Introduction
Epithelial ovarian cancer (EOC) is the sixth leading
type cancer in women worldwide and the most lethal gynecological
malignancy in the Western world (1). One of the greatest obstacles to
improving outcome is the poor understanding of the molecular
underpinnings of EOC pathogenesis and progression. MicroRNAs
(miRNAs) are a large family of 21- to 23-nucleotide long,
non-protein-coding RNAs that regulate gene expression by mediating
target mRNA cleavage or translational inhibition (2). miRNAs participate in essential
processes, such as cell differentiation, growth, apoptosis and
invasion (3). Evidence
demonstrates that aberrant miRNA expression patterns exist in most,
if not all human malignancies (4). A number of studies have shown that
miRNA deregulation is of critical importance in EOC tumorigenesis
(5), metastasis (6), recurrence (7) and chemoresistance (8).
Ovarian serous carcinoma (OSC) is the most common
type of ovarian cancer, accounting for 68% of ovarian cancers and
88% of stage III and IV ovarian cancers (9). It has been reported that microRNA-9
(miR-9) is downregulated in OSC, as well as in endometrioid and
clear cell ovarian carcinoma (5).
Moreover, a further reduction in miR-9 levels has been found in
recurrent OSC compared with the primary tumor (7). A recent study identified miRNAs with
altered expression in a tumor-initiating subpopulation of OVCAR3
OSC cells, among which miR-9 was downregulated (10). Taken together, these data suggest
that miR-9 acts as a tumor suppressor in OSC, although conflicting
results have been shown in other types of cancer (11). To date, several targets of miR-9
associated with cancer have been identified: chromobox protein
homolog 7 (CBX7) in human glioma (12), caudal type homeobox 2 (CDX2) in
gastric cancer (8), nuclear
factor (NF)-κB1 in clear cell ovarian carcinoma (13), E-cadherin (11) and methylenetetrahydrofolate
dehydrogenase (NADP+ dependent) 2,
methenyltetrahydrofolate cyclohydrolase (MTHFD2) (14) in breast cancer. However, the
precise role of miR-9 in cancer development and progression has not
yet been elucidated.
In the present study, we investigated the functions
of miR-9 in OSC cells and identified a novel target of miR-9. We
demonstrate that miR-9 exerts its tumor suppressor activity by
downregulating the expression of talin 1 (TLN1), a focal adhesion
protein. TLN1 immunoprofiling in human OSC specimens revealed its
overexpression in primary tumors compared with normal tissues, and
an even higher expression in metastatic lesions compared with
primary tumors.
Materials and methods
Clinical specimens and
immunohistochemistry
Tissue microarrays (BC11115; US Biomax, Inc.,
Rockville, MD, USA) containing formalin-fixed, paraffin-embedded
primary and lymph node metastatic OSC tissues, normal ovarian
tissue and tumor-adjacent ovarian tissues were subjected to
immunoprofiling for TLN1 expression. In addition, OSC tissues were
obtained at Shanghai Fengxian District Central Hospital between
2008 and 2011 with informed patient consent for TLN1
immunostaining. The study was approved by the Ethics Committee of
Shanghai Fengxian District Central Hospital. H&E-stained
sections of primary OSC tissues were reviewed by an experienced
gynecological pathologist to grade the tumors using a two-tier
system (15). In total, 14 normal
ovarian tissues, 13 tumor-adjacent ovarian tissues, 67 primary (18
low- and 39 high-grade) lesions and 26 metastatic lesions of OSC
were evaluated for TLN1 expression. Immunostaining was performed
using an EnVision™ + System peroxidase kit (Dako, Glostrup,
Denmark) and a monoclonal mouse antibody against TLN1 (1:200
dilution; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Fromowitz’s standard was used to semiquantitatively assess the
staining intensity as previously described (16). Briefly, the final scoring of TLN1
staining was judged by the positive range score plus the positive
extent score, so that the TLN1 level was analyzed comprehensively
by the extent and range of staining.
Cell lines
The human OSC cell lines, SKOV3, CAOV3 and OVCAR3,
were purchased from the Cell Bank of the Chinese Academy of
Sciences (Shanghai, China). SKOV3 cells were maintained in Roswell
Park Memorial Institute (RPMI)-1640 medium (Invitrogen, Carslbad,
CA, USA) supplemented with 10% fetal bovine serum (FBS). OVCAR3
cells were maintained in RPMI-1640 (Invitrogen) supplemented with
20% FBS. CAOV3 cells were maintained in Dulbecco’s modified Eagle’s
medium in high glucose (Invitrogen) supplemented with 10% FBS.
HOSEpiC human ovarian surface epithelial cells were purchased from
ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in
ovarian epithelial cell medium (ScienCell Research
Laboratories).
Transient transfection of miRNA and
siRNA
The overexpression or knockdown of miR-9 was carried
out by transfecting the OSC cells (SKOV3, CAOV3 and OVCAR3) with
100 nM miR-9 mimic or 50 nM antagomir-9 (an anti-miR-9 molecule)
using Lipofectamine 2000 reagent following the manufacturer’s
instructions. In order to inhibit TLN1 expression in the OSC cells,
specific siRNA for TLN1 was similarly transfected into the OSC
cells at a concentration of 100 nM. Forty-eight hours after
transfection, western blot analysis and/or quantitative reverse
transcription-PCR (qRT-PCR) were conducted to confirm the
transfection efficiencies. The miRNAs and siRNAs (including
respective negative controls) were all purchased from RiboBio Co.,
Ltd., Guangzhou, China.
Vector construction and luciferase
reporter assay
The full-length 3′ untranslated region (3′UTR) of
TLN1 (454 bp) was amplified from the genomic DNA of SKOV3 cells by
PCR (forward primer, 5′-CGAGCTCGATAGAAGAAGCCTCTTCT ATTT-3′; reverse
primer, 5′-ACGCGTCGACTCTAGATTGTA GGTAGAATCAT-3′) and cloned
downstream of the firefly luciferase gene into the pmirGLO
dual-luciferase miRNA target expression vector (pmirGLO Vector;
Promega, Madison, WI, USA) at the SacI and SalI
sites. The fragment of the TLN1-3′UTR mutant (TLN1-3′UTRmu)
construct, which contained a mutational miR-9 binding site, was
generated from the TLN1-3′UTR construct using the QuickChange™
Site- Directed Mutagenesis kit (Agilent Technologies, Inc., Santa
Clara, CA, USA). The TLN1-3′UTR or TLN1-3′UTRmu construct (100 ng)
was co-transfected with miR-9 or the negative control (50 nM) into
the OSC cells for luciferase reporter assays. Twenty-four hours
after transfection, cells were collected for Luciferase activity
detection using the Dual-Luciferase Reporter Assay System
(Promega).
qRT-PCR
To determine the relative level of miR-9 or TLN1
expression, total cellular RNA from the OSC cells was extracted
using TRIzol reagent (Invitrogen). cDNA was then synthesized with a
miRNA-specific stem-loop primer or oligo(dT) using the Script™
First-Strand Synthesis System for RT-PCR (Invitrogen).
Subsequently, qRT-PCR was performed with SYBR-Green PCR master mix
(Applied Biosystems, Inc. Foster City, CA, USA) on an ABI 7500
System. The U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
gene was used as an endogenous control. RT primers were as follows:
miR-9 RT, 5′-TCGTATCCAGTGCAGGGTCCGAGG TGCACTGGATACGACTCATACAG-3′;
U6 RT, 5′-GTCGTAT CCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC
AAAATATGGAAC-3′; TLN1/GAPDH RT, oligo(dT). PCR primers were: miR-9
forward, 5′-GCCCGCTCTTTGGTTAT CTAG-3′; U6 forward,
5-TGCGGGTGCTCGCTTCGGC AGC-3′; miR-9/U6 reverse, 5′-CCAGTGCAGGGTCCG
AGGT-3. TLN1 forward, 5′-AGTGACGGACAGCATCAA CCAG-3′, TLN1 reverse,
5′-GGATTCTCCAGGAGTTCTC GGA-3′. GAPDH forward, 5′-ACCCACTCCTCCACCT
TTG-3′, GAPDH reverse, 5′-CACCACCCTGTTGCTG TAG-3′. The relative
gene expression between multiple samples was quantified by
normalization against endogenous U6 or GAPDH using the ΔCt method.
Fold-changes were calculated as 2−ΔΔC(t).
Western blot analysis
Total proteins from the OSC cells were extracted
using RIPA lysis buffer (Beyotime Biotech, Haimen, China). Proteins
were quantified (bicinchoninic acid assay; Bio-Rad), and separated
on 6% SDS-PAGE gels and subsequently transferred onto
nitrocellulose membranes. The membranes were then blocked in 5%
bovine serum albumin (BSA) for 1 h at room temperature, followed by
incubation with each primary antibody overnight at 4°C. The
following primary antibodies were used: rabbit or mouse monoclonal
antibodies to TLN1, FAK, phospho-FAK (Y397), AKT, phospho-Akt
(S473) and GAPDH (Abcam, Cambridge, MA, USA). The membranes were
then incubated with species-specific horseradish peroxidase-labeled
secondary antibodies. Immunoreactive proteins were visualized using
enhanced chemiluminescence detection system (Amersham/GE
Healthcare, San Diego, CA, USA). Protein levels were normalized to
GAPDH expression.
Transwell migration and invasion
assays
Forty-eight hours after transfection,
1×105 OSC cells suspended in 100 μl 0.1% FBS medium were
added to the upper chamber of a Transwell chamber (8 μm pore size,
6.5 mm in diameter; Corning Inc., Corning, NY, USA). For invasion
assay, the filter membrane was coated with Matrigel (BD
Biosciences, Franklin Lakes, NJ, USA) prior to the experiment. A
total of 600 μl of 10% FBS medium was added to the lower chamber.
After incubation for 18 h, the migrated/invaded cells were fixed,
stained with 0.5% crystal violet and counted under an inverted
microscope.
MTT and colony formation assays
The OSC cells were transfected with miRNA or siRNA
as described above, and then seeded into 96-well plates at
2×103 cells/well. The numbers of viable cells were
determined by MTT assay at 1, 2, 3, 4 and 5 days after
transfection.
For colony formation assay, cells were seeded into
35-mm culture dishes at 200 cells/dish 24 h after transfection. The
medium was replaced with fresh culture medium every 3–4 days. When
the cells formed visible colonies (after approximately 2 weeks),
the cells were stained with crystal violet and colonies containing
>50 cells were counted.
Statistical analysis
One-way ANOVA or the Student’s t-test was performed
using SPSS version 17.0 software to determine the statistical
significance between groups. A P-value <0.05 was considered to
indicate a statistically significant difference. Data are presented
as the means ± standard deviation (SD) of measurement.
Results
TLN1 overexpression correlates with OSC
development and progression
Immunohistochemical staining was performed to
determine the significance of TLN1 expression in human OSC. A
representative image of normal ovarian tissue, tumor-adjacent
ovarian tissue, primary OSC tissue and a metastatic tumor to the
lymph node is shown in Fig. 1A.
Cytoplasmic TLN1 immunoreactivity was comparably weak or
undetectable in the normal and tumor-adjacent ovarian tissues
(P=0.886) (Table I). By contrast,
TLN1 expression was markedly elevated in the primary OSC tissue and
lymph node metastatic lesions compared with normal ovarian tissue
(P<0.001, P<0.001, respectively) (Table I). TLN1 expression in the
metastatic lesions was significantly higher compared with that in
primary tumors (P=0.022) (Table
I). In addition, TLN1 was differentially expressed between low-
and high-grade OSC (Fig. 1B),
i.e., the intensity of TLN1 immunoreactivity was significantly
higher in the high- compared with the low-grade tumors (P<0.001)
(Table I).
 | Table IExpression of TLN1 in human ovarian
serous carcinoma. |
Table I
Expression of TLN1 in human ovarian
serous carcinoma.
| A, Quantitative
analysis of TLN1 expression in human ovarian tissue |
|---|
|
|---|
| Specimen | n | Score (mean ±
SD) | P-value |
|---|
| Normal | 14 | 0.43±0.514 | |
| Adjacent | 13 | 0.46±0.660 | 0.886 |
| Primary tumor | 67 | 3.54±1.869 |
<0.001a |
| Metastasis | 26 | 4.50±1.530 |
<0.001a;
0.022b,c |
| B, TLN1 expression
in primary tumor |
|---|
|
|---|
| Tumor grade | n | Score (mean ±
SD) | P-value |
|---|
| Low | 18 | 1.67±1.237 | |
| High | 49 | 4.20±1.554 |
<0.001a |
miR-9 inhibits OSC cell growth
Quantitative analysis of mature miR-9 expression
levels in the OSC cell lines, SKOV3, CAOV3 and OVCAR3, and the
normal ovarian surface epithelial cell line, HOSEpiC, revealed
lower miR-9 levels in the OSC cells compared with the normal
ovarian surface epithelial cells (Fig. 2A). The effect of miR-9
overexpression on OSC cell growth was investigated. Twenty-four
hours after miRNA transfection, miR-9 levels were determined by
qRT-PCR to confirm the transfection efficiencies (Fig. 2B). MTT assay revealed that miR-9
suppressed OSC cell growth in a time-dependant manner (Fig. 2C-E). Although there was no
significant difference in cell growth between the miR-9-transfected
cells and those in the control groups during the first 2 days,
miR-9 overexpression significantly inhibited cell growth at days 3
and 4 post-transfection. By day 5, the growth of SKOV3, CAOV3 and
OVCAR3 cells was reduced by 73.5 (P<0.01), 71.6 (P<0.01) and
61.2% (P<0.01), respectively. To further validate the
anti-proliferative effects of miR-9 on OSC cells, a colony
formation assay was performed. Less colony formation was observed
in the miR-9-transfected cells compared with the
control-transfected cells (Fig.
2F-H), indicating that miR-9 suppressed the colony forming
ability of OSC cells.
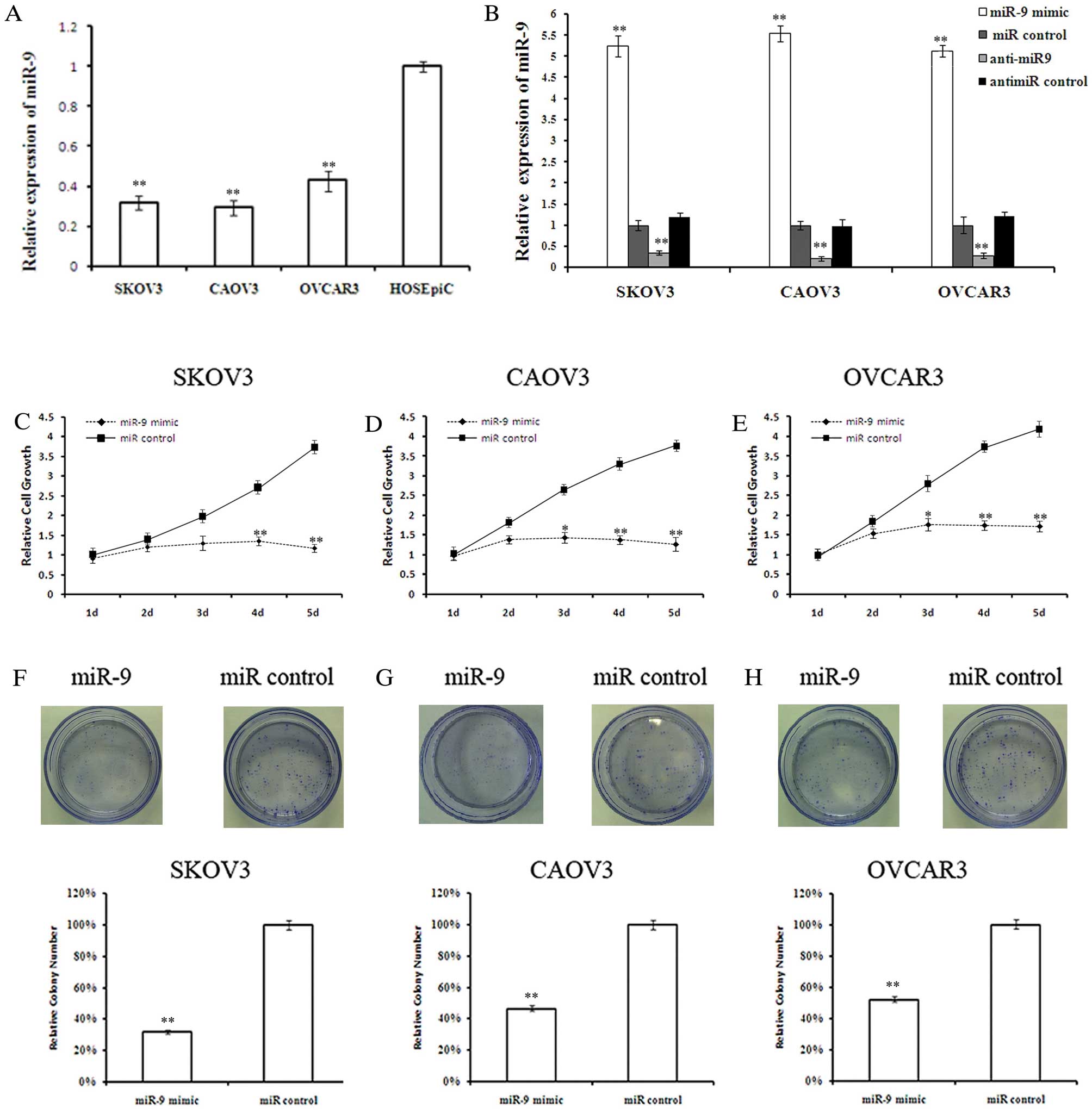 | Figure 2Overexpression of miR-9 suppresses
ovarian serous carcinoma (OSC) cell growth. (A) Quantitative RT-PCR
(qRT-PCR) analysis of miR-9 in the OSC cell lines, SKOV3, CAOV3 and
OVCAR3, and the normal ovarian surface epithelial cell line,
HOSEpiC, represented as normalized relative expression. (B) Levels
of miR-9 in OSC cells analyzed 48 h after miRNA transfection. (C-E)
Cell viability was measured by MTT assay on days 1–5 after miR-9
mimic or control transfection in SKOV3, CAOV3 and OVCAR3 cells,
respectively. (F-H) Cell proliferation was measured by colony
formation assay after miR-9 mimic or control transfection in SKOV3,
CAOV3 and OVCAR3 cells, respectively. (n=3; *P<0.05,
**P<0.01). |
miR-9 impairs SKOV3, CAOV3 and OVCAR3
cell migration and invasion
Previous studies have demonstrated that miR-9
inhibits the migration and invasion of hepatocellular carcinoma
(HCC) cells and uveal melanoma cells (17,18). We wished to determine whether
miR-9 affects the migratory and invasive capacity of OSC cells. As
miR-9 overexpression did not cause a significant difference in OSC
cell proliferation until 72 h after transfection, we performed
Transwell migration and invasion assays at 48 h after miRNA
transfection. The migratory and invasive ability of the OSC cells
was significantly impaired by miR-9 overexpression (Fig. 3A).
miR-9 suppresses TLN1 mRNA and protein
expression by binding to its 3′UTR
Using miRecords (http://mirecords.biolead.org), TargetScan (http://www.targetscan.org), PicTar (http://www.ncRNA.org/) and miRanda (http://www.microrna.org), we predicted that TLN1 is a
candidate target gene of miR-9 since it carries a putative miR-9
binding site within its 3′UTR. To confirm that miR-9 binds to this
region and induces translational repression, we constructed 2 TLN1
3′UTR luciferase reporter vectors bearing either a wild-type or a
mutated sequence of the predicted miR-9 binding site (Fig. 4A). Co-transfection experiments
revealed that exogenous miR-9 decreased the luciferase activity of
TLN1-3′UTR (P<0.01), but had little effect on TLN1-3′UTRmu
(Fig. 4B), indicating that miR-9
directly interacts with the predicted target site in TLN1-3′UTR. To
further confirm that miR-9 plays a functional role in TLN1
downregulation, OSC cells were transfected with miR-9 mimic or
anti-miR-9. Subsequent RT-PCR and western blot analysis indicated
that miR-9 overexpression led to a decline in TLN1 mRNA and protein
levels (Fig. 4C); by contrast,
the knockdown of endogenous miR-9 resulted in an increase in TLN1
mRNA and protein expression (Fig.
4D). These findings suggest that the miR-9-mediated suppression
of TLN1 expression may be associated with mRNA degradation.
Inhibition of TLN1 expression mimics the
anti-proliferative, anti-migratory and anti-invasive effects of
miR-9 overexpression on OSC cells
As we found that TLN1 was upregulated in OSC and
correlated with OSC aggressiveness and metastasis, and that miR-9
directly suppressed the expression of TLN1, we wished to determine
whether a reduction in TLN1 expression may provide an explanation
for the observed effects of miR-9 overexpression. TLN1 knockdown
with siRNA transfection was performed, followed by MTT, Transwell
migration and invasion assays. The successful knockdown of TLN1 in
OSC cells was confirmed by qRT-PCR and western blot analysis
(Fig. 5A). As with miR-9
overexpression, the decrease in TLN1 expression significantly
inhibited OSC cell growth, migration and invasion (Fig. 5B-D). These results indicate that
TLN1 is involved in the miR-9-mediated suppression of OSC, and TLN1
inhibition may explain, at least in part, the anti-proliferative,
anti-migratory and anti-invasive effects of miR-9 on OSC cells.
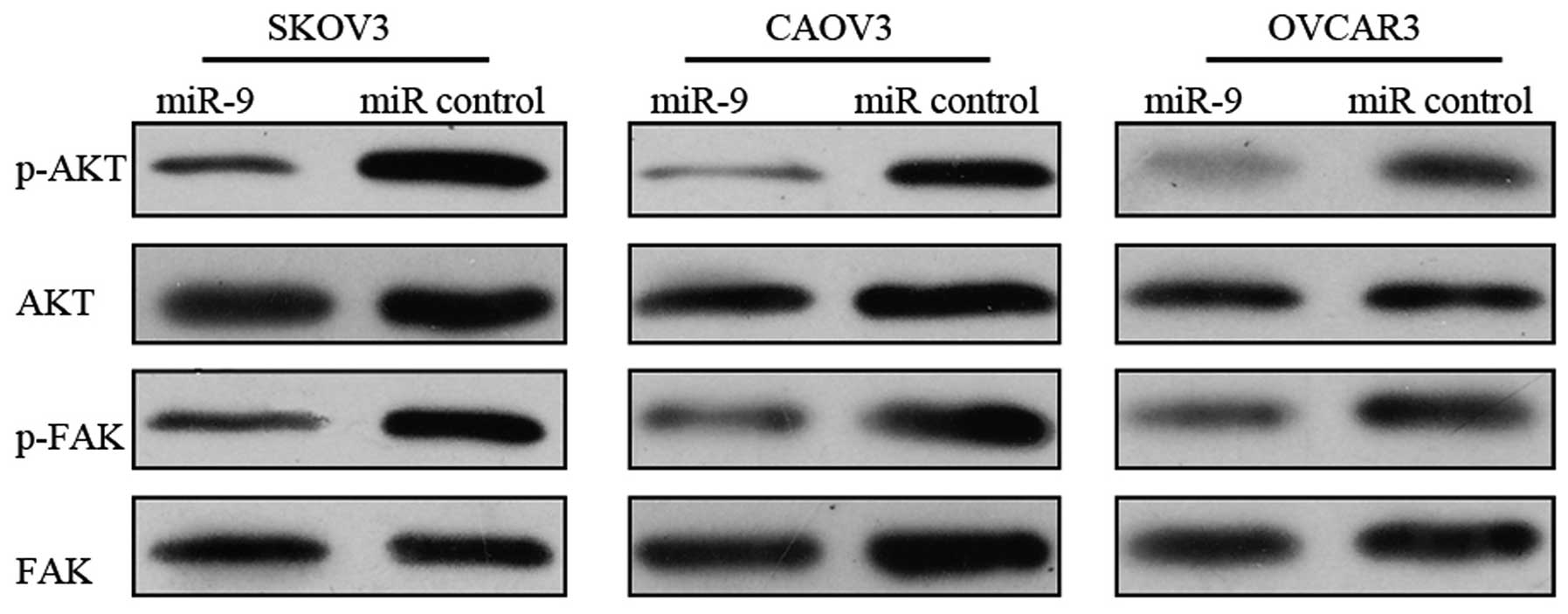
mir-9 inhibits the activation of the
FAK/AKT signaling pathway
Since we demonstrated that TLN1 is a direct and
functional target of miR-9, we further investigated the effects of
miR-9 overexpression on the TLN1-regulated FAK/AKT signaling
pathway (19). Forty-eight hours
after tranfeciton with miR-9 or miR control, the OSC cells were
subjected to western blot analysis to evaluate the total and
phosphorylated protein levels of FAK and AKT. miR-9 overexpression
in the OSC cells resulted in a marked reduction in the
phosphorylation levels of FAK and AKT, suggesting the inhibtion of
the FAK/AKT pathway (Fig. 6).
Discussion
miRNAs have emerged as a novel pathway of
tumorigenesis (20). Evidence
shows that miRNAs function as either tumor suppressor or oncogenes.
Recent studies have demonstrate that miR-9 is downregulated by
aberrant CpG island methylation in a variety of human cancers,
including breast (21), gastric
(22) and lung cancer (19). Moreover, evidence suggests that
miR-9 downregulation is associated with cancer metastasis (23) and the ‘cancer stem cell’ phenotype
(10,24). miR-9 is downregulated in human
ovarian serous, endometrioid and clear cell carcinoma (5). In addition, a previous study
demonstrated that the miR-9 expression level was significantly
lower in recurrent OSC compared with primary tumors (7). Therefore, miR-9 may act as a tumor
suppressor in OSC, and its exact role in OSC biology needs to be
further elucidated.
In this study, we first demonstrated that endogenous
miR-9 levels are lower in OSC cells compared with normal ovarian
surface epithelial cells (HOSEpiC), consistent with previous
reports of lower miR-9 levels in OSC tissues (5). We then used a gain-of-function
approach by transfecting miR-9 mimics into OSC cells to determine
the function of miR-9. Changes in cell proliferation are key
phenotypes observed in malignant transformation; uncontrolled
migration and invasion lead to metastasis, which causes the human
cancer mortality rate to reach as high as 90% (25,26). Our results revealed that the
overexpression of miR-9 inhibited OSC cell proliferation, migration
and invasion, supporting a tumor suppressor-like role for miR-9 in
OSC development and progression.
A few biological targets of miR-9 have been
identified. For example, miR-9 downregulates MTHFD2 expression,
resulting in the inhibition of cell proliferation in breast cancer
(14); miR-9 suppresses uveal
melanoma cell migration and invasion by targeting NF-κB1 (18). In this study, we demonstrate that
miR-9 inhibits TLN1 expression by targeting its 3′UTR, indicating
that TLN1 is a direct target of miR-9. Furthermore, the knockdown
of TLN1 expression mimicked the anti-proliferative, anti-migratory
and anti-invasive effects of miR-9 overexpression on OSC cells.
Therefore, TLN1 may be one of the key mediators of OSC suppression
by miR-9.
Recent studies have suggested that TLN1 is involved
in the progression of cancer (27–29). Our previous study demonstrated
that serum TLN1 was upregulated during the early and advanced phase
of a rat model of induced OSC, suggesting a role for TLN1 in the
onset and progression of this malignancy (30). Consistent with these observations,
the present study revealed a higher expression of TLN1 in OSC
tissues as compared with tumor-adjacent normal ovarian tissues. In
addition, the TLN1 expression level was significantly higher in the
lymph node metastatic lesions compared with primary tumors,
suggesting the involvement of TLN1 in the metastatic process. As
evidence shows that there are 2 distinct types of OSC, namely, low-
and high-grade OSC, which exhibit different pathogenesis and
clinicopathological features (15), we analyzed TLN1 expression after
grading the primary OSC specimens using the two-tier system. Of
note, the high-grade OSC samples exhibited a significantly higher
TLN1 expression compared with the low-grade OSC samples. Thus, TLN1
expression in OSC may be used as a promising predictor of
aggressiveness, as high-grade cancer is associated with more
aggressive behavior and is an independent predictor of poor
survival (31). Given that TLN1
is a target gene of miR-9, the suppression of miR-9 expression may
account for the overexpression of TLN1 in OSC, although the
involvement of other mechanisms cannot be excluded.
TLN1 is a component of the focal adhesion complex
that is responsible for mediating the interaction between the actin
cytoskeleton and integrins. In addition to its structural role,
TLN1 plays an essential role in integrin activation (32). TLN1-mediated integrin activation
is key to focal adhesion signaling and the induction of downstream
pathways that ultimately regulate adhesion, proliferation, anoikis,
survival and tumor progression (27,28). Previous studies have indicated
that the activation of the FAK/AKT pathway is associated with
increased proliferation, migration and invasion in a variety of
tumors (29,33–35). TLN1 overexpression in prostate
cancer cells has been shown to result in a significant enhancement
in the phosphorylation of FAK and AKT (36). In the present study, miR-9
overexpression led to a marked reduction in FAK and AKT
phosphorylation levels. The miR-9-mediated suppression of TLN1
expression may provide an explanation for the observed inhibition
of the FAK/AKT pathway; however, since miR-9 regulates multiple
gene expressions, the involvement of other mechanisms cannot be
excluded. The fact that miR-9 inhibits the activation of the
FAK/AKT signaling pathway, and that NF-κB1 is a downstream effector
of the AKT survival pathway, has led to the hypothesis that the
inhibition of the FAK/AKT pathway may be the mechanism through
which miR-9 targets NF-κB1 expression in certain tumors (13,18).
In conclusion, miR-9 is downregulated in OSC cells
compared with normal ovarian surface epithelial cells. miR-9 acts
as a tumor suppressor by inhibiting OSC cell growth, migration and
invasion. A direct and functional target of miR-9, TLN1, is
overexpressed and associated with aggressiveness and metastasis in
OSC. Our results indicate that the downregulation of miR-9 in OSC
may contribute to the malignant phenotype by maintaining a high
level of TLN1. Thus, the elucidation of the roles of miR-9 and its
target gene, TLN1, in OSC may aid in the development of promising
strategies for the targeted therapy of OSC in the future.
References
|
1
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mo YY: MicroRNA regulatory networks and
human disease. Cell Mol Life Sci. 69:3529–3531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zamore PD and Haley B: Ribo-gnome: the big
world of small RNAs. Science. 309:1519–1524. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Olson P, Lu J, Zhang H, et al: MicroRNA
dynamics in the stages of tumorigenesis correlate with hallmark
capabilities of cancer. Genes Dev. 23:2152–2165. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Laios A, O’Toole S, Flavin R, et al:
Potential role of miR-9 and miR-223 in recurrent ovarian cancer.
Mol Cancer. 7:352008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rotkrua P, Akiyama Y, Hashimoto Y, Otsubo
T and Yuasa Y: MiR-9 downregulates CDX2 expression in gastric
cancer cells. Int J Cancer. 129:2611–2620. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Moore RG, Chung M, Granai CO, Gajewski W
and Steinhoff MM: Incidence of metastasis to the ovaries from
nongenital tract primary tumors. Gynecol Oncol. 93:87–91. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo R, Wu Q, Liu F and Wang Y: Description
of the CD133+ subpopulation of the human ovarian cancer
cell line OVCAR3. Oncol Rep. 25:141–146. 2011.
|
|
11
|
Ma L, Young J, Prabhala H, et al: miR-9, a
MYC/MYCN-activated microRNA, regulates E-cadherin and cancer
metastasis. Nat Cell Biol. 12:247–256. 2010.PubMed/NCBI
|
|
12
|
Chao TF, Zhang Y, Yan XQ, et al: MiR-9
regulates the expression of CBX7 in human glioma. Zhongguo Yi Xue
Ke Xue Yuan Xue Bao. 30:268–274. 2008.(In Chinese).
|
|
13
|
Guo LM, Pu Y, Han Z, et al: MicroRNA-9
inhibits ovarian cancer cell growth through regulation of
NF-kappaB1. FEBS J. 276:5537–5546. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Selcuklu SD, Donoghue MT, Rehmet K, et al:
MicroRNA-9 inhibition of cell proliferation and identification of
novel miR-9 targets by transcriptome profiling in breast cancer
cells. J Biol Chem. 287:29516–29528. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Malpica A, Deavers MT, Lu K, et al:
Grading ovarian serous carcinoma using a two-tier system. Am J Surg
Pathol. 28:496–504. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kobel M, Kalloger SE, Huntsman DG, et al:
Differences in tumor type in low-stage versus high-stage ovarian
carcinomas. Int J Gynecol Pathol. 29:203–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tan HX, Wang Q, Chen LZ, et al: MicroRNA-9
reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell.
Med Oncol. 27:654–660. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liu N, Sun Q, Chen J, et al: MicroRNA-9
suppresses uveal melanoma cell migration and invasion through the
NF-κB1 pathway. Oncol Rep. 28:961–968. 2012.PubMed/NCBI
|
|
19
|
Heller G, Weinzierl M, Noll C, et al:
Genome-wide miRNA expression profiling identifies miR-9–3 and
miR-193a as targets for DNA methylation in non-small cell lung
cancers. Clin Cancer Res. 18:1619–1629. 2012.
|
|
20
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lehmann U, Hasemeier B, Christgen M, et
al: Epigenetic inactivation of microRNA gene hsa-mir-9–1 in human
breast cancer. J Pathol. 214:17–24. 2008.
|
|
22
|
Du Y, Liu Z, Gu L, et al: Characterization
of human gastric carcinoma-related methylation of 9 miR CpG islands
and repression of their expressions in vitro and in vivo. BMC
Cancer. 12:2492012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lujambio A, Calin GA, Villanueva A, et al:
A microRNA DNA methylation signature for human cancer metastasis.
Proc Natl Acad Sci USA. 105:13556–13561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wan HY, Guo LM, Liu T, Liu M, Li X and
Tang H: Regulation of the transcription factor NF-kappaB1 by
microRNA-9 in human gastric adenocarcinoma. Mol Cancer. 9:162010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
26
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J and
Thun MJ: Cancer statistics, 2009. CA Cancer J Clin. 59:225–249.
2009. View Article : Google Scholar
|
|
27
|
Vachon PH: Integrin signaling, cell
survival, and anoikis: distinctions, differences, and
differentiation. J Signal Transduct. 2011:7381372011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Desiniotis A and Kyprianou N: Significance
of talin in cancer progression and metastasis. Int Rev Cell Mol
Biol. 289:117–147. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fong YC, Liu SC, Huang CY, et al:
Osteopontin increases lung cancer cells migration via activation of
the alphavbeta3 integrin/FAK/Akt and NF-kappaB-dependent pathway.
Lung Cancer. 64:263–270. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang Y, Zhang X, Jiang W, et al:
Discovery of serum biomarkers implicated in the onset and
progression of serous ovarian cancer in a rat model using iTRAQ
technique. Eur J Obstet Gynecol Reprod Biol. 165:96–103. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vang R, Shihle M and Kurman RJ: Ovarian
low-grade and high-grade serous carcinoma: pathogenesis,
clinicopathologic and molecular biologic features, and diagnostic
problems. Adv Anat Pathol. 16:267–282. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Critchley DR and Gingras AR: Talin at a
glance. J Cell Sci. 121:1345–1347. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hwang YP, Yun HJ, Choi JH, et al:
Suppression of EGF-induced tumor cell migration and matrix
metalloproteinase-9 expression by capsaicin via the inhibition of
EGFR-mediated FAK/Akt, PKC/Raf/ERK, p38 MAPK, and AP-1 signaling.
Mol Nutr Food Res. 55:594–605. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ren K, Jin H, Bian C, et al: MR-1
modulates proliferation and migration of human hepatoma HepG2 cells
through myosin light chains-2 (MLC2)/focal adhesion kinase
(FAK)/Akt signaling pathway. J Biol Chem. 283:35598–35605. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sakamoto S, McCann RO, Dhir R and
Kyprianou N: Talin1 promotes tumor invasion and metastasis via
focal adhesion signaling and anoikis resistance. Cancer Res.
70:1885–1895. 2010. View Article : Google Scholar : PubMed/NCBI
|