc.,
England, UK), was added to one of the primers. A Taqman detection
kit for β-actin was purchased from Perkin-Elmer. The reaction was
carried out using hot-start Q-master mix (Abgene), 10 pmol of
specific forward primer, 10 pmol reverse primer which has the Z
sequence, 100 pmol of 6-carboxyfluorescein FAM-tagged probe
(Intergen Inc.) and cDNA from ∼50 ng RNA. The reaction was carried
out using Icycler IQ™ (Bio-Rad) which is equipped with an optic
unit that allows real-time detection of 96 reactions, using the
conditions: 94°C for 12 min, 50 cycles of 94°C for 15 sec, 55°C for
40 sec and 72°C for 20 sec (13,15,16).
Primers used for quantification of the oestrogen receptor (ER) and
ER-β were as previously reported (17). Cytokeratin-19 (CK19) was used to
compare cellularity during the analysis (18). Transcript levels were generated
from an internal standard (12)
that was simultaneously amplified with the samples. The transcript
levels were based on equal amounts of RNA and as a target/CK19
ratio.
 | Table II.Primer sequences (underlined sections
are the Z-sequence for quantification). |
Table II.
Primer sequences (underlined sections
are the Z-sequence for quantification).
| Sense (5′-′3) | Antisense
(5′-′3) |
|---|
| Ezrin |
agatgagcagtctgccttt | actgaacctgaccgtacacaacatgagagattgggaaaga |
| Moesin |
cgacagaagaaggagagtga |
Actgaacctgaccgtacaggtgtactcatggcagtctt |
| Radixin |
gaccagatgaagaatcagga | Actgaacctgaccgtacaagttgcttccctcttcctttt |
| CK19
quantification |
caggtccgaggttactgac | actgaacctgaccgtacacactttctgccagtgtgtcttc |
| ER |
cctactacctggagaacgag |
Ctcttcggtcttttcgtatg |
| ER-β |
aaaagaatcattcaatgaca |
Attaacacctccatccaaca |
| β-actin |
atgatatcgccgcgctcg |
cgctcgtgtaggatcttca |
Immunohistochemical staining of ezrin,
radixin and moesin
Immunohistochemical staining was based on a method
described earlier (16). Frozen
sections of breast tumours and background tissues were cut at 6 μm
using a cryostat. The sections were mounted on Superfrost plus
microscope slides, air dried and fixed in a mixture of 50% acetone
and 50% methanol. The staining procedure was completed
simultaneously for the section, to ensure comparison. The sections
were then placed in Optimax wash buffer for 5–10 min to rehydrate.
Sections were incubated for 20 min in a 0.6% BSA blocking solution
and probed with the primary antibody (diluted at 1:100 for
anti-moesin and anti-radixin and 1:50 for anti-ezrin). Following
extensive washings, sections were incubated for 30 min in the
secondary biotinylated antibody (Multilink Swine
anti-goat/mouse/rabbit immunoglobulin, Dako Inc.). Following
washings, the Avidin-Biotin Complex (Vector Laboratories) was
applied to the sections followed by extensive washings.
Diaminobenzidine chromogen (Vector Laboratories) was added to the
sections which were incubated in the dark for 5 min. Sections were
counterstained in Gill’s haematoxylin and dehydrated in ascending
grades of methanol before they were cleared in xylene and mounted
under a cover slip. The staining intensity of the respective
proteins in the cytoplasmic area and nucleus was semi-quantified
from non-counterstained images, using a method established in our
laboratory (18,19). Briefly, grayscale digitized images
were imported into the Optimas software (Optimas 6.0). The staining
intensity was analysed in the nucleus and cytoplasmic compartments.
Control staining (without primary antibody) was used for extraction
of the background staining. Intensity data were exported to Excel
for statistical analysis and are shown here as the mean intensity
of either the cytoplasmic region or nucleus staining.
Statistical analysis was carried out using the
Mann-Whitney U and Kruskal-Wallis tests, and the Kaplan-Meier
survival and Cox proportional analysis where appropriate, using an
SPSS (version 12) package.
Results
Distribution of ezrin, radixin and moesin
family in mammary tissues
The three molecules were positively stained in
mammary tissues (Fig. 1). In
normal tissues, ezrin prominently stained in epithelial cells. Its
location is primarily on the luminal side and intercellular region
of the epithelial cells. The luminal side of distribution is of
particular interest, as ezrin is known to be rich in the microvilli
area of the cells. The same distribution pattern was seen with
radixin in normal mammary epithelial cells (Fig. 1, right panel), while that of moesin
is less distinct.
Of note is that breast cancer cells have lost the
staining pattern for the three molecules as was observed in normal
epithelium.
Differential pattern of expression of the
ezrin, radixin and moesin family in non-neoplastic and tumour
tissues
Transcript levels of the three members were
quantified using real-time quantitative PCR. As shown in Fig. 2, there were marginally low levels
of ezrin (p=0.54) and high levels of moesin (p=0.12) in tumour
tissues compared with normal tissues. A similar trend was observed
after normalisation with CK19 (Fig. 2,
right panel). However, radixin transcript levels were
significantly higher in tumour tissues (p=0.028), as was the
radixin CK19 ratio (Fig. 2, right
panel).
Correlation between ezrin, radixin and
moesin proteins, nodal status and tumour grade
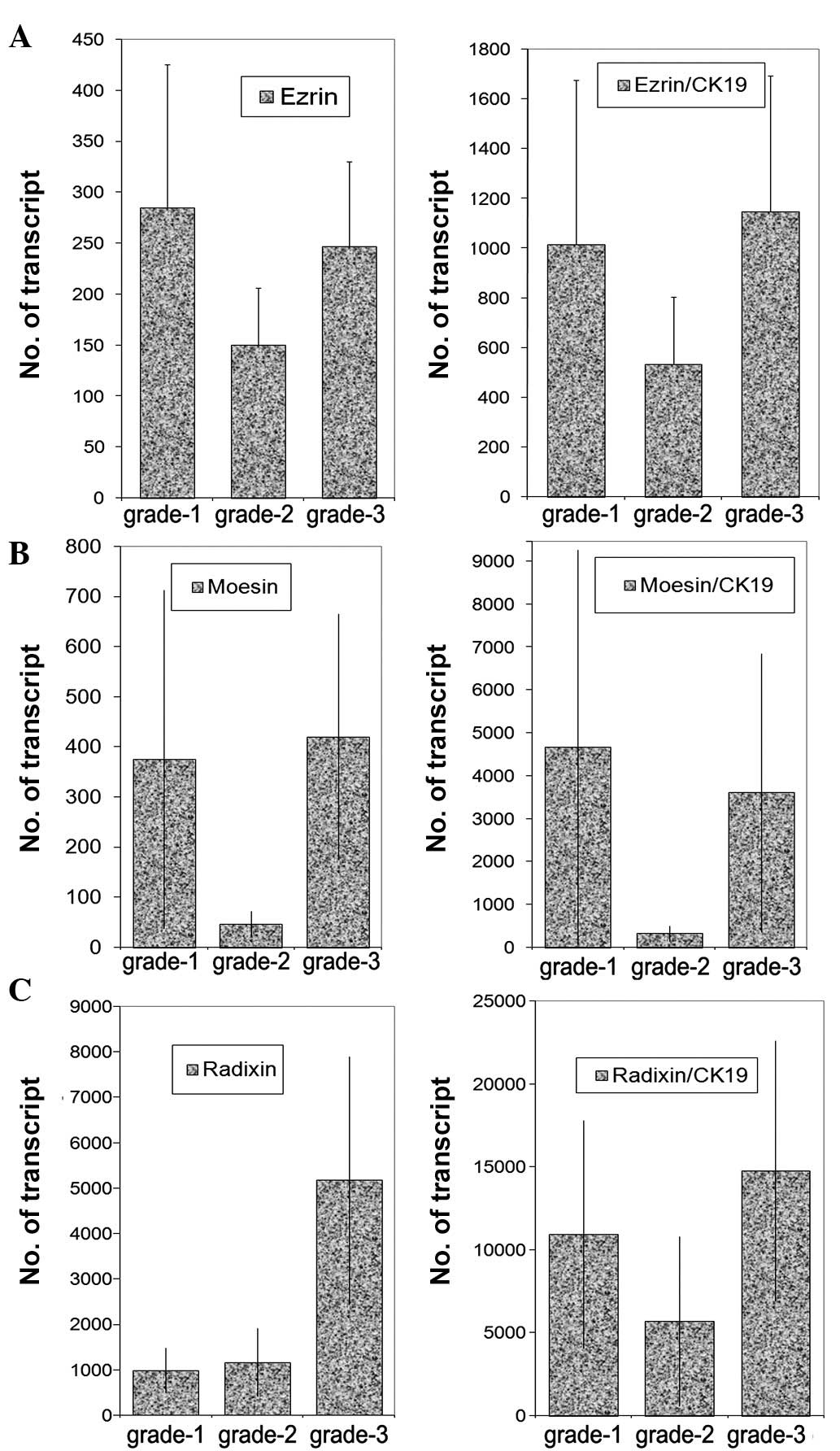
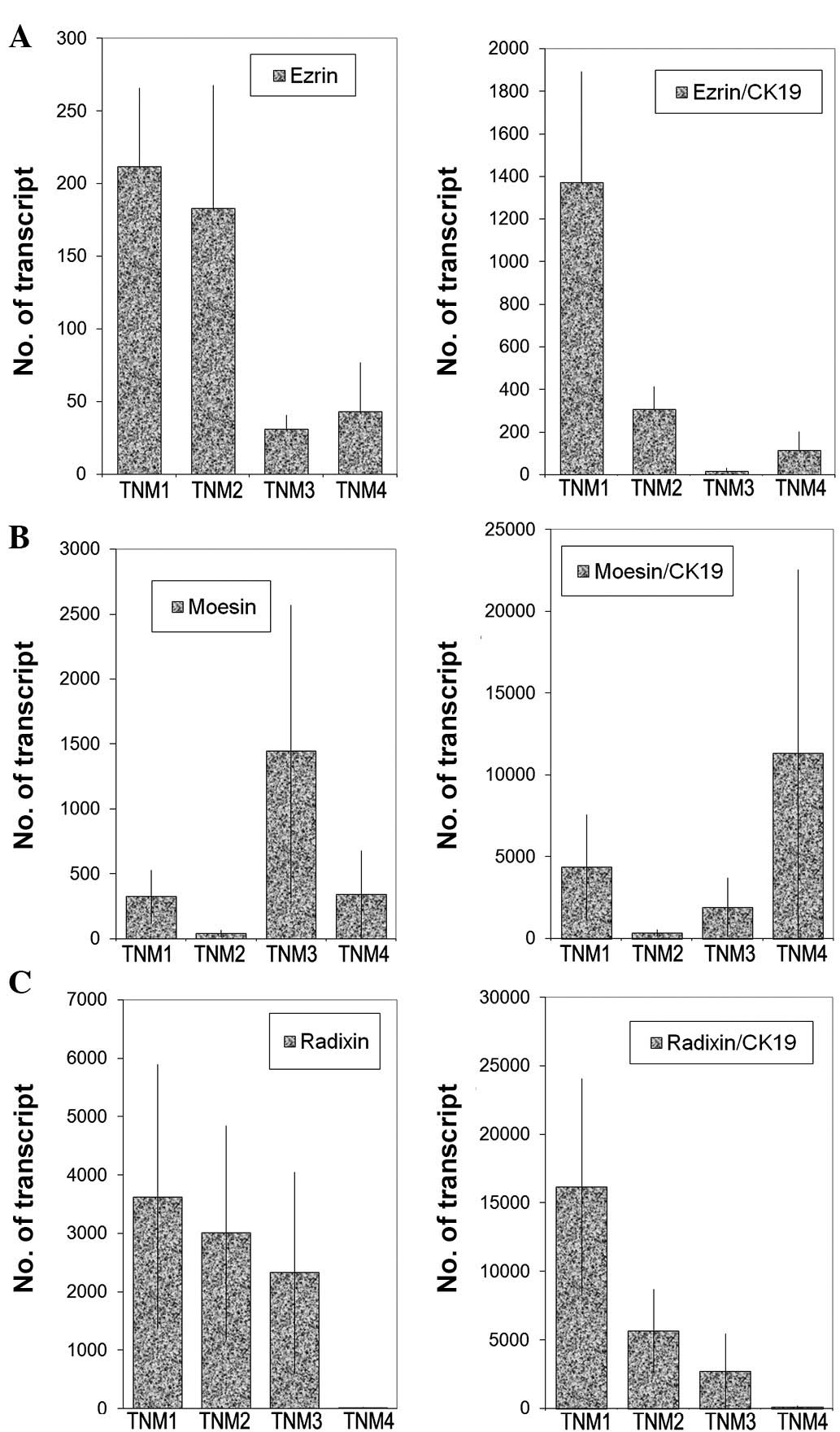
Transcript levels were analysed against tumour grade
(Fig. 3), nodal status (Fig. 4) and predicted clinical prognosis,
based on the Nottingham Prognostic Index (Fig. 5), but no significant difference was
noted. However, there appear to be significant low levels of ezrin
transcripts in TNM3 and TNM4 tumours (p=0.0017 and p=0.006,
respectively) (Fig. 6). No
significant difference was noted for moesin and radixin.
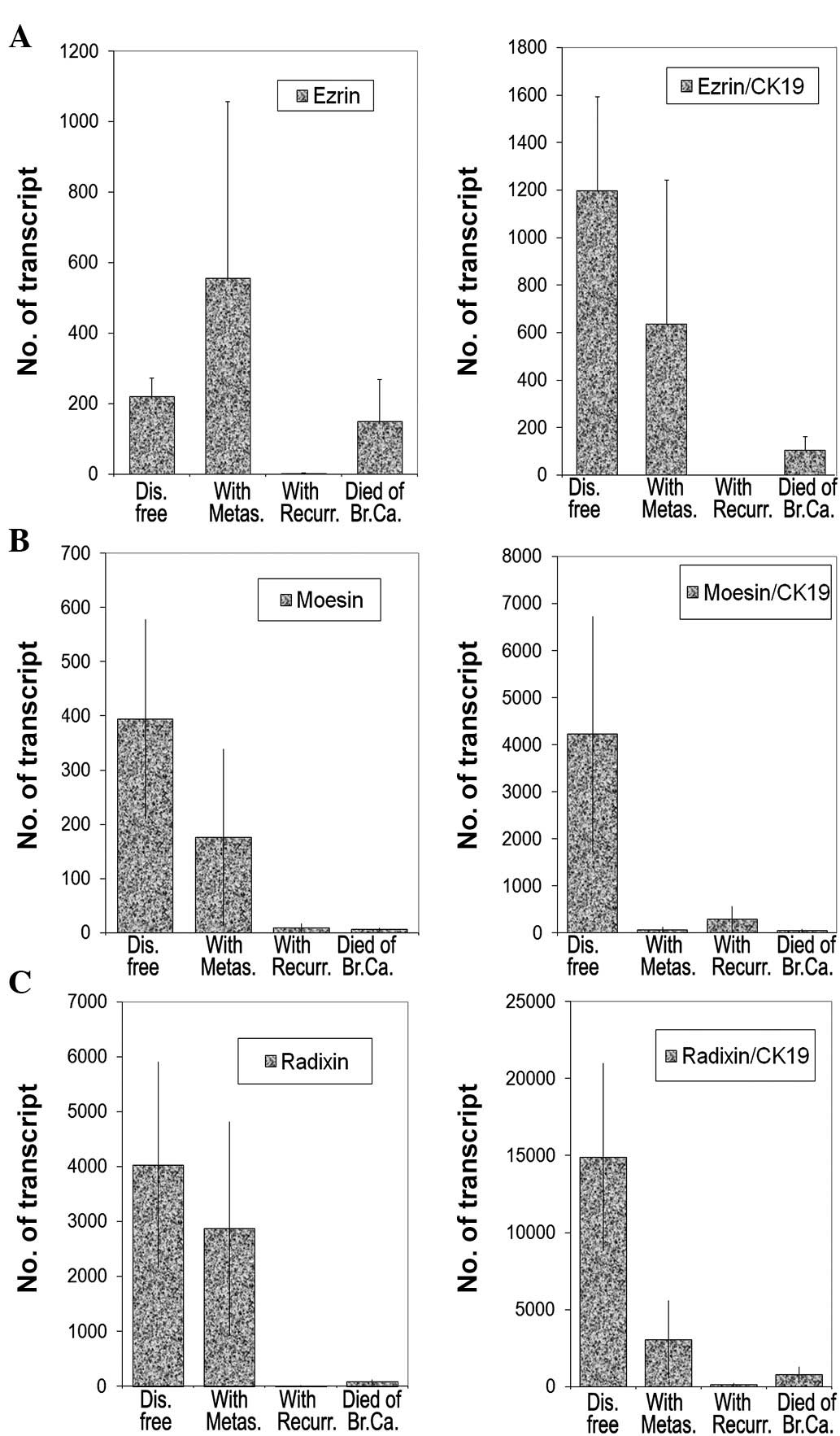
Relationship between transcript levels of
ezrin, radixin and moesin family members with clinical
outcomes
As shown in Fig. 7,
moesin and radixin levels together with their respective ratios to
CK19 were inversely correlated with the patient outcome. Low levels
of moesin and radixin transcripts and lower ratios to CK19 were
noted in tumours obtained from patients with metastasis, local
recurrence and in patients who succumbed to the disease (moesin:
p=0.039, p=0.037 and p=0.066, respectively, and radixin: p=0.039,
p=0.039 and p=0.04, respectively) (Fig. 7B and C). Although the levels of
ezrin transcripts appear to be marginally higher in tumours
associated with metastasis (p=0.53), the levels were significantly
lower in tumour recurrence and in patients who succumbed to the
disease (p=0.0001 and p=0.59) (Fig.
7). Interestingly, the ezrin:CK19 ratio has shown a low level
of survival in patients with metastasis, recurrence or mortality,
compared with those who remained disease-free.
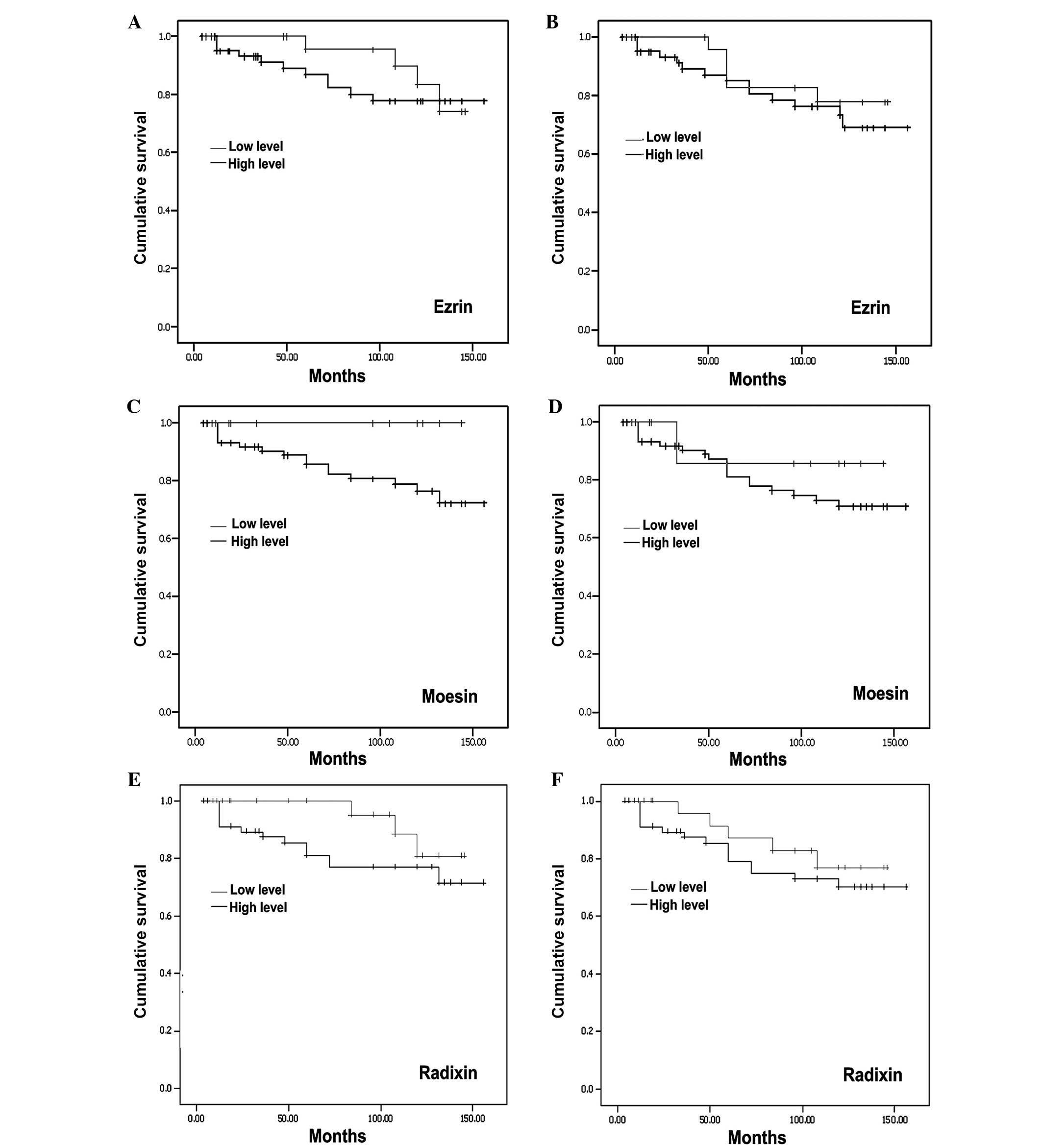
Using the Kaplan-Meier survival analysis, a general
trend of higher levels of ERM was observed with marginal long
overall (Fig. 8, left panel) and
disease-free survival (Fig. 8, right
panel). However, no statistical significance was noted.
Discussion
The present study is the first to report the
association between the proteins belonging to the band 4.1
superfamily and breast cancer. The ERM subgroup of the superfamily
has consistently been found to be co-expressed in the majority of
cultured cells (1). In humans, the
proteins have been found to have a tissue-specific expression
pattern (1). High concentrations
of ezrin are found in the lung, gut and the kidney while high
levels of radixin occur in the liver and intestine (21). While endothelial cells express
moesin, ezrin is expressed in epithelial and mesothelial cells
(1). ERM molecules are found in
high levels of actin-rich surface structures such as microvilli,
filopodia and membrane ruffles (22). These are the vital structures
required for the microinvasion and metastasis of cancer cells. The
suppression of ERM expression by anti-sense oligonucleotides
results in the destruction of the cell-substrate and cell-cell
adhesion in colorectal cell lines (5) and thymoma cells. Similarly it has
been shown that the suppression of ERM expression resulted in the
disappearance of microvilli from the cell surface (23). Therefore, if a link is found
between the expression of these molecules and metastasis of breast
cancer in patients, appropriate therapy may be employed to arrest
the metastasis of the disease.
This study has revealed interesting associations
between the ERM family and the clinical course of breast cancer.
Such associations are observed between a higher incidence of
recurrence and metastasis to low expression levels of the ERM
family in the tumour tissues. The Kaplan-Meier analysis showed that
a marginal overall improved survival occured in patients who had a
higher expression of these proteins in their tumour tissue.
Therefore, these results suggest an inverse relationship between
ERM expression and survival benefit. We previously reported that
following the inhibition of colorectal cancer cell lines using
antisense nucleotides, these cells displayed a reduced cell-cell
adhesion together with an increase in their motile and invasive
behaviour. A correlation of the two studies shows that this
mechanism also extends to breast cancer; thus, a higher incidence
of recurrence and metastasis occurred in those cases with a low
expression. Therefore, these results are important in predicting
the tumour behaviour of these patients post-surgery.
The present study, together with that of Bruce et
al (9) indicated that ezrin
and other members of the ERM family are aberrant in their
expression in breast cancer. This occurs at the protein [the
present and Bruce et al (9)] and messenger level (the present
study). A clinical connection exists since aberrant levels are
associated with disease progression and clinical outcome. However,
this connection appears to be weak in adenocarcinomas when compared
to that observed in other tumour types, i.e. sarcomas and head and
neck squamous cell carcinomas. Ezrin was found to be overexpressed
in high-grade prostatic intraepithelial neoplasia (HGPIN) and
prostate cancer compared with the adjacent benign prostatic
epithelium. In addition, HGPIN has a greater expression level of
ezrin compared with that of prostate cancer. The above studies also
indicated that the aberrant expression of ezrin is involved in the
pathogenesis of prostate cancer; thus, ezrin expression may be
useful for the diagnosis of HGPIN (7). Ezrin represents a promising
therapeutic target for patients with advanced-stage
rhabdomyosarcoma (24). A high
ezrin expression in the primary tumour of initially metastasis-free
STS patients is strongly associated with the development of
metastases during follow-up, and consequently with poor survival
(8). A high ezrin expression in
dog tumours was associated with early development of metastases.
Consistent with these data, a significant association between a
high ezrin expression and poor outcome was found in paediatric
osteosarcoma patients (25). The
expression pattern of ezrin in particular, and of the ERM family
members in general, is contrary to the clinical pattern of disease
progression in adenocarcinomas as compared to soft tissue sarcomas
and other mesenchymal tumours.
In conclusion, an inverse relationship between ERM
expression and tumour behaviour of breast cancer patients was
noted. Further studies, however, need to be conducted in other
cancer types in clinical situations to obtain consistent
results.
Acknowledgements
We would like to thank Cancer Research
Wales for supporting this work.
References
|
1.
|
Sato N, Funayama N, Nagafuchi A, Yonemura
S and Tsukita S: A gene family consisting of ezrin, radixin and
moesin. Its specific localization at actin filament/plasma membrane
association sites. J Cell Sci. 103:131–143. 1992.
|
|
2.
|
Pataky F, Pironkova R and Hudspeth AJ:
Radixin is a constituent of stereocilia in hair cells. Proc Natl
Acad Sci USA. 101:2601–2606. 2004.
|
|
3.
|
Faure S, Salazar-Fontana LI, Semichon M,
Tybulewicz VL, Bismuth G, Trautmann A, Germain RN and Delon J: ERM
proteins regulate cytoskeleton relaxation promoting T cell-APC
conjugation. Nat Immunol. 5:272–279. 2004.
|
|
4.
|
Vaheri A, Carpén O, Heiska L, Helander TS,
Jääskeläinen J, Majander-Nordenswan P, Sainio M, Timonen T and
Turunen O: The ezrin protein family: membrane-cytoskeleton
interactions and disease associations. Curr Opin Cell Biol.
9:659–666. 1997.
|
|
5.
|
Hiscox S and Jiang WG: Ezrin regulates
cell-cell and cell-matrix adhesion, a possible role with
E-cadherin/β-catenin. J Cell Sci. 112:3081–3090. 1999.
|
|
6.
|
Mhawech-Fauceglia P, Dulguerov P, Beck A,
Bonet M and Allal AS: Value of ezrin, maspin and nm23-H1 protein
expressions in predicting outcome of patients with head and neck
squamous-cell carcinoma treated with radical radiotherapy. J Clin
Pathol. 60:185–189. 2007.
|
|
7.
|
Valdman A, Fang X, Pang ST, Nilsson B,
Ekman P and Egevad L: Ezrin expression in prostate cancer and
benign prostatic tissue. Eur Urol. 48:852–857. 2005.
|
|
8.
|
Weng WH, Ahlen J, Astrom K, Lui WO and
Larsson C: Prognostic impact of immunohistochemical expression of
ezrin in highly malignant soft tissue sarcomas. Clin Cancer Res.
11:6198–6204. 2005.
|
|
9.
|
Bruce B, Khanna G, Ren L, Landberg G,
Jirstrom K, Powell C, Borczuk A, Keller ET, Wojno KJ, Meltzer P,
Baird K, McClatchey A, Bretscher A, Hewitt SM and Khanna C:
Expression of the cytoskeleton linker protein ezrin in human
cancers. Clin Exp Metastasis. 24:69–78. 2007.
|
|
10.
|
Hanavadi S, Martin T, Mansel R and Jiang
WG: Interleukin-11 and its receptor expression in human breast
cancer. Ann Surg Oncol. 13:802–808. 2006.
|
|
11.
|
Martin T, Goyal A, Mansel R, Watkins G and
Jiang W: The transcription factor for the E-cadherin complex,
Twist, Slug and Snail, in human breast cancer. Ann Surg Oncol.
12:1–9. 2005.
|
|
12.
|
Jiang WG, Watkins G, Lane J, Cunnick G,
Douglas-Jones A, Mokbel K and Mansel R: Prognostic value of Rho
family and rho-GDIs in breast cancer. Clin Cancer Res. 9:6432–6440.
2003.
|
|
13.
|
Jiang WG, Douglas-Jones A and Mansel R:
Level of expression of PPAR-gamma and its co-activator (PPAR-GCA)
in human breast cancer. Int J Cancer. 106:752–757. 2003.
|
|
14.
|
Nazarenko A, Bhatnagar S and Hohman R: A
closed tube format for amplification and detection of DNA based on
energy transfer. Nucleic Acids Res. 25:2516–2521. 1997.
|
|
15.
|
Parr C, Watkins G and Jiang WG: The
possible correlation of Notch-1 and Notch-2 with clinical outcome
and tumor clinicopathological parameters in human breast cancer.
Int J Mol Med. 14:779–786. 2004.
|
|
16.
|
Jiang WG, Grimshaw D, Lane J, Martin T,
Abounader R, Laterra J and Mansel RE: A hammerhead ribozyme
suppresses expresion of hepatocyte growth factor/scatter factor
receptor c-MET and reduces migration and invasiveness of breast
cancer cells. Clin Cancer Res. 7:2555–2562. 2001.
|
|
17.
|
Ye L, Martin T, Parr C, Harrison G, Mansel
R and Jiang WG: Biphasic effects of 17-β-oestradiol on expression
of occludin and transendothelial resistance and paracellular
permiability in human vascular endothelial cells. J Cell Physiol.
196:362–369. 2003.
|
|
18.
|
King J, Ofori-Acquah SF, Stevens T,
Al-Mehdi A-B, Fodstad O and Jiang WG: Activated leukocyte adhesion
molecule in breast cancer: prognostic indicator. Breast Cancer Res.
6:478–487. 2004.
|
|
19.
|
Davies G, Jiang WG and Mason MD: Cell-cell
adhesion and signalling intermediates in human prostate cancer. J
Urol. 163:985–992. 2000.
|
|
20.
|
Berryman M, Franck Z and Bretscher A:
Ezrin is concentrated in the apical microvilli of a wide variety of
epithelial cells whereas moesin is found primarily in endothelial
cells. J Cell Sci. 105:1025–1043. 1993.
|
|
21.
|
Tsukita S and Hieda Y: A new 82-kD barbed
end-capping protein (radixin) localized in the cell-to-cell
adherens junction: purification and characterization. J Cell Biol.
108:2369–2382. 1989.
|
|
22.
|
Yonemura S and Tsukita S: Direct
involvement of ezrin/radixin/moesin (ERM)-binding membrane proteins
in the organization of microvilli in collaboration with activated
ERM proteins. J Cell Biol. 145:1497–1509. 1999.
|
|
23.
|
Takeuchi K, Sato N, Kasahara H, Funayama
N, Nagafuchi A, Yonemura S and Tsukita S: Perturbation of cell
adhesion and microvilli formation by antisense oligonucleotides to
ERM family members. J Cell Biol. 125:1371–1384. 1994.
|
|
24.
|
Yu Y, Davicioni E, Triche TJ and Merlino
G: The homeo-protein six1 transcriptionally activates multiple
protumorigenic genes but requires ezrin to promote metastasis.
Cancer Res. 66:1982–1989. 2006.
|
|
25.
|
Khanna C, Wan X, Bose S, Cassaday R, Olomu
O, Mendoza A, Yeung C, Gorlick R, Hewitt SM and Helman LJ: The
membrane-cytoskeleton linker ezrin is necessary for osteosarcoma
metastasis. Nat Med. 10:182–186. 2004.
|