Introduction
Genome-wide association studies (GWAS) have
identified numerous single nucleotide polymorphisms (SNPs) that are
associated with human diseases, such as type 2 diabetes mellitus
(1). However, the majority of
these SNPs are located in non-coding regions of the genome, and the
mechanisms through which they promote disease risk have not been
completely elucidated. Potassium voltage-gated channel, KQT-like
subfamily Q, member 1 (KCNQ1) is one of the genes most
significantly associated with type 2 diabetes in different ethnic
groups, particularly in East Asian populations, and type 2
diabetes-associated SNPs are also located mainly in an intron of
KCNQ1 (2–4). The KCNQ1 gene encodes the α
subunit of the voltage-gated potassium channel Kv7.1, and
KCNQ1 is also expressed in pancreatic β cells, although the
Kv7.1 channel is best characterized for its contribution to cardiac
function (5). A recent study
indicated that increased KCNQ1 protein expression limits insulin
secretion from pancreatic β cells by regulating the potassium
channel current (6). However, to
date, and to the very best of our knowledge, no data have been
presented regarding the association between the disease-associated
variants and the expression of KCNQ1.
Recent studies from the ENCODE project have
confirmed that disease-associated variants are enriched in
regulatory DNA and that promoters and distal elements engage in
multiple long-range interactions to form complex networks (7–10).
Therefore, these SNPs in the KCNQ1 gene may affect the
expression of nearby or long-distance genes. Among the candidate
genes close to KCNQ1, the loss of cyclin-dependent kinase
inhibitor 1C (CDKN1C) in focal lesions in infants with
hyperinsulinism has been shown to correlate with the increased
proliferation of pancreatic β cells (11), and targeting CDKN1C
promotes adult human β cell replication (12). Furthermore, the long non-coding
RNA, KCNQ1 opposite strand/antisense transcript 1
(KCNQ1OT1), regulates the expression of CDKN1C
through imprinting (13–15). Transient receptor potential cation
channel, subfamily M, member 5 (TRPM5), another nearby gene,
has been reported to regulate glucose-stimulated insulin secretion
in pancreatic β cells (16–19). Again, however, to the best of our
knowledge, no data to date have been presented regarding the
association between the disease-associated variants and the
expression of these nearby genes.
In order to analyze the molecular mechanisms linking
these non-coding variants with type 2 diabetes mellitus, it is
crucial to identify DNA-binding proteins that recognize the
nucleotide sequence surrounding disease-associated variants in an
allele-specific manner; however, to the best of our knowledge,
there have only been a few reports that have been successful in the
identificatoin of such proteins. We and our colleagues have
previously developed novel nanobeads composed of glycidyl
methacrylate and styrene, and demonstrated that these nanobeads
enable the rapid and efficient purification of ‘bait’ binding
proteins, including transcription factors using DNA fragments as
bait (20–22). In the present study, we applied
further-improved magnetic nanobeads (23), and aimed to identify the proteins
that bind the KCNQ1 intronic locus containing
disease-associated SNPs in an allele-specific manner. We also
investigated the function of these proteins in order to clarify the
association between genetic variants and the risk of type 2
diabetes.
Materials and methods
Cells
The rat pancreatic β-cell line, INS-1 (24), which had kindly been provided by
Dr C.B. Wollheim and Dr N. Sekine (University of Geneva, Geneva,
Switzerland), was cultured at 37°C in a 5% CO2
atmosphere in RPMI-1640 medium which was supplemented with 10%
fetal bovine serum, 10 mM HEPES, 1 mM sodium pyruvate and 50 mM
2-mercaptoeth-anol. KP-3 (human pancreatic cancer), KP-4
(pancreatic ductal cell carcinoma), QGP-1 (human pancreatic
endocrine) and SUIT-2 (human pancreatic cancer) cells were obtained
from the Health Science Research Resources Bank (Osaka, Japan).
AsPC-1 (human pancreatic adenocarcinoma), Capan-1 (human pancreatic
ductal adenocarcinoma), 293T and HeLa cells were obtained from the
American Type Culture Collection (ATCC, Manassas, VA, USA). The
KP-3, QGP-1, SUIT-2 and AsPC-1 cells were cultured in RPMI-1640
medium which was supplemented with 10% fetal bovine serum. The KP-4
and Capan-1 cells were cultured in Iscove’s modified Dulbecco’s
medium (IMDM) supplemented with 10% fetal bovine serum. The 293T
and HeLa cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) which was supplemented with 10% fetal bovine serum.
Preparation of DNA-immobilized
nanobeads
The oligonucleotides used in the present study were
synthesized at Japan BioServices Co. Ltd. (Saitama, Japan). These
oligonucleotides contained either risk or non-risk alleles through
single-nucleotide substitution (Table
I). DNA-immobilized nanobeads were prepared as previously
described with slight modifications (21,25). Two complementary oligonucleotides
were phosphorylated, annealed and then ligated to produce oligomers
that ranged mainly from 100 to 3,000 bp. These oligomers containing
tandemly repeated SNP regions were applied to the NICK Column
according to the manufacturer’s instructions (GE Healthcare,
Buckinghamshire, UK). The second fractions of the NICK Column were
ready for coupling onto FG beads (Tamagawa Seiki Co., Ltd., Nagano,
Japan) (23). One milligram of FG
beads was then mixed with 100 µg of prepared DNA, and the
coupling reaction was carried out at 50°C for 24 h.
 | Table IOligonucleotides used for the
preparation of the DNA-immobilized nanobeads. |
Table I
Oligonucleotides used for the
preparation of the DNA-immobilized nanobeads.
| SNP | Allele | | Primer sequence
(5′→3′) |
|---|
| rs2237892 | Risk | Forward |
GGGGGTTTGCCACC[C]GGGGTGAGG |
| | Reverse |
CCCCCCCTCACCCC[G]GGTGGCAAA |
| Non-risk | Forward |
GGGGGTTTGCCACC[T]GGGGTGAGG |
| | Reverse |
CCCCCCCTCACCCC[A]GGTGGCAAA |
| rs2237895 | Risk | Forward |
GGGGGAGTGGTCCC[C]GGGGTCGGG |
| | Reverse |
CCCCCCCCGACCCC[G]GGGACCACT |
| Non-risk | Forward |
GGGGGAGTGGTCCC[A]GGGGTCGGG |
| | Reverse |
CCCCCCCCGACCCC[T]GGGACCACT |
| rs151290 | Risk | Forward |
GGGGGTGAGCCCAG[C]CCCCTGGGC |
| | Reverse |
CCCCCGCCCAGGGG[G]CTGGGCTCA |
| Non-risk | Forward |
GGGGGTGAGCCCAG[A]CCCCTGGGC |
| | Reverse |
CCCCCGCCCAGGGG[T]CTGGGCTCA |
| rs2074196 | Risk | Forward |
GGGGGGGGGCTTCA[G]TGGAGCCCG |
| | Reverse |
CCCCCCGGGCTCCA[C]TGAAGCCCC |
| Non-risk | Forward |
GGGGGGGGGCTTCA[T]TGGAGCCCG |
| | Reverse |
CCCCCCGGGCTCCA[A]TGAAGCCCC |
| rs2283228 | Risk | Forward |
GGGGGGCTGAAAGC[A]CTGGTTAAA |
| | Reverse |
CCCCCTTTAACCAG[T]GCTTTCAGC |
| Non-risk | Forward |
GGGGGGCTGAAAGC[C]CTGGTTAAA |
| | Reverse |
CCCCCTTTAACCAG[G]GCTTTCAGC |
| rs2237897 | Risk | Forward |
GGGGGAGCTGGGGA[C]GAGGGGCCT |
| | Reverse |
CCCCCAGGCCCCTC[G]TCCCCAGCT |
| Non-risk | Forward |
GGGGGAGCTGGGGA[T]GAGGGGCCT |
| | Reverse |
CCCCCAGGCCCCTC[A]TCCCCAGCT |
Affinity purification
Nuclear extracts were prepared from the INS-1 cells
according to the method described in the study by Dignam et
al (26). DNA-immobilized
nanobeads (200 µg) were equilibrated with binding buffer (20
mM HEPES-NaOH, pH 7.9, 10% glycerol, 100 mM KCl, 1 mM
MgCl2, 0.2 mM CaCl2, 0.2 mM EDTA, 1 mM DTT
and 0.2 mM PMSF). INS-1 nuclear extracts (200 µg) were mixed
with 100 µg of single-stranded DNA (Ambion/Life
Technologies, Austin, TX, USA) and 10 µg of
poly(dI-dC):poly(dI-dC) (Sigma-Aldrich, St. Louis, MO, USA), and
then incubated with equilibrated DNA-immobilized nanobeads for 4 h
at 4°C using a RT-50 rotator (Taitec Corp., Saitama, Japan). After
washing with binding buffer 3 times, the bound proteins were eluted
with binding buffer supplemented with 1 M KCl (final
concentration).
Mass spectrometry
Affinity-purified proteins were separated using
SDS-PAGE and subjected to silver staining as described in the study
by Shevchenko et al (27).
The specific protein bands were excised and mass spectrometry was
then performed as previously described (28). Band slices were reduced with 10 mM
dithiothreitol, alkylated with 55 mM iodoacetamide and digested
with sequencing grade modified trypsin (12 µg/ml; Promega
Corp., Madison, WI, USA) overnight. The extracted peptides were
desalted with ZipTip C18 (Millipore, Billerica, MA, USA) and
separated by nano-flow liquid chromatography (LC) (Paradigm MS4;
Michrom BioResources, Inc., Auburn, CA, USA) using a reverse phase
C18 column (Magic C18, 0.2 ×50mm; Michrom BioResources, Inc.). The
LC eluent was coupled to a micro-ionspray source attached to an LCQ
Advantage MAX mass spectrometer (Thermo Fisher Scientific Inc.,
Waltham, MA, USA). All MS/MS spectra were searched using the
TurboSEQUEST algorithm within the BioWorks 3.2 software (Thermo
Fisher Scientific Inc.).
Immunoblot analysis
Nuclear extracts and affinity-purified fractions
were fractionated by SDS-PAGE and electrotransferred onto a
polyvinylidene difluoride membrane (Millipore). The membrane was
incubated with 5% non-fat milk for 1 h and incubated with antibody
against nuclear transcription factor Y, beta (NF-YB) overnight
(sc-13045; Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
followed by incubation with peroxidase-conjugated anti-rabbit IgG
(no. 7074; Cell Signaling Technology, Inc., Danvers, MA, USA) for 1
h. Visualization was accomplished using enhanced chemiluminescense
reagents (Millipore) and analyzed using the ChemiDoc XRS+ imaging
system (Bio-Rad, Hercules, CA, USA).
Preparation of radiolabeled recombinant
proteins
Complementary DNA (cDNA) for nuclear transcription
factor Y, alpha (NF-YA), NF-YB, nuclear transcription
factor Y, gamma (NF-YC) and cut-like homeobox 1
(CUX1) were amplified by RT-PCR from the total RNA prepared
from HeLa cells, inserted into the pUC119 vector, and sequenced
using an ABI PRISM 3700 sequence analyzer (Applied Biosystems,
Foster City, CA, USA). The preparation of radiolabeled recombinant
proteins was then performed as previously described (28). T7 promoter-tagged DNA fragments of
these cDNAs were amplified using PCR with two primers: T7 promoter
fused to a 5′ sequence of each cDNA and polyT fused to a 3′
complementary sequence of each cDNA. These amplified DNA fragments
were used to synthesize 35S-radiolabeled recombinant proteins in a
coupled transcription/translation system according to the protocol
of the manufacturer (Promega Corp.).
Binding assay
DNA-immobilized nanobeads or control nanobeads (200
µg) were equilibrated with binding buffer as described above
and incubated with 200 µl of the radiolabeled proteins
(NF-YA, NF-YB, NF-YC and CUX1) at 4°C for 4 h using a RT-50
rotator. After washing with binding buffer, the bound proteins were
eluted by boiling for 5 min with SDS-PAGE sample buffer. The
eluates and inputs were subjected to SDS-PAGE. The gels were dried,
and autoradiography was then performed to visualize the
radiolabeled proteins.
Construction of plasmids
Reporter plasmids that contain the luciferase gene
under the control of the SNP rs2074196 region were constructed as
follows: complementary oligonucleotides containing 3 copies of the
SNP region were synthesized. These oligonucleotides were annealed
and cloned into the pGL4.10 vector (Promega Corp.) along with the
thymidine kinase promoter derived from pGL4.74 (Promega Corp.).
To construct mammalian expression plasmids, cDNA of
NF-YA, NF-YB, NF-YC and CUX1 were
amplified by PCR and subcloned into the pcDNA6 vector (Invitrogen
Life Technologies, Carlsbad, CA, USA) to allow the expression of
these cDNAs under the control of a CMV promoter. During PCR
amplification, FLAG-tag was fused at the C-terminus of NF-YA;
HA-tag was fused at the C-terminus of NF-YB; and Myc-tag was fused
at the C-terminus of NF-YC. All the sequences were confirmed using
an ABI PRISM 3700 sequence analyzer.
Luciferase assay
One of the constructed luciferase reporter plasmids
(100 ng) and the control Renilla luciferase reporter vector
pGL4.74 (10 ng) were co-transfected into the 293T cells
(2x105 cells) along with mammalian expression vectors:
NF-Ys (NF-YA-FLAG, NF-YB-HA, and NF-YC-Myc, 300 ng each), CUX1 (900
ng), or empty pcDNA6 (900 ng) using Lipofectamine 2000 (Invitrogen
Life Technologies) and seeded into a 24-well plate. Twelve hours
after transfection, the medium was changed. Forty-eight hours after
transfection, cell extracts were prepared, and Firefly luciferase
activities and Renilla luciferase activities were determined
using a Dual-Glo Luciferase assay system (Promega Corp.) and a
CentroPRO LB 962 Microplate Luminometer (Berthold Technologies, Bad
Wildbad, Germany).
Chromatin immunoprecipitation assay
The preparation of the samples was performed using a
Magna ChIP G kit according to the manufacturer’s instructions
(Millipore, Temecula, CA, USA). The samples were sonicated for 10
rounds of 30 pulses each (0.5 sec on and 0.5 sec off) using a
Branson Sonifier S-450 digital ultrasonic cell
disruptor/homogenizer (Branson Ultrasonics Corp., Danbury, CT, USA)
at 15% amplitude. The efficiency of sonication was assessed to
ensure that the majority of chromatin fragments were in the range
of 200–1,000 bp. The sonicated samples were immunoprecipitated
overnight at 4°C with 2.5 µg of anti-Myc-tag antibody (no.
2276; Cell Signaling Technology, Inc.) or normal mouse IgG
(Millipore). The immunoprecipitated DNA was quantified by qPCR
performed on the input and bound fractions, using the Fast
SYBR-Green Master Mix and a StepOne Real-Time PCR system (both from
Applied Biosystems). The primers used for qPCR were as follows:
5′-GAGCCAGTTGTTCCCAAACC-3′ and 5′-TAGGCTTGTGTCCCCAGTCC-3′ for SNP
rs2074196, and 5′-CGCAAAAGGAGGGGAGAG-3′ and
5′-GCCGCTGGGTTTTATAGGG-3′ for the ACTB promoter.
Statistical analysis
All data are expressed as the means ± SD.
Statistical analysis was performed using an unpaired two-tailed
Student’s t-test. A value of P<0.01 was considered to indicate a
statistically significant difference.
Results
Purification of allele-specific
DNA-binding proteins by novel magnetic nanobeads
In this study, we aimed to identify proteins that
bind the genomic region encompassing the type 2 diabetes-associated
SNPs: rs151290, rs2074196, rs2237892, rs2283228, rs2237895 and
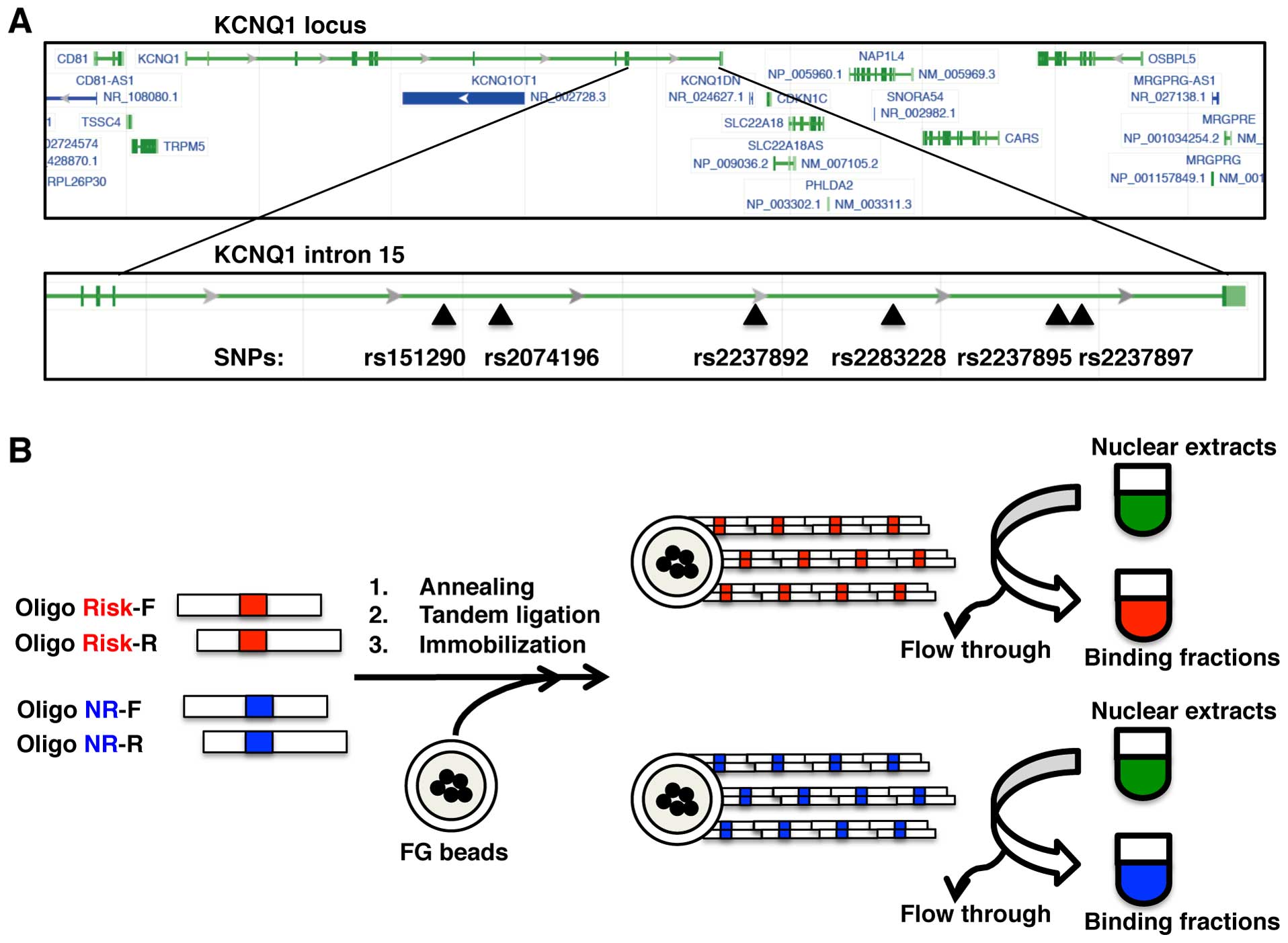
rs2237897 (Fig. 1A). In order to
identify the allele-specific binding proteins, we first prepared 2
types of beads, risk and non-risk allele beads, by immobilizing
each allelic DNA onto magnetic nanobeads (Table I and Fig. 1B). The lengths of the immobilized
DNA fragments were almost the same (100–3,000 bp) for the risk and
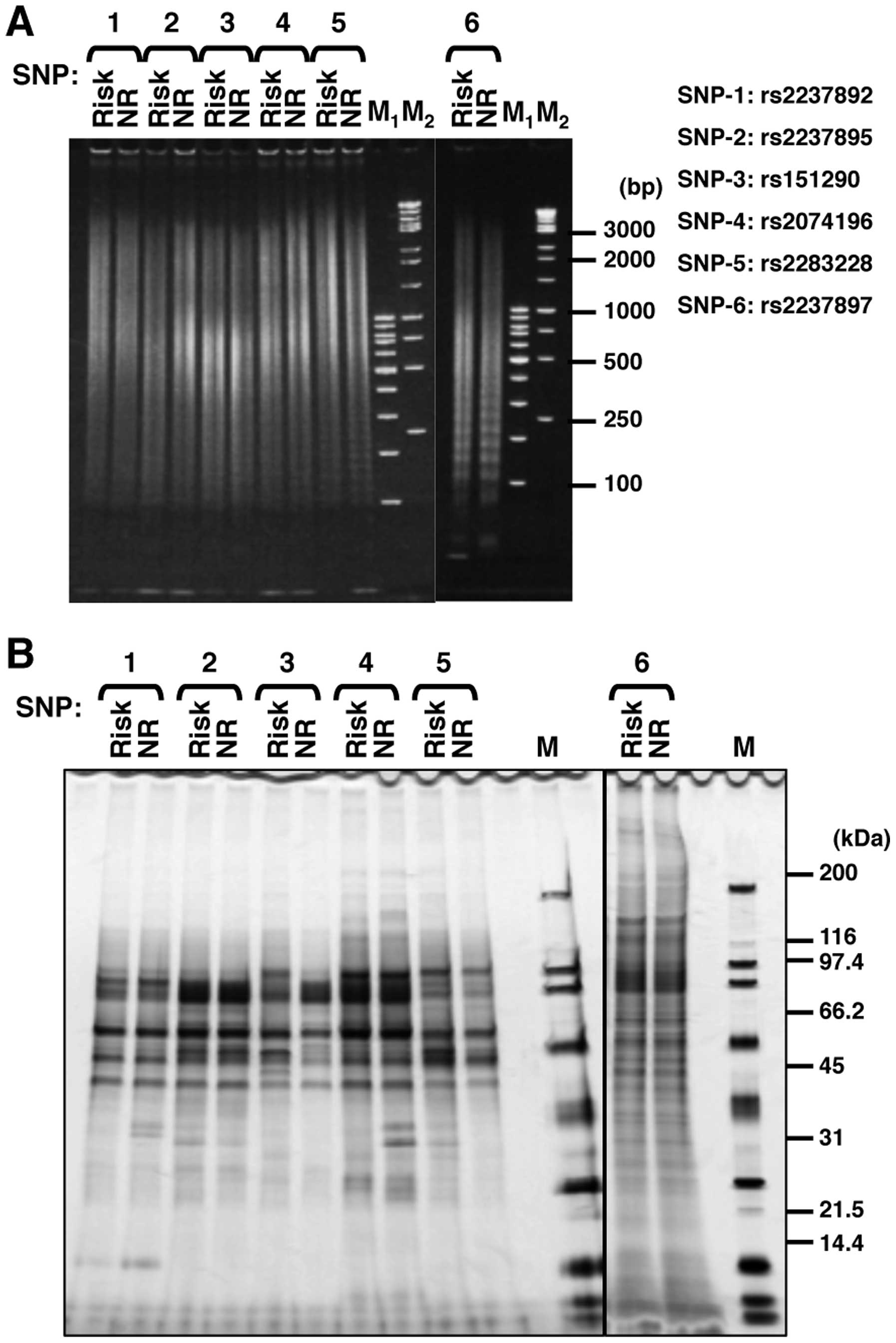
non-risk alleles (Fig. 2A). Using
these nanobeads, we purified DNA-binding proteins from the nuclear
extract of INS-1 pancreatic β cells and analyzed the bound proteins
using SDS-PAGE (Fig. 2B). We
found that the affinities of several proteins for each examined SNP
differed between alleles. There were obvious allele-specific bands,
particularly in the SNP rs151290 and SNP rs2074196 regions. We
decided to perform the following analysis with SNP rs2074196, as
this was among the 3 SNPs (rs2074196, rs2237892 and rs2237895)
which showed the best association P-values with type 2 diabetes in
our previous study (2).
Identification of allele-specific
DNA-binding proteins
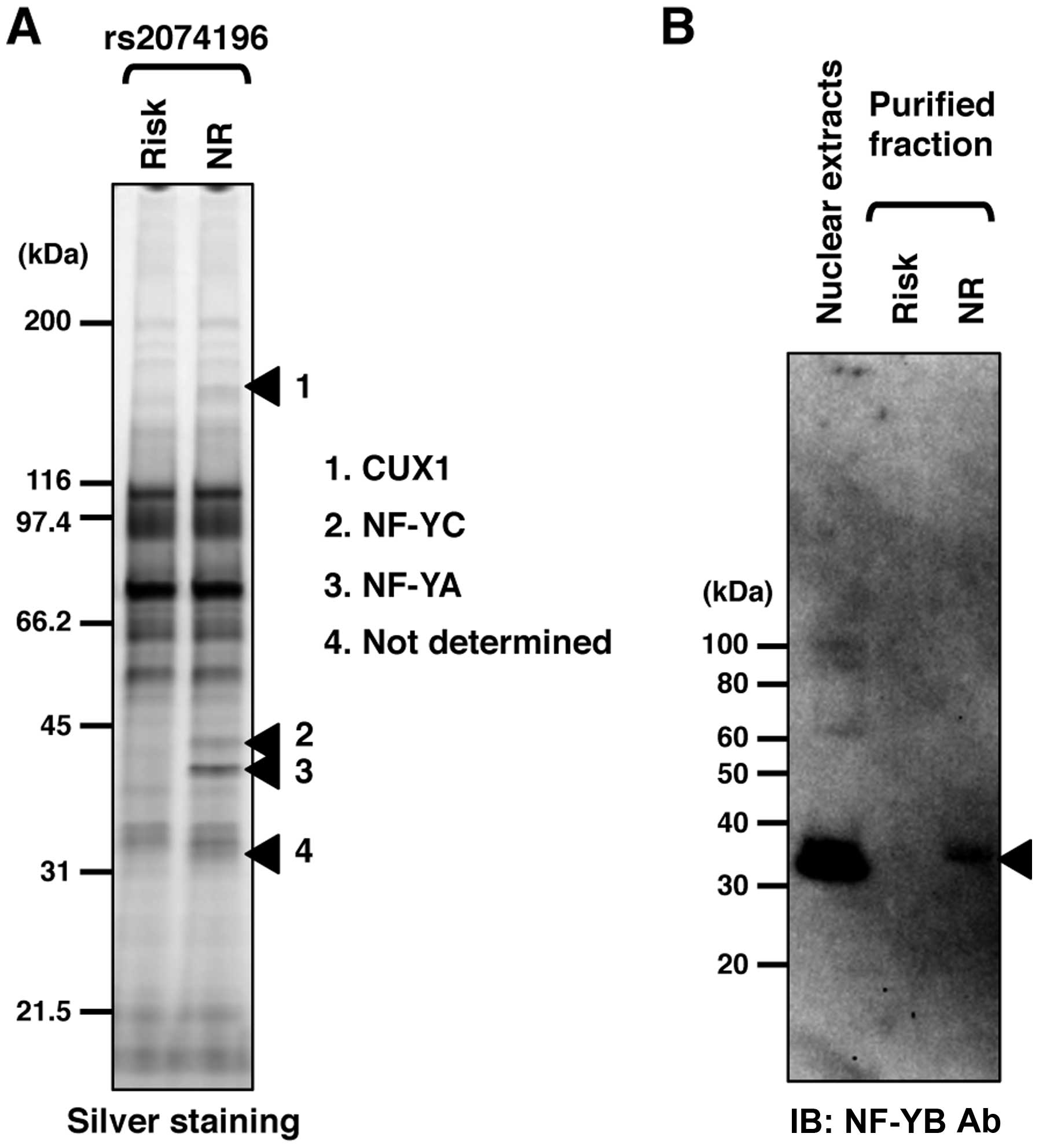
We compared the bound proteins of the risk and
non-risk alleles and selected 4 differentially bound protein bands
(Fig. 3A). Following mass
spectrometry, we identified 3 proteins, CUX1, NF-YC and NF-YA. One
protein band was not determined using mass spectrometry. However,
we speculated that the band was NF-YB, due to the size of the
protein band and the lack of a partner protein for the NF-Y ternary
complex. Immunoblot analysis using a specific antibody against
NF-YB confirmed that NF-YB had been purified using DNA-immobilized
nano-beads in an allele-specific manner (Fig. 3B).
Evaluation of allele-specific DNA-binding
proteins
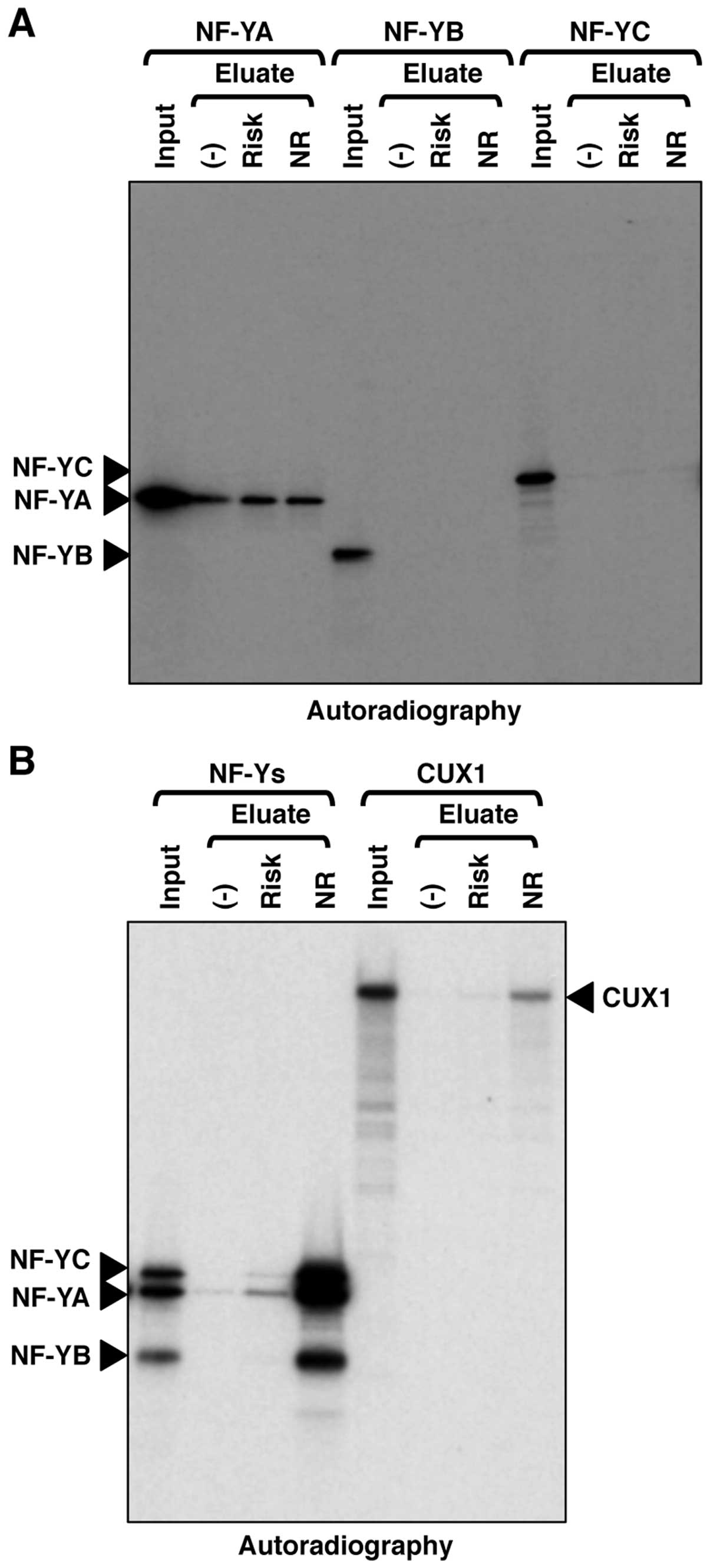
We then confirmed the DNA-binding activities of NF-Y
and CUX1 in vitro using recombinant proteins. NF-YA, NF-YB
or NF-YC alone exhibited no binding activity or only non-specific
binding activity against the genomic region encompassing SNP
rs2074196 (Fig. 4A). The ternary
complex of NF-YA, NF-YB and NF-YC exhibited specific binding
activity against the non-risk allele of the genomic region
encompassing SNP rs2074196 (Fig.
4B). CUX1 also exhibited specific binding activity against the
non-risk allele of the genomic region encompassing SNP rs2074196
(Fig. 4B). These DNA-binding
specificities against the non-risk allele of recombinant NF-Y and
CUX1 were consistent with those observed in the silver staining of
purified proteins from the INS-1 cell nuclear extracts (Figs. 2B and 3A).
SNP rs2074196 modulates NF-Y-dependent
transcriptional activity
We then examined the transcriptional activities of
NF-Y and CUX1 using luciferase reporter assay. As illustrated in
Fig. 5A, we prepared 4 reporter
constructs: risk allele and forward orientation, non-risk allele
and forward orientation, risk allele and reverse orientation, and
non-risk allele and reverse orientation. The basal luciferase
activities, which are mediated by endogenous transcription factors
possibly including NF-Y and CUX1, differed among the constructs
(Fig. 5B): non-risk-forward,
1.60-fold; risk-reverse, 0.57-fold; and non-risk-reverse, 0.84-fold
higher compared to risk-forward. Hence, the basal luciferase
activities of non-risk-forward were the highest for these 4
constructions in 293T cells.
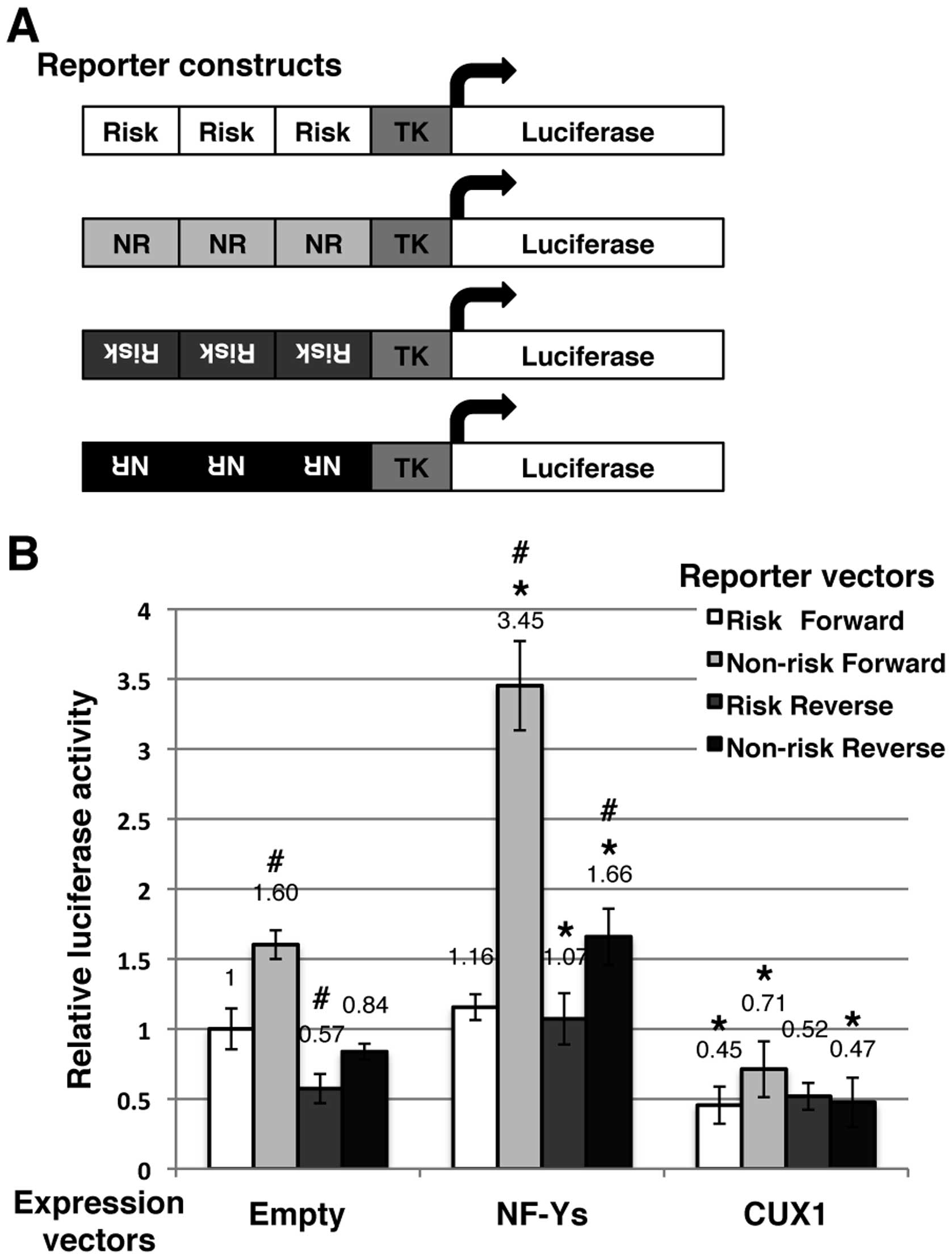 | Figure 5Effects of nuclear transcription
factor Y (NF-Y) or cut-like homeobox 1 (CUX1) on transcriptional
activity under the control of the genomic region encompassing
single nucleotide polymorphism (SNP) rs2074196. (A) Construction of
4 types of Firefly luciferase reporter vectors. (B) 293T cells were
transfected with one of each firefly luciferase reporter constructs
and control Renilla luciferase reporter vector pGL4.74 along
with the mammalian expression vectors, NF-Ys [nuclear transcription
factor Y, alpha (NF-YA)-FLAG, nuclear transcription factor Y, beta
(NF-YB)-HA, and nuclear transcription factor Y, gamma (NF-YC)-Myc],
CUX1, or empty. Firefly luminescence was normalized using renilla
luminescence. Data are the means ± SD calculated from 4 independent
replicates. Statistical significance: *P<0.01,
relative to the empty expression vector in each reporter vector;
#P<0.01, relative to the risk-forward reporter vector
in each expression vector calculated by an unpaired Student’s
t-test, two-tailed. |
When the expression constructs of NF-YA, NF-YB and
NF-YC were co-transfected, the luciferase activities of each
reporter construct were upregulated (Fig. 5B): risk-forward, 1.16-fold;
non-risk-forward, 3.45-fold, risk-reverse, 1.07-fold; and
non-risk-reverse, 1.66-fold compared to basal risk-rorward; in
addition non-risk-forward, 2.16-fold, risk-reverse, 1.87-fold, and
non-risk-reverse, 1.98-fold compared to the basal value of each
construction. These results confirmed that the NF-Y complex
upregulated the transcription under the control of the genomic
region encompassing SNP rs2074196, particularly for the non-risk
allele and forward orientation.
When the expression construct of CUX1 was
co-trans-fected, the luciferase activities of each reporter
construct were downregulated (Fig.
5B). These results indicated that CUX1 downregulated the
transcription under the control of the genomic region encompassing
SNP rs2074196; however, no allele specificity was observed.
SNP rs2074196 modulates the affinity of
the locus for NF-Y
We further examined the DNA-binding activities of
NF-Y in cell lines using chromatin immunoprecipitation assay using
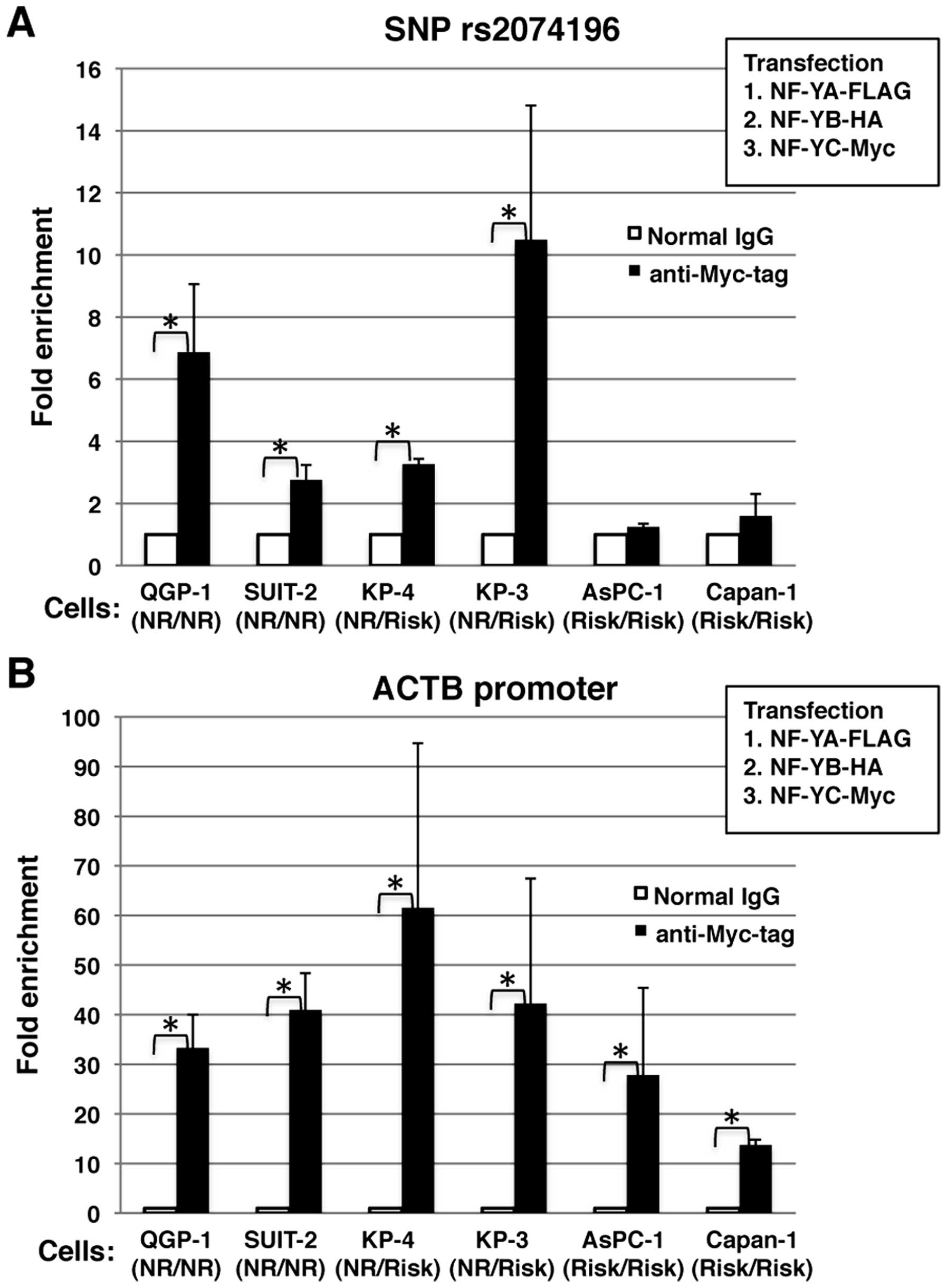
6 cell lines: QGP-1, SUIT-2, KP-4, KP-3, AsPC-1 and Capan-1. The
QGP-1 and SUIT-2 cells were found to be homozygous for the non-risk
allele, the KP-4 and KP-3 were heterozygous for the risk and
non-risk allele, and the AsPC-1 and Capan-1 cells were homozygous
for the risk allele. We transfected these cells with the expression
vectors, NF-YA-FLAG, NF-YB-HA, and NF-YC-Myc. As illustrated in
Fig. 6A, the genomic region
encompassing SNP rs2074196 was enriched in the QGP-1, SUIT-2, KP-4
and KP-3 cells, but not in the AsPC-1 and Capan-1 cells. As a
positive control, the genomic region of the ACTB promoter
containing NF-Y-binding CCAAT-box elements was enriched in all the
examined cell lines (Fig. 6B).
These results confirmed that NF-Y specifically bound the non-risk
allele of the endogenous KCNQ1 intronic region encompassing
SNP rs2074196.
Discussion
GWAS have indicated that SNPs in the KCNQ1
locus are associated with type 2 diabetes (2–4).
Since the majority of these SNPs are located in the intronic region
of the KCNQ1 gene and are associated with an impaired
insulin secretion, we hypothesized that some of these
SNP-containing regions may participate in regulating gene
expression in pancreatic β cells as cis-regulatory elements,
such as enhancers and that SNPs may affect such activity of this
locus.
Novel nanobeads, as we and our colleague have
previously reported (20–23,25,28), have been proven to be useful for
screening proteins that bind to a variety of baits. Since one of
the major features of the beads was a very low background, we
hypothesized that they may be also applied to purify DNA-binding
proteins in an allele-specific manner. Indeed, we successfully
found that the affinities of several proteins for the examined SNP
regions in the KCNQ1 gene locus differed between alleles.
NF-Y specifically bound the non-risk allele of the SNP rs2074196
region and stimulated the transcriptional activity of the
artificial promoter containing SNP rs2074196 in an allele-specific
manner. These results suggest that SNP rs2074196 modulates the
affinity of the locus for NF-Y and possibly induces subsequent
changes in gene expression, which may provide important clues as to
how type 2 diabetes-associated SNPs in the KCNQ1 gene locus
promote disease risk.
Recent studies have confirmed that NF-Y is necessary
for hematopoietic stem cell proliferation and survival.
NF-YA deletion creates an accumulation of hematopoietic stem
cells in the G2/M phase, accompanied by the dysregulation of
multiple genes that influence cell cycle control and apoptosis
(29). Another group reported
that the inactivation of NF-YA in postnatal liver causes
hepatocellular degeneration, lipid deposition and endoplasmic
reticulum stress (30),
indicating that NF-Y is a key transcription factor controlling
endoplasmic reticulum function and metabolic processes in mature
hepatocytes. Although the function of NF-Y in pancreatic β cells
has not been well studied, these observations suggest that NF-Y
participates in the regulation of β cell development and function.
Future identification of the target genes regulated by this locus
encompassing SNP rs2074196 and NF-Y are required to provide further
information.
In addition, we cannot exclude the possibility that
other genomic regions harboring intronic SNPs in the KCNQ1
gene may also be functional and contribute to susceptibility to
type 2 diabetes as gene expression may be regulated by several
independent or correlated regions containing respective SNPs.
Although SNP rs2074196 is the only variant extensively examined
thus far, we have already detected allele-specific binding proteins
for other SNP regions (Fig. 2B).
Further experiments on each allele-specific binding protein and the
interaction between these SNPs should clarify the molecular
association between SNPs in the KCNQ1 locus and
susceptibility to type 2 diabetes.
Although recent GWAS have been extremely successful
(1–4), it remains a big challenge to
functionally annotate SNPs in susceptibility genes that are very
often localized in non-coding regions. For this purpose, it is
essential to identify DNA-binding proteins that recognize the
nucleotide sequence surrounding disease-associated variants. Our
comparative analysis between alleles using DNA-immobilized
nanobeads enabled systematic identification of allele-specific
DNA-binding proteins in multiple SNP regions. We believe that this
system is also very useful for investigating the molecular
mechanisms linking non-coding genetic variants to the risk of
common diseases in general.
Acknowledgments
We would like to thank Mr. Dai Suzuki and Ms. Kazuko
Nagase for providing technical assistance. This study was supported
by grants from the Japan Society for the Promotion of Science
(JSPS) Grants-in-Aid for Scientific Research (KAKENHI), grant no.
22510216 (to M.H.), the Japan Diabetes Foundation (to M.H.), the
Leading Research Project of Ministry of Education, Culture, Sports,
Science and Technology, Japan (to K.Y.), National Institute of
Biomedical Innovation (to K.Y.) and National Center for Global
Health and Medicine (to K.Y.).
Abbreviations:
|
SNP
|
single nucleotide polymorphism
|
|
KCNQ1
|
potassium voltage-gated channel,
KQT-like subfamily Q, member 1
|
|
NF-Y
|
nuclear transcription factor Y
|
|
CDKN1C
|
cyclin-dependent kinase inhibitor
1C
|
|
KCNQ1OT1
|
KCNQ1 opposite strand/antisense
transcript 1
|
|
TRPM5
|
transient receptor potential cation
channel, subfamily M, member 5
|
|
CUX1
|
cut-like homeobox 1
|
References
|
1
|
Imamura M and Maeda S: Genetics of type 2
diabetes: The GWAS era and future perspectives (Review). Endocr J.
58:723–739. 2011. View Article : Google Scholar
|
|
2
|
Yasuda K, Miyake K, Horikawa Y, Hara K,
Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y, et al:
Variants in KCNQ1 are associated with susceptibility to type 2
diabetes mellitus. Nat Genet. 40:1092–1097. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Unoki H, Takahashi A, Kawaguchi T, Hara K,
Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K,
Jørgensen T, et al: SNPs in KCNQ1 are associated with
susceptibility to type 2 diabetes in East Asian and European
populations. Nat Genet. 40:1098–1102. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Voight BF, Scott LJ, Steinthorsdottir V,
Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS,
Thorleifsson G, et al: MAGIC investigators; GIANT Consortium:
Twelve type 2 diabetes susceptibility loci identified through
large-scale association analysis. Nat Genet. 42:579–589. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Neyroud N, Tesson F, Denjoy I, Leibovici
M, Donger C, Barhanin J, Fauré S, Gary F, Coumel P, Petit C, et al:
A novel mutation in the potassium channel gene KVLQT1 causes the
Jervell and Lange-Nielsen cardioauditory syndrome. Nat Genet.
15:186–189. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yamagata K, Senokuchi T, Lu M, Takemoto M,
Fazlul Karim M, Go C, Sato Y, Hatta M, Yoshizawa T, Araki E, et al:
Voltage-gated K+ channel KCNQ1 regulates insulin
secretion in MIN6 β-cell line. Biochem Biophys Res Commun.
407:620–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sanyal A, Lajoie BR, Jain G and Dekker J:
The long-range interaction landscape of gene promoters. Nature.
489:109–113. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Maurano MT, Humbert R, Rynes E, Thurman
RE, Haugen E, Wang H, Reynolds AP, Sandstrom R, Qu H, Brody J, et
al: Systematic localization of common disease-associated variation
in regulatory DNA. Science. 337:1190–1195. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smemo S, Tena JJ, Kim KH, Gamazon ER,
Sakabe NJ, Gómez-Marín C, Aneas I, Credidio FL, Sobreira DR,
Wasserman NF, et al: Obesity-associated variants within FTO form
long-range functional connections with IRX3. Nature. 507:371–375.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pasquali L, Gaulton KJ, Rodríguez-Seguí
SA, Mularoni L, Miguel-Escalada I, Akerman I, Tena JJ, Morán I,
Gómez-Marín C, van de Bunt M, et al: Pancreatic islet enhancer
clusters enriched in type 2 diabetes risk-associated variants. Nat
Genet. 46:136–143. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kassem SA, Ariel I, Thornton PS, Hussain
K, Smith V, Lindley KJ, Aynsley-Green A and Glaser B: p57(KIP2)
expression in normal islet cells and in hyperinsulinism of infancy.
Diabetes. 50:2763–2769. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Avrahami D, Li C, Yu M, Jiao Y, Zhang J,
Naji A, Ziaie S, Glaser B and Kaestner KH: Targeting the cell cycle
inhibitor p57Kip2 promotes adult human β cell replication. J Clin
Invest. 124:670–674. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Horike S, Mitsuya K, Meguro M, Kotobuki N,
Kashiwagi A, Notsu T, Schulz TC, Shirayoshi Y and Oshimura M:
Targeted disruption of the human LIT1 locus defines a putative
imprinting control element playing an essential role in
Beckwith-Wiedemann syndrome. Hum Mol Genet. 9:2075–2083. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fitzpatrick GV, Soloway PD and Higgins MJ:
Regional loss of imprinting and growth deficiency in mice with a
targeted deletion of KvDMR1. Nat Genet. 32:426–431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Arima T, Kamikihara T, Hayashida T, Kato
K, Inoue T, Shirayoshi Y, Oshimura M, Soejima H, Mukai T and Wake
N: ZAC, LIT1 (KCNQ1OT1) and p57KIP2 (CDKN1C) are in an imprinted
gene network that may play a role in Beckwith-Wiedemann syndrome.
Nucleic Acids Res. 33:2650–2660. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Colsoul B, Schraenen A, Lemaire K,
Quintens R, Van Lommel L, Segal A, Owsianik G, Talavera K, Voets T,
Margolskee RF, et al: Loss of high-frequency glucose-induced
Ca2+ oscillations in pancreatic islets correlates with
impaired glucose tolerance in Trpm5−/− mice. Proc Natl
Acad Sci USA. 107:5208–5213. 2010. View Article : Google Scholar
|
|
17
|
Brixel LR, Monteilh-Zoller MK, Ingenbrandt
CS, Fleig A, Penner R, Enklaar T, Zabel BU and Prawitt D: TRPM5
regulates glucose-stimulated insulin secretion. Pflugers Arch.
460:69–76. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kyriazis GA, Soundarapandian MM and
Tyrberg B: Sweet taste receptor signaling in beta cells mediates
fructose-induced potentiation of glucose-stimulated insulin
secretion. Proc Natl Acad Sci USA. 109:E524–E532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Krishnan K, Ma Z, Björklund A and Islam
MS: Role of transient receptor potential melastatin-like subtype 5
channel in insulin secretion from rat β-cells. Pancreas.
43:597–604. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kawaguchi H, Asai A, Ohtsuka Y, Watanabe
H, Wada T and Handa H: Purification of DNA-binding transcription
factors by their selective adsorption on the affinity latex
particles. Nucleic Acids Res. 17:6229–6240. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Inomata Y, Kawaguchi H, Hiramoto M, Wada T
and Handa H: Direct purification of multiple ATF/E4TF3 polypeptides
from HeLa cell crude nuclear extracts using DNA affinity latex
particles. Anal Biochem. 206:109–114. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wada T, Takagi T, Yamaguchi Y, Kawase H,
Hiramoto M, Ferdous A, Takayama M, Lee KA, Hurst HC and Handa H:
Copurification of casein kinase II with transcription factor
ATF/E4TF3. Nucleic Acids Res. 24:876–884. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nishio K, Masaike Y, Ikeda M, Narimatsu H,
Gokon N, Tsubouchi S, Hatakeyama M, Sakamoto S, Hanyu N, Sandhu A,
et al: Development of novel magnetic nano-carriers for
high-performance affinity purification. Colloids Surf B
Biointerfaces. 64:162–169. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Asfari M, Janjic D, Meda P, Li G, Halban
PA and Wollheim CB: Establishment of 2-mercaptoethanol-dependent
differentiated insulin-secreting cell lines. Endocrinology.
130:167–178. 1992.PubMed/NCBI
|
|
25
|
Wada T, Watanabe H, Kawaguchi H and Handa
H: DNA affinity chromatography. Methods Enzymol. 254:595–604. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dignam JD, Lebovitz RM and Roeder RG:
Accurate transcription initiation by RNA polymerase II in a soluble
extract from isolated mammalian nuclei. Nucleic Acids Res.
11:1475–1489. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shevchenko A, Wilm M, Vorm O and Mann M:
Mass spectrometric sequencing of proteins silver-stained
polyacrylamide gels. Anal Chem. 68:850–858. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hiramoto M, Maekawa N, Kuge T, Ayabe F,
Watanabe A, Masaike Y, Hatakeyama M, Handa H and Imai T:
High-performance affinity chromatography method for identification
of L-arginine interacting factors using magnetic nanobeads. Biomed
Chromatogr. 24:606–612. 2010.
|
|
29
|
Bungartz G, Land H, Scadden DT and Emerson
SG: NF-Y is necessary for hematopoietic stem cell proliferation and
survival. Blood. 119:1380–1389. 2012. View Article : Google Scholar :
|
|
30
|
Luo R, Klumpp SA, Finegold MJ and Maity
SN: Inactivation of CBF/NF-Y in postnatal liver causes
hepatocellular degeneration, lipid deposition, and endoplasmic
reticulum stress. Sci Rep. 1:1362011. View Article : Google Scholar :
|