Introduction
A large number of secreted glycoproteins involved in
organogenesis, stem cell homeostasis and tumor formation encoded by
the Wnt gene have been verified from the Hydra to the human
genome (1). The enhanced
activation of Wnt/β-catenin signaling and β-catenin nuclear
translocation have been shown to play a role in podocyte injury
in vivo and in vitro (3). The administration of puromycin to
cultured podocytes has been demonstrated to induce the nuclear
translocation of β-catenin (2).
The Wnt/β-catenin signaling pathway may also be regulated by
transforming growth factor-β (TGF-β) and adriamycin (ADR) in
vitro (3,4). Increased podocyte Wnt/β-catenin
signaling has also been observed in podocytes in murine models of
diabetic nephropathy and focal segmental glomerulosclerosis (FSGS)
(3,5). These results indicate a high
consistency in the activation of Wnt/β-catenin signaling in
podocytes in response to various types of injury and various
diseases. However, the potential mechanisms involved remain poorly
understood.
MicroRNAs (miRNAs or miRs) are a class of small
non-coding RNAs which play indispensible roles in the regulation of
gene expression through translational repression or transcript
degradation (6). Recently,
studies have indicated that miRNAs play a key role in kidney
diseases. miR-93 has been shown to facilitate glomerular injury
through the activation of vascular endothelial growth factor
(7). By targeting Bcl-2, miR-195
aggravates podocyte apoptosis (8). The downregulation of miR-30 has also
been shown to promote podocyte injury (9). Studies have demonstrated that
miR-192 accelerates collagen formation in glomerular mesangial
cells in models of diabetic nephropathy (10) and promotes TGF-β/Smad3-induced
tubulointerstitial fibrosis (11). The loss of Dicer in podocytes has
been shown to lead to the development of proteinuria and
glomerulosclerosis (12). These
studies indicate that miRNAs play indispensable roles in the
development of glomerular diseases, particularly
podocyte-associated disorders. However, the underlying mechanisms
have not yet been fully delineated.
The miR-135 family is highly conserved among mammals
and consists of 2 members, miR-135a and miR-135b. It has been
reported that miR-135a and miR-135b function as oncogenes and play
prominent roles in the development of various types of cancer,
including the pathogenesis of colorectal cancer (13), a role in the promotion of
paclitaxel resistance in non-small cell lung cancer (14) and in the facilitation of growth
and invasion in colorectal cancer (15). However, other studies have
demonstrated that miR-135a is a tumor suppressor gene that inhibits
cell proliferation in renal cancer (16) and selectively kills malignant
glioma cells (17). Additionally,
miR-135a determines the size of the midbrain during its development
(18) and promotes renal fibrosis
in diabetic nephropathy (19).
Despite these findings, the exact function of the two miR-135
family members remains largely unknown, particularly their function
in podocyte injury-associated renal diseases.
In the present study, we aimed to determine the
roles and mechanisms of action of miR-135a and miR-135b in podocyte
injury, and to elucidate the mechanisms underlying podocyte injury.
We found that miR-135a and miR-135b were overexpressed in patients
with FSGS and in models of podocyte injury, and that the ectopic
expression of these miRNAs promoted podocyte injury by activating
Wnt/β-catenin signaling through the suppression of glycogen
synthase kinase 3β (GSK3β) expression. Our findings demonstrate
that miR-135a and miR-135b play an important role in podocyte
injury. Our findings may provide new insight into the understanding
of the molecular mechanisms underlying podocyte injury, which may
be crucial for the development of novel therapeutic agents for the
treatment of podocytopathy.
Materials and methods
Ethics statement
Approval for human subject research in the present
study was granted by the Ethics Committee of the First Affiliated
Hospital of Chongqing Medical University, Chongqing, China. Each
patient enrolled in this study provided written informed consent.
For the animal experiments, all surgical procedures performed on
mice were carried out under sodium pentobarbital anesthesia (5
mg/100 g). The animals were treated according to the
recommendations listed in the Guide for the Care and Use of
Laboratory Animals of the National Institutes of Health (NIH). The
experimental procedures were approved by the Animal Experimental
Ethics Committee of Chongqing Medical University.
Animal experiments
BALB/c mice (6–7 weeks old) were purchased from the
Animal Center Management, Chongqing Medical University. The mouse
models of podocyte injury were created by the administration of an
intravenous injection of ADR (10.5 mg/kg; Sigma, St. Louis, MO,
USA), as previously described (20,21). The mice in the control group
received an equivalent volume of normal saline (NS). The mice were
sacrificed by cervical vertebra dislocation on days 4, 7, 11, 15
and 20 following the administration of ADR or NS. The glomeruli
were isolated from the kidneys using a sieving technique as
previously described (22). Mice
were anesthetized and the kidneys were removed, minced into 1
mm3 sections and digested in collagenase (1 mg/ml
collagenase A, 100 U/ml deoxyribonuclease I in HBSS) at 37°C for 30
min with gentle agitation. The collagenase-digested tissue was
gently pressed through a 100-µm cell strainer using a
flattened pestle and the cell strainer was then washed with 5 ml of
HBSS. The filtered cells were passed through a new cell strainer
without pressing and the cell strainer washed with 5 ml of HBSS.
The cell suspension was then centrifuged at 200 × g for 5 min. The
supernatant was discarded and the cell pellet was resuspended in 2
ml of HBSS. Finally, glomeruli were gathered by centrifugation (at
200 × g for 5 min). They were then stored in liquid nitrogen until
further analysis.
Human samples
Three patients diagnosed as having FSGS by a renal
biopsy at the Department of Nephrology, the First Affiliated
Hospital, Chongqing Medical University were enrolled in the present
study. The control group included 3 patients with kidney rupture
following incidents of violence or traffic accidents. The glomeruli
were isolated from the renal tissues using the aforementioned
sieving technique and were then snap-frozen in liquid nitrogen for
RNA extraction and molecular analysis. Podocyte damage was
determined by analysis of the pathological sections and albuminuria
(27).
Cell culture and treatment
293T cells were cultured in Dulbecco's modified
Eagle's medium (DMEM) supplemented with 10% fetal bovine serum
(Gibco-BRL, Carlsbad, CA, USA) at 37°C with 5% CO2. The
conditionally immortalized mouse podocyte cell line 5 (MPC5) was
cultured as previously described (23). MPC-5 cells were cultured at 33°C
(10 U/ml of interferon-γ) with 5% CO2 and were
differentiated at 37°C (without interferon-γ) with 5%
CO2 in RPMI Dutch-modified medium (Invitrogen, Carlsbad,
CA, USA) supplemented with 10% v/v fetal calf serum. Depending on
the exact experimental setup, the cultured cells were treated with
different doses of adriamycin and transfected with microRNA mimics
or plasmid using Lipofectamine 3000 reagent (Invitrogen, Grand
Island, NY, USA). NT group cells were treated without any
agents.
Bioinformatics analysis
The miR-135 candidate genes were predicted using the
following websites: miRwalk (www.umm.uni-heidelberg.de/apps/zmf/mirwalk),
miRanda (www.microrna.org/microrna/home.do) and TargetScan
(www.targetscan.org/).
Constructs and luciferase assays
The 3′ untranslated region (3′UTR) of GSK3β was
obtained by the amplification of genomic DNA using a forward primer
(5′-GCTCCTCACCAAATTCAAGC-3′) and a reverse primer
(5′-TGAAGTGAGACTGCCAACATG-3′). The PCR product was cloned into the
pcDNA3.1-Luc reporter vector and verified by sequencing. For the
wild type construct, the seed sequence was wild type. For the
mutant construct, the seed sequence was mutated. miR-135 inhibitor
(Inh-135) was from RiboBio Co., Ltd., Guangzhou, China. The
microRNA control inhibitor (Inh-NC) was from RiboBio Co., Ltd.
Luciferase assays were conducted according to a method described in
a previous study (24). In brief,
the 293T and MPC5 cells were cultured in 24-well plates and
transfected with 500 ng of the repoter vector, 50 ng of pRL-CMV and
50 nM of miR-135a/miR-135b mimics or the miRNA mimic negative
control (RiboBio Co., Ltd.) using Lipofectamine 3000 reagent
(Invitrogen, Grand Island, NY, USA). The firefly and Renilla
luciferase activities were measured at 36 h after transfection
using a dual-luciferase assay system (Promega, Madison, WI, USA)
according to the manufacturer's instructions. Dishevelled regulates
GSK3β activity. This results in the stabilization and accumulation
of cytosolic β-catenin, a GSK-3β substrate, which is when
phosphorylated by GSK-3β, and is targeted for ubiquitin-mediated
proteolysis. β-catenin then translocates to the nucleus, where in
complexes with members of the TCF family of transcription
regulators, activates the transcription of TCF-responsive genes.
This TCF-reporter plasmid enables the quantification of Wnt
signaling in cells transfected with these constructs. LiCl can
repress GSK-3β expression, this results in the stabilization and
accumulation of cytosolic β-catenin.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
RNA was extracted using TRIzol reagent (Ambion,
Austin, TX, USA) according to the manufacturer's instructions.
RT-qPCR for the detection of miR-135a and miR-135b was performed
using miR-135a- and miR-135b-specific PCR primers (RiboBio Co.,
Ltd.) with the RevertAid First Strand cDNA Synthesis kit
(Fermentas, Burlington, ON, Canada) and SYBR Premix Ex Taq™ II
(Takara, Dalian, China) according to the manufacturers'
instructions. For the mRNA quantification of protein-encoding
genes, the RNA was reverse transcribed with a random primer and the
mRNA levels were determined by RT-qPCR. All the sequences of the
primers using in RT-qPCR primers are presented in Table I.
 | Table ISequences of the primers used in
RT-qPCR. |
Table I
Sequences of the primers used in
RT-qPCR.
| Name | GenBank accession
no. | Sense | Antisense | Product size
(bp) |
|---|
| Desmin | NM_010043 |
TGCAGCCACTCTAGCTCGTA |
GACATGTCCATCTCCACCTG | 150 |
| Snail | NM_011427 |
AGCCCAACTATAGCGAGCTG |
CCAGGAGAGAGTCCCAGATG | 150 |
| Nephrin | NM_019459 |
CGCCACCTGGTCGTAGATTC |
ACCCCTCTATGATGAAGTAC | 94 |
| E-cadherin | NM_009864 |
GGCCAGTGCATCCTTCAAAT |
CAGGTCTCCTCATGGCTTTG | 148 |
| WT1 | NM_144783 |
CCGTGTGGTTCTCACTCTCA |
CAAATGACCTCCCAGCTTGA | 151 |
| 18s | NR_003278 |
GTAACCCGTTGAACCCCATT |
CCATCCAATCGGTAGTAGCG | 151 |
Protein extraction and western blot
analysis
Total protein was extracted from the MPC5 cells
using RIPA lysis buffer (Beyotime, Jiangsu, China) following the
manufacturer's instructions. Western blot analysis was performed as
previously described (25). The
antibodies used were the following: rabbit anti-desmin (1:1,000;
sc-14026), rabbit anti-snail (1:1,000; sc-28199) (all from Santa
Cruz Biotechnology, Inc., Santa Cruz, CA, USA), rabbit anti-nephrin
(1:2,000; ab136894; Abcam, Cambridge, MA, USA), rabbit anti-Wilms
tumor 1 (WT1; 1:500; sc-192), rabbit anti-E-cadherin (1:1,000l;
sc-7870) (both from Santa Cruz Biotechnology, Inc.), rabbit
anti-GSK3β (1:2,000; ab18893; Abcam), rabbit anti-β-catenin
[activated and unphosphorylated, 1:2,000; Netherlands Cancer
Institute (NKI), Amsterdam, The Netherlands], mouse anti-β-tubulin
(1:3,000; sc-80011), goat anti-rabbit IgG-HRP (1:3,000; sc-2004)
and goat anti-mouse IgG-HRP (1:3,000; sc-2005) (all from Santa Cruz
Biotechnology, Inc.). For the quantitative analysis of the western
blots, the protein band intensities were quantified using Image J
software and β-actin was used for normalization.
F-actin cytoskeleton staining and
immunofluorescence staining
F-actin was stained using rhodamine-labeled
phalloidin (diluted in PBS containing 5% BSA, 1:1,000; Sigma)
according to the manufacturer's instructions. Immunostaining for
the determination of the location of β-catenin was performed as
previously described (26). The
following antibodies were used: rabbit anti-β-catenin (1:1,000;
ab32572; Abcam) and goat anti-rabbit IgG-CFL 488 (1:2,000;
sc-362262; Santa Cruz Biotechnology, Inc.).
Statistical analysis
All the results are presented as the means ± SD. The
t-test was employed to determine whether 2 sets of data (groups)
differed significantly from each other using GraphPad Prism 5
software (GraphPad Software, Inc., La Jolla, CA, USA). Values of
P<0.05 and P<0.01 were considered to indicate statistically
significant and highly statistically significant differences,
respectively. All experiments were performed at least 3 times.
Results
miR-135a and miR-135b are upregulated in
injured podocytes
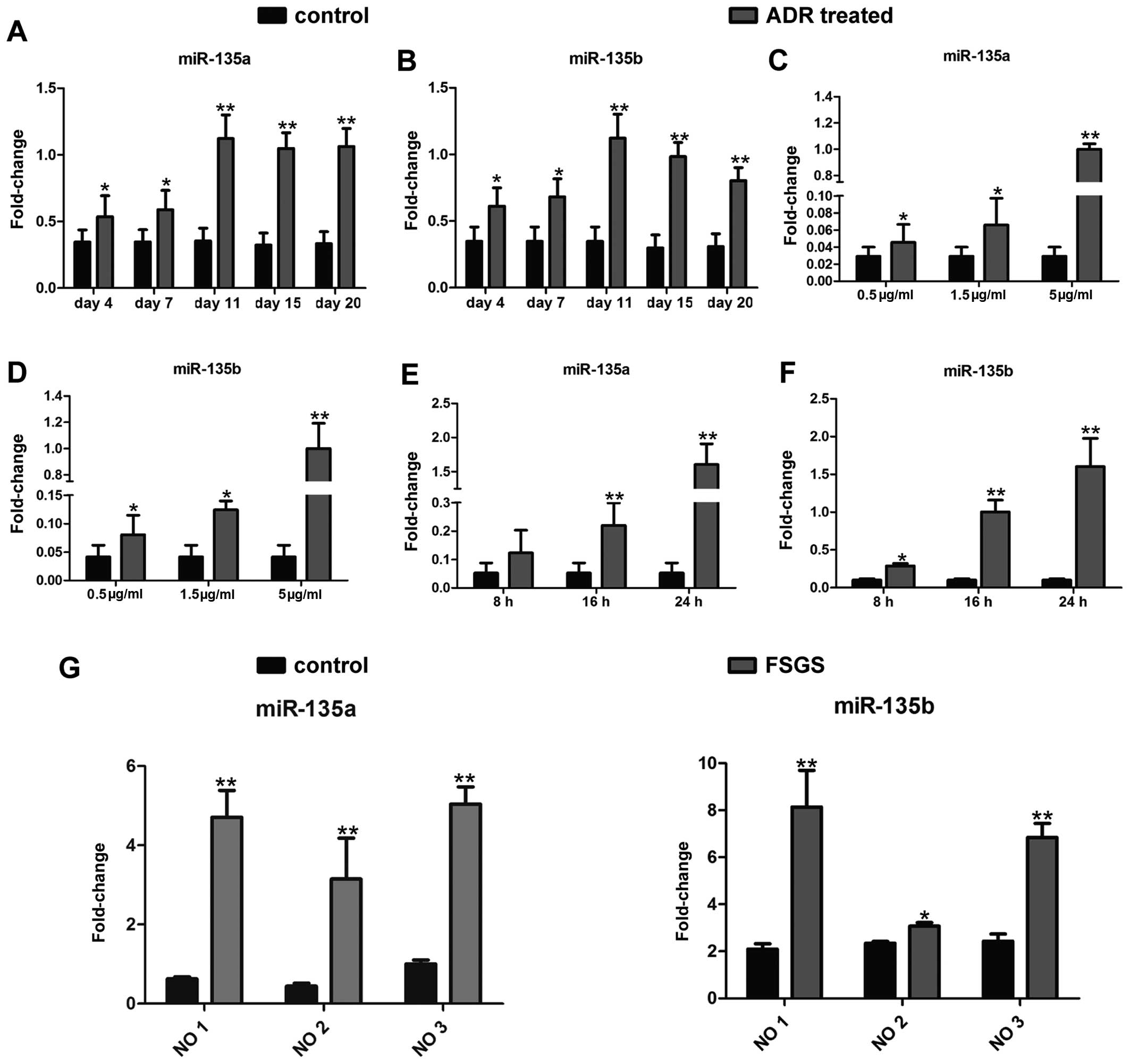
To investigate the role of miR-135a and miR-135b in
podocyte injury, firstly, a mouse model of ADR-induced nephropathy,
a widely recognized model of podocyte injury (20,21), was established by an intravenous
injection of ADR (10.5 mg/kg). The expression levels of both
members of the miR-135 family were detected in the isolated
glomeruli by RT-qPCR as indicated in Fig. 1A and B. The expression levels of
miR-135a and miR-135b increased significantly as early as day 4
following the administration of ADR. Eleven days following the
administration of ADR, the miR-135a and miR-135b expression levels
increased by approximately 3-fold. Furthermore, the expression
levels of both miRNAs were measured in an in vitro model of
ADR-induced podocyte injury. As demonstrated in Fig. 1C–F, treatment with ADR enhanced
the expression of miR-135a and miR-135b in the cultured mouse
podocyte cell line (MPC5) in a dose-dependent (Fig. 1C and D; 24 h after ADR treatment)
and time-dependent manner (Fig. 1E
and F; 5 µg/ml ADR treatment). Additionally, in order to
further investigate the clinical significance of miR-135a and
miR-135b in podocyte injury, glomeruli were isolated from 3
patients with FSGS whose podocytes were severely damaged and from 3
patients with kidney rupture caused by incidents of violence or
traffic accidents. The renal samples from patients with kidney
rupture were used as controls. miR-135a and miR-135b expression in
the glomeruli from these patients was examined by RT-qPCR. As
expected, miR-135a and miR-135b were highly expressed in the
glomeruli of the patients with FSGS compared with those of the
controls (Fig. 1G). Thus, the
aforementioned results clearly indicate that miR-135a and miR-135b
are upregulated in injured podocytes.
Overexpression of miR-135 family members
leads to podocyte injury
To determine whether the increase in the expression
of miR-135a and miR-135b is functionally significant in podocyte
injury, miR-135a and miR-135b mimics and miRNA mimic negative
control (miR-NC) were transfected into the MPC5 cells. Forty-eight
hours after transfection, the protein expression levels of markers
related to podocyte injury were measured by western blot analysis.
The expression of desmin and snail, two markers of podocyte injury
(3,28), significantly increased in the
cells transfected with miR-135a and miR-135b mimics compared with
the miR-NC group (Fig. 2A and B).
However, the expression of the podocyte functional markers, WT1,
nephrin and E-cadherin, was suppressed by transfection with
miR-135a and miR-135b mimics (Fig. 2A
and B). Furthermore, the mRNA expression of these genes was
determined by RT-qPCR, the results of which were consistent with
those of the protein expression (Fig.
2C).
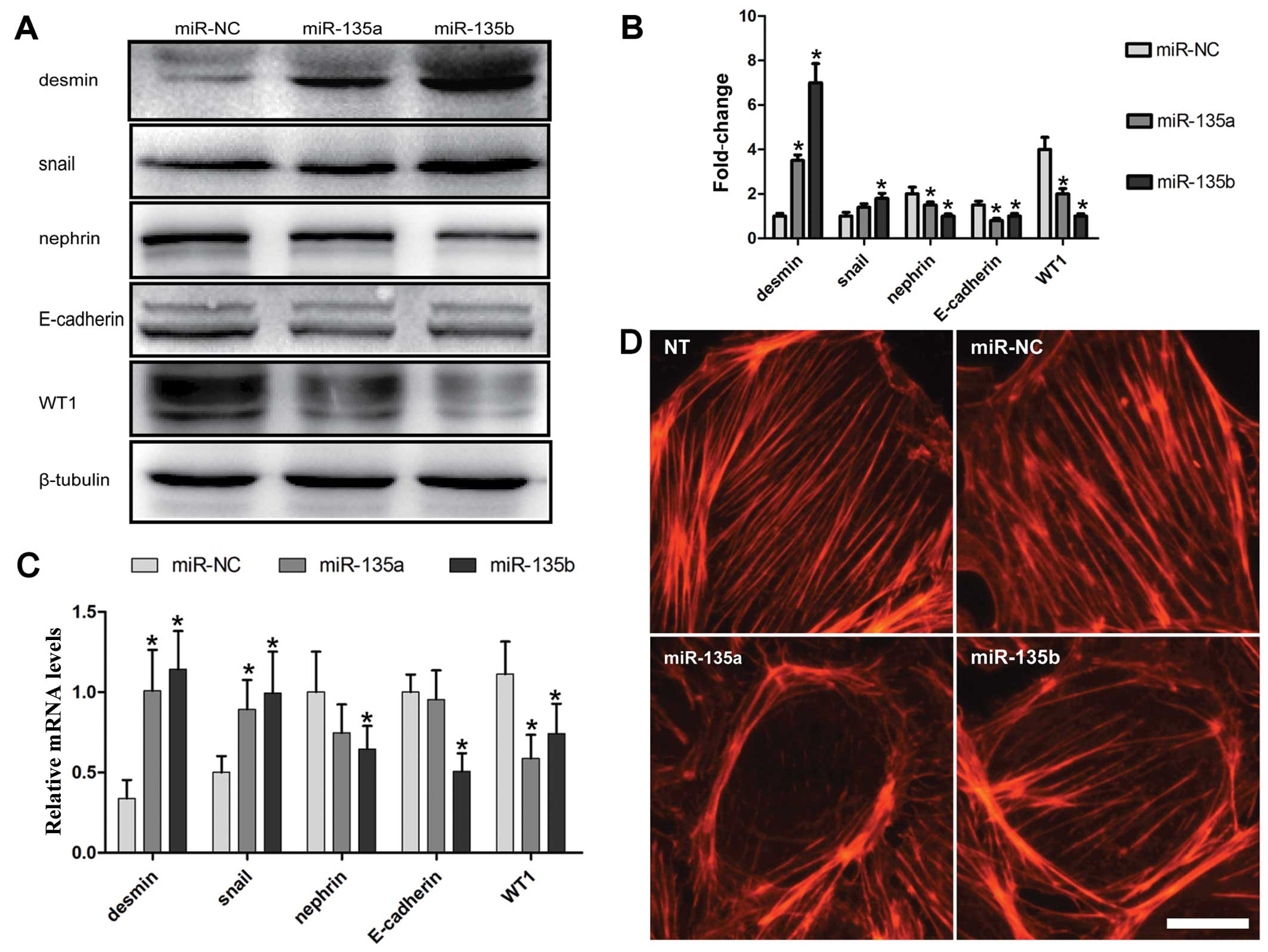 | Figure 2miR-135a and miR-135b promote
podocyte damage and cytoskeletal injury. (A) Western blot analysis
was applied to determine the changes in the expression levels of
desmin, snail, nephrin, E-cadherin and WT1 at 48 h following
transfection of the cultured mouse podocyte cell line 5 (MPC5) with
miR-135a and miR-135b mimics of the negative control (miR-NC). (B)
Quantitative analysis of the results of western blot analysis. The
intensities of the protein bands were quantified and normalized to
β-tubulin. (C) Quantification by RT-qPCR of the mRNA expression of
desmin, E-cadherin, nephrin, WT1 and snail following transfection
of the MPC5 cells with miR-135a and miR-135b mimics or miR-NC. (D)
Effects of miR-135a and miR-135b on cytoskeletal stabilization in
podocytes. Scale bar, 10 µm. NT, not treated.
*P<0.05 and **P<0.01 indicate
statistically significant and highly statistically significant
differences, respectively, compared with the miR-NC group. |
Any factor that can cause the disorder or
rearrangement of the podocyte cytoskeleton may result in podocyte
dysfunction and injury (29).
Therefore, in this study, the effects of miR-135a and miR-135b on
the podocyte cytoskeleton were examined. The podocyte cytoskeleton
was examined by means of fluorescein-conjugated phalloidin staining
at 36 h following transfection with miR-135a and miR-135b mimics or
miR-NC. The results revealed that miR-135a and miR-135b promoted
the rearrangement of stress fibers in podocytes, causing them to
localize around the cytomembrane compared with the
control-transfected and untreated groups (NT) (Fig. 2D).
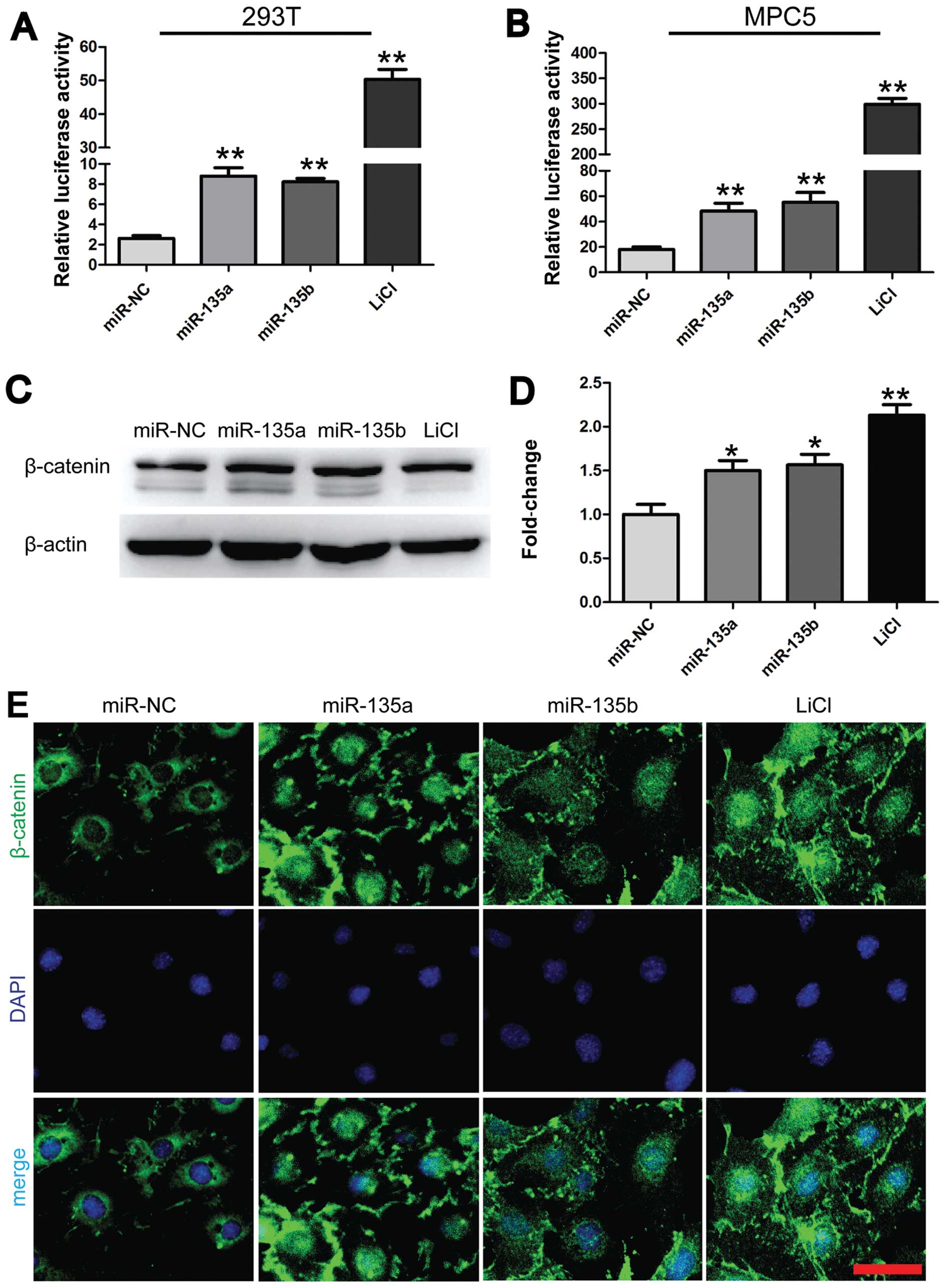
miR-135a and miR-135b promote
Wnt/β-catenin signaling in MPC5 cells
As is well known, desmin and snail are potential
downstream targets of the Wnt/β-catenin signaling pathway (30). As shown in Fig. 2A, desmin and snail were
overexpressed in the injured podocytes. Therefore, we hypothesized
that miR-135a and miR-135b mediate podocyte injury through
Wnt/β-catenin signaling. To verify this hypothesis, we first
examined the effects of miR-135a and miR-135b on T-cell factor
(TCF) transcriptional activity. The 293T and MPC5 cells were
co-transfected with miR-135a and miR-135b mimics or miR-NC and
luciferase reporter constructs harboring 3 optimal TCF binding
sites (TOPflash). The lithium chloride (LiCl; 10 mM) group was used
as a positive control. The results revealed that transfection with
either miR-135a or miR-135b significantly increased TOPflash
reporter activity by approximately 3-fold compared with the
negative control (Fig. 3A and B).
Subsequently, the effects of miR-135a and miR-135b on the
expression levels of activated and unphosphorylated β-catenin were
examined in the MPC5 cells by western blot analysis. In accordance
with previous findings (13), we
found that the overexpression of miR-135a and miR-135b increased
the levels of activated and unphosphorylated β-catenin by
approximately 1.5-fold compared with the negative control (Fig. 3C and D). This effect was very
similar to that observed in the group treated with LiCl (10 mM).
Furthermore, immunofluorescence staining for β-catenin was
conducted to examine the effects of miR-135a and miR-135b on the
subcellular location of β-catenin in the MPC5 cells. The nuclear
translocation of β-catenin occurred following transfection of the
cells with miR-135a and miR-135b mimics and was comparable to that
in the group treated with LiCl (10 mM) (Fig. 3E). By contrast, β-catenin was
predominantly localized at the cell-cell adhesion sites and around
the nucleus in the MPC5 cells in the control group (Fig. 3E). Thus, the aforementioned
findings indicate that miR-135a and miR-135b activate Wnt/β-catenin
signaling.
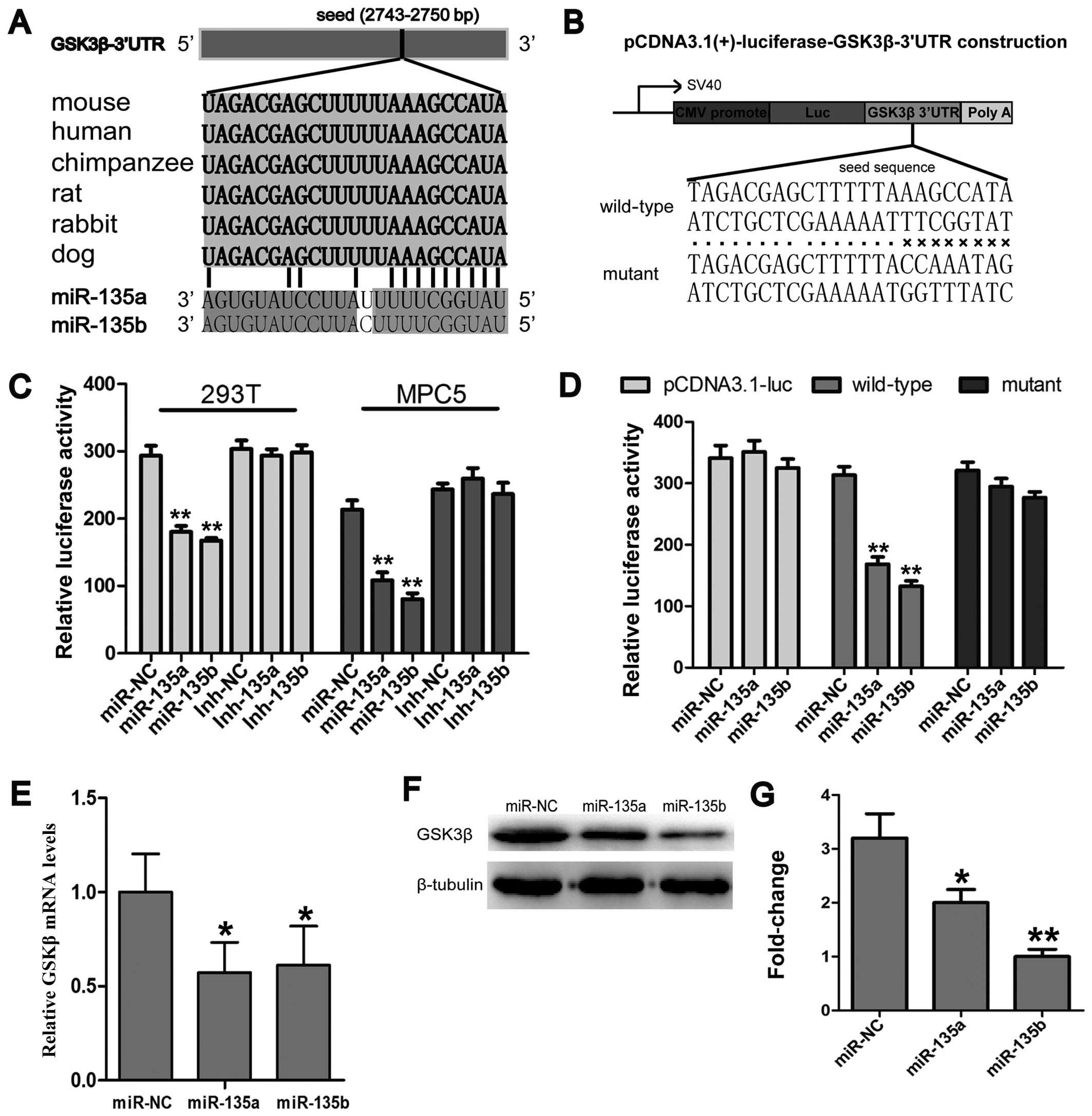
miR-135a and miR-135b target the 3′UTR of
GSK3β and inhibit GSK3β expression
In order to further elucidate the molecular
mechanisms underlying the miR-135a- and miR-135b-induced activation
of Wnt/β-catenin signaling, the targets of miR-135a and miR-135b
involved in Wnt/β-catenin signaling were identified by online miRNA
target prediction programmes, including miRwalk, miRanda and
TargetScan release 6.2. The analysis revealed that an
evolutionarily conserved region in the 3′UTR of GSK3β mRNA had a
perfect complementary sequence (5′-UGUUUACA-3′) to the seed
sequence (3′-ACAAAUGU-5′) of the miR-135 family (Fig. 4A). In order ro further validate
the results of bioinformatics analysis, reporter vectors bearing
the GSK-3β 3′UTR were constructed (Fig. 4B). Luciferase reporter assays were
conducted to determine whether miR-135a and miR-135b directly
target the 3′UTR of GSK3β. The results revealed that luciferase
expression was significantly suppressed in the groups transfected
with the miR-135a and miR-135b mimics compared with the miR-NC
group, while mutations of the miR-135a and miR-135b binding sites
abrogated the suppression of luciferase expression (Fig. 4C and D), which suggests that
miR-135a and miR-135b directly bind to the 3′UTR of GSK3β.
Furthermore, the effects of miR-135a and miR-135b on GSK3β
expression at the protein and mRNA level were also determined. The
results revealed that miR-135a and miR-135b inhibited the
expression of GSK3β at both the protein and mRNA level (Fig. 4E–G). Taken together, these results
indicate that both members of the miR-135 family activate
Wnt/β-catenin signaling through the inhibition of GSK3β
expression.
Discussion
It is widely accepted that glomerular visceral
epithelial cells, also known as podocytes, are a critical target of
injury in a wide variety of kidney diseases (31). The present study primarily
demonstrated that miR-135a and miR-135b were significantly
upregulated in the glomeruli of mice and in cultured podocytes
treated with ADR, and that miR-135a and miR-135b are potential
diagnostic biomarkers for glomerular and podocyte injury. A number
of signaling pathways have been demonstrated to be involved in
podocyte injury and apoptosis (32); however, the molecular mechanisms
underlying podocyte injury remain poorly understood. The present
study demonstrates that the ectopic expression of miR-135a and
miR-135b promotes Wnt/β-catenin signaling and results in podocyte
injury, which suggests that miR-135a and miR-135b are novel
detrimental factors for podocytes and cause podocyte injury through
the activation of Wnt/β-catenin signaling. Thus, we propose a novel
mechanism underlying podocyte damage and suggest that podocyte
injury is mediated by Wnt/β-catenin signaling.
Thus far, a great deal of understanding on podocyte
biology, particularly in the regulation of the actin cytoskeleton,
a main determinant of the complex architecture for podocyte
function, that facilitates the identification of therapeutic
targets for podocyte-associated diseases, has been achieved. For
example, cyclosporine A (33) and
rituximab (34) have been shown
to protect podocytes from injury and dysfunction through the
stabilization of the podocyte cytoskeleton. In the present study,
we found that miR-135a and miR-135b are the detrimental factors for
the stabilization of the podocyte cytoskeleton and that they
disrupt the location of the actin skeleton. Thus, this suggests
that the disruption of the cytoskeleton arrangement may be the main
mechanism underlying podocyte dysfunction induced by miR-135a and
miR-135b.
miRNAs are a class of short non-coding RNAs. In this
study, using bioinformatics software, we found that GSK3β is a
potential target gene of miR-135a and miR-135b. As is well known
(35), GSK3β is an inhibitor of
Wnt/β-catenin signaling and, thus, it was speculated that miR-135a
and miR-135b may promote Wnt/β-catenin signaling through the
suppression of GSK3β. Using luciferase reporter assays, western
blot analysis and RT-qPCR, GSK3β was found to be a target gene of
miR-135a and miR-135b. Adenomatous polyposis coli (APC), another
inhibitor of Wnt/β catenin signaling, has been identified as a
target of miR-135a and miR-135b in humans (13,14). However, the target sites of APC
for miR-135a and miR-135b are not conserved between humans and
mice. Therefore, we did not find that miR-135a and miR-135b
inhibits APC expression in mice (data not shown), which is
consistent with the results of the study by Nagel et al
(13).
Despite a better understanding of the
pathophysiological mechanisms of podocytes in kidney disease, only
a few findings may be translated into more effective treatments due
to difficulties in identifying functionally relevant
podocyte-associated molecules. Hence, the need for the
identification of the molecular mechanisms responsible for podocyte
injury is imperative. The miR-135 family has 2 members, miR-135a
and miR-135b. In the present study, we found that these 2 members
were upregulated in models of podocyte injury and in glomeruli from
patients with FSGS, indicating that there may be a common signal
promoting high levels of miR-135a and miR-135b expression. The
identification of this signal would decisively contribute to the
development of novel drugs for the treatement of renal diseases
associated with podocyte injury.
In conclusion, in the present study, we primarily
found that miR-135a and miR-135b were upregulated in models of
podocyte injury and in glomeruli from patients with FSGS. To the
best of our knowledge, the present study is the first to
demonstrate that miR-135a and miR-135b activate Wnt/β-catenin
signaling through the inhibition of GSK3β in podocytes, suggesting
a novel mechanism underlying miR-135a- and miR-135b-mediated
podocyte injury. Our findings also suggests that miR-135a and
miR-135b are potential diagnostic biomarkers for podocyte injury
and that their inhibition may be a potential clinical therapeutic
strategy for the treatment of podocytopathy. Although the findings
of the current study demonstrate that miR-135a and miR-135b are
upregulated by ADR in podocytes, the exact mechanisms through which
ADR regulates miR-135a and miR-135b expression in podocytes remain
unknown; thus, further research is required on this matter. In the
future, our intention is to generate the ectopic expression and
knockdown of these miRNAs in a mouse model in order to investigate
the functions of miR-135a and miR-135b and the association between
miR-135 family members and podocyte injury in vivo.
Acknowledgments
We wish to acknowledge support from the National
Basic Research Program of China (grant no. 2011CB944002) awarded to
Q.Z. The present study was also supported by a grant from the
National Natural Science Foundation of China (no. 31271563) awarded
to Q.Z.
References
|
1
|
Nelson WJ and Nusse R: Convergence of Wnt,
beta-catenin, and cadherin pathways. Science. 303:1483–1487. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Teixeira VP, Blattner SM, Li M, Anders HJ,
Cohen CD, Edenhofer I, Calvaresi N, Merkle M, Rastaldi MP and
Kretzler M: Functional consequences of integrin-linked kinase
activation in podocyte damage. Kidney Int. 67:514–523. 2005.
View Article : Google Scholar
|
|
3
|
Dai C, Stolz DB, Kiss LP, Monga SP,
Holzman LB and Liu Y: Wnt/beta-catenin signaling promotes podocyte
dysfunction and albuminuria. J Am Soc Nephrol. 20:1997–2008. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cai J, Schleidt S, Pelta-Heller J,
Hutchings D, Cannarsa G and Iacovitti L: BMP and TGF-β pathway
mediators are critical upstream regulators of Wnt signaling during
midbrain dopamine differentiation in human pluripotent stem cells.
Dev Biol. 376:62–73. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kato H, Gruenwald A, Suh JH, Miner JH,
Barisoni-Thomas L, Taketo MM, Faul C, Millar SE, Holzman LB and
Susztak K: Wnt/β-catenin pathway in podocytes integrates cell
adhesion, differentiation, and survival. J Biol Chem.
286:26003–26015. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jackson RJ and Standart N: How do
microRNAs regulate gene expression? Sci STKE. 2007:re12007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Long J, Wang Y, Wang W, Chang BH and
Danesh FR: Identification of microRNA-93 as a novel regulator of
vascular endothelial growth factor in hyperglycemic conditions. J
Biol Chem. 285:23457–23465. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen YQ, Wang XX, Yao XM, Zhang DL, Yang
XF, Tian SF and Wang NS: MicroRNA-195 promotes apoptosis in mouse
podocytes via enhanced caspase activity driven by BCL2
insufficiency. Am J Nephrol. 34:549–559. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu J, Zheng C, Fan Y, Zeng C, Chen Z, Qin
W, Zhang C, Zhang W, Wang X, Zhu X, et al: Downregulation of
microRNA-30 facilitates podocyte injury and is prevented by
glucocorticoids. J Am Soc Nephrol. 25:92–104. 2014. View Article : Google Scholar :
|
|
10
|
Kato M, Zhang J, Wang M, Lanting L, Yuan
H, Rossi JJ and Natarajan R: MicroRNA-192 in diabetic kidney
glomeruli and its function in TGF-beta-induced collagen expression
via inhibition of E-box repressors. Proc Natl Acad Sci USA.
104:3432–3437. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chung AC, Huang XR, Meng X and Lan HY:
miR-192 mediates TGF-beta/Smad3-driven renal fibrosis. J Am Soc
Nephrol. 21:1317–1325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi S, Yu L, Chiu C, Sun Y, Chen J,
Khitrov G, Merkenschlager M, Holzman LB, Zhang W, Mundel P and
Bottinger EP: Podocyte-selective deletion of dicer induces
proteinuria and glomerulosclerosis. J Am Soc Nephrol. 19:2159–2169.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nagel R, le Sage C, Diosdado B, van der
Waal M, Oude Vrielink JA, Bolijn A, Meijer GA and Agami R:
Regulation of the adenomatous polyposis coli gene by the miR-135
family in colorectal cancer. Cancer Res. 68:5795–5802. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Holleman A, Chung I, Olsen RR, Kwak B,
Mizokami A, Saijo N, Parissenti A, Duan Z, Voest EE and Zetter BR:
miR-135a contributes to paclitaxel resistance in tumor cells both
in vitro and in vivo. Oncogene. 30:4386–4398. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou W, Li X, Liu F, Xiao Z, He M, Shen S
and Liu S: MiR-135a promotes growth and invasion of colorectal
cancer via metastasis suppressor 1 in vitro. Acta Biochim Biophys
Sin (Shanghai). 44:838–846. 2012. View Article : Google Scholar
|
|
16
|
Yamada Y, Hidaka H, Seki N, Yoshino H,
Yamasaki T, Itesako T, Nakagawa M and Enokida H: Tumor-suppressive
microRNA-135a inhibits cancer cell proliferation by targeting the
c-MYC oncogene in renal cell carcinoma. Cancer Sci. 104:304–312.
2013. View Article : Google Scholar
|
|
17
|
Wu S, Lin Y, Xu D, Chen J, Shu M, Zhou Y,
Zhu W, Su X, Zhou Y, Qiu P, et al Yan G: MiR-135a functions as a
selective killer of malignant glioma. Oncogene. 31:3866–3874. 2012.
View Article : Google Scholar
|
|
18
|
Anderegg A, Lin HP, Chen JA, Caronia-Brown
G, Cherepanova N, Yun B, Joksimovic M, Rock J, Harfe BD, Johnson R
and Awatramani R: An Lmx1b-miR135a2 regulatory circuit modulates
Wnt1/Wnt signaling and determines the size of the midbrain
dopaminergic progenitor pool. PLoS Genet. 9:e10039732013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He F, Peng F, Xia X, Zhao C, Luo Q, Guan
W, Li Z, Yu X and Huang F: MiR-135a promotes renal fibrosis in
diabetic nephropathy by regulating TRPC1. Diabetologia.
57:1726–1736. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fogo AB: Animal models of FSGS: Lessons
for pathogenesis and treatment. Semin Nephrol. 23:161–171. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Wang YP, Tay YC and Harris DC:
Progressive adriamycin nephropathy in mice: Sequence of histologic
and immunohistochemical events. Kidney Int. 58:1797–1804. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ni L, Saleem M and Mathieson PW: Podocyte
culture: Tricks of the trade. Nephrology (Carlton). 17:525–531.
2012. View Article : Google Scholar
|
|
23
|
Mundel P, Reiser J, Zúñiga Mejía Borja A,
Pavenstädt H, Davidson GR, Kriz W and Zeller R: Rearrangements of
the cytoskeleton and cell contacts induce process formation during
differentiation of conditionally immortalized mouse podocyte cell
lines. Exp Cell Res. 236:248–258. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kolfschoten IG, van Leeuwen B, Berns K,
Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM and Agami
R: A genetic screen identifies PITX1 as a suppressor of RAS
activity and tumorigenicity. Cell. 121:849–858. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bae GU, Lee JR, Kim BG, Han JW, Leem YE,
Lee HJ, Ho SM, Hahn MJ and Kang JS: Cdo interacts with APPL1 and
activates Akt in myoblast differentiation. Mol Biol Cell.
21:2399–2411. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bae GU, Kim BG, Lee HJ, Oh JE, Lee SJ,
Zhang W, Krauss RS and Kang JS: Cdo binds Abl to promote
p38alpha/beta mitogen-activated protein kinase activity and
myogenic differentiation. Mol Cell Biol. 29:4130–4143. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pollak MR: Inherited podocytopathies: FSGS
and nephrotic syndrome from a genetic viewpoint. J Am Soc Nephrol.
13:3016–3023. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang Y, Jarad G, Tripathi P, Pan M,
Cunningham J, Martin DR, Liapis H, Miner JH and Chen F: Activation
of NFAT signaling in podocytes causes glomerulosclerosis. J Am Soc
Nephrol. 21:1657–1666. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Welsh GI and Saleem MA: The podocyte
cytoskeleton - key to a functioning glomerulus in health and
disease. Nat Rev Nephrol. 8:14–21. 2012. View Article : Google Scholar
|
|
30
|
Wang D, Dai C, Li Y and Liu Y: Canonical
Wnt/β-catenin signaling mediates transforming growth
factor-β1-driven podocyte injury and proteinuria. Kidney Int.
80:1159–1169. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Asanuma K: The role of podocyte injury in
chronic kidney disease. Nihon Rinsho Meneki Gakkai Kaishi.
38:26–36. 2015.In Japanese. View Article : Google Scholar
|
|
32
|
Kato H and Susztak K: Repair problems in
podocytes: Wnt, Notch, and glomerulosclerosis. Semin Nephrol.
32:350–356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Faul C, Donnelly M, Merscher-Gomez S,
Chang YH, Franz S, Delfgaauw J, Chang JM, Choi HY, Campbell KN, Kim
K, Reiser J and Mundel P: The actin cytoskeleton of kidney
podocytes is a direct target of the antiproteinuric effect of
cyclosporine A. Nat Med. 14:931–938. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fornoni A, Sageshima J, Wei C,
Merscher-Gomez S, Aguillon-Prada R, Jauregui AN, Li J, Mattiazzi A,
Ciancio G, Chen L, et al: Rituximab targets podocytes in recurrent
focal segmental glomerulosclerosis. Sci Translat Med.
3:85ra462011.
|
|
35
|
Nakamura T, Hamada F, Ishidate T, Anai K,
Kawahara K, Toyoshima K and Akiyama T: Axin, an inhibitor of the
Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and
APC and reduces the beta-catenin level. Genes Cells. 3:395–403.
1998. View Article : Google Scholar : PubMed/NCBI
|