Introduction
Lower back pain (LBP) is a common orthopedic problem
in aged populations, which has a significant socio-economic effect
on the lives of patients (1,2).
Intervertebral disc (IVD) degeneration (IDD) is considered to be
the primary cause of LBP (3). IDD
is characterized by histomorphological changes, including nucleus
pulposus (NP) fibrosis, annulus fibrosus (AF) disorganization and
cartilage endplate calcification (4-6).
It has been found to be associated with several factors, including
mechanical loading, aging, infection, nutrition and genetics
(4). Mechanical stress is one of
the important factors affecting IDD (7,8).
One of the major events in IDD is the decrease of
functional IVD cells (9-11). Programmed cell death (PCD) has an
important role in the decrease of IVD cells. Apoptosis, also known
as type I PCD, relies on the activation of caspase and is featured
by DNA damage, karyopyknosis, cell shrinkage and the formation of
apoptotic bodies. It has been demonstrated that apoptosis occurs in
NP cells and that mechanical stress is an important factor
affecting disc cell apoptosis and IDD (12,13).
At present, autophagy has been widely acknowledged
to be involved in several physiological and pathological processes,
including maintaining cell internal homeostasis, survival,
proliferation and differentiation (14). The major role of autophagy is
considered to be an adaptive response of the cells to survival when
faced with different stimuli, including aging, and oxidative or
mechanical stress (15-17). Autophagy maintains cellular
homeostasis by degrading and recycling damaged and dysfunctional
organelles, proteins and other macromolecules. Various studies have
demonstrated that both apoptosis and autophagy exist in IDD
(15,18-20). The conversion and coordination
between apoptosis and autophagy remains to be fully elucidated and
warrants further investigation.
Cyclic mechanical tension (CMT), a type of
mechanical loading, has been shown to influence IVD cell apoptosis
and autophagy (15,19). These two events share the same
stimuli and regulatory proteins, but have different threshold
responses to the same stimulus. The association between apoptosis
and autophagy in the response of NP cells under cyclic tension has
not been clarified.
In the present study, the Flexercell Tension system
(Flexcell International Corporation, Hillsborough, NC, USA), a
widely used system that exerts cyclic tension on cultured cells
in vitro, was used for cyclic tension loading. The study
illustrates how apoptosis interacts with autophagy in NP cells
under cyclic tension, providing novel insight into the functions of
mechanical loading in the pathophysiological significance of
IDD.
Materials and methods
Ethics
The present study was approved by the Ethics
Committee of Xinqiao Hospital (Chongqing, China). All protocols
were subject to the standards set forth in the 8th edition of the
Guide for the Care and Use of Laboratory Animals published by the
National Academy of Sciences (Washington, DC, USA).
Antibodies
Rabbit monoclonal anti-rat microtubule-associated
protein light chain 3 (LC3)B (cat. no. 3868, 1:1,000 dilution),
Beclin-1 (cat. no. 3495, 1:1,000 dilution) and cleaved caspase-3
(cat. no. 9661, 1:1,000 dilution) were purchased from Cell
Signaling Technology, Inc. (Danvers, MA, USA). Mouse monoclonal
anti-rat glyceraldehyde3-phosphate dehydrogenase (GAPDH; cat. no.
sc-47724, 1:1,000 dilution) was purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, CA, USA). Goat anti-rabbit
horseradish peroxidase (HRP)-conjugated secondary antibody (cat.
no.. ZB2301, 1:400 dilution) were purchased from ZSGB-BIO (Beijing,
China).
Isolation and culture of rat NP
cells
A total of 60 male Sprague-Dawley rats (age, 10-12
weeks; weight, 200-240 g; specific pathogen-free grade) were
supplied by the Laboratory Animal Research Center of Daping
Hospital (Chongqing, China). Animals were all housed at 20-24°C
under a 12-h light/dark cycle, and provided with food and water
ad libitum. The caudal IVDs of 10-12-week-old male
Sprague-Dawley rats were aseptically excised immediately following
sacrifice. The NP tissues were carefully separated from the discs
under aseptic conditions and digested in 0.2% type II collagenase
(Merck KGaA, Darmstadt, Germany) at 37°C for 2 h to release the NP
cells. The cell suspension was passed through a 70-µm cell
strainer and centrifuged at 200 × g for 5 min at room temperature.
The supernatant was removed and the cells were suspended in
DMEM/F12 medium (Thermo Fisher Scientific, Inc., Waltham, MA, USA)
with 10% fetal bovine serum (Thermo Fisher Scientific, Inc.) and 1%
penicillin-streptomycin (Thermo Fisher Scientific, Inc.). The NP
cells were cultured in 5% CO2 at 37°C. The medium was
replaced every 3 days. When the cells grew to 90% confluence, they
were digested using 0.25% trypsin and subcultured in culture
flasks. The cells of passages 2-5 were used in the subsequent
experiments.
Application of CMT on NP cells
The NP cells were seeded on a 6-well BioFlex™ plate
(Flexcell International Corporation, McKeesport, PA, USA) in
DMEM/F12 medium containing 10% fetal bovine serum. On reaching 80%
confluence, the NP cells were stretched with CMT (20% elongation)
at a frequency of 1 Hz for 2, 4, 6, 12, 24 or 48 h using a FX-5000T
Flexercell Tension Plus system (Flexcell International Corporation)
at 37°C and 5% CO2. Cells cultured in the same plates
under the same conditions were used as the control. Following
cyclic tension loading, the morphology of NP cells was observed
using a phase contrast microscope (Olympus Corporation, Tokyo,
Japan).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from the NP cell using TRIzol
reagent (Takara Bio, Inc., Otsu, Japan). The RNA quality and
quantity were determined using a NanoDrop ND-1000 spectrophotometer
(Thermo Fisher Scientific, Inc). RNA (1 µg) was reverse
transcribed using a PrimeScript RT Reagent kit (Takara Bio). The
PrimeScript RT Reagent kit (Takara Bio, Inc.) was used for RT-qPCR
according to the manufacturer's instructions. RT-qPCR was performed
using a ViiA™ 7 Real-Time PCR system using SYBR® Premix
Ex Taq™ II (Takara Bio, Inc.), according to the manufacturer's
instructions. The 20-µl reaction volume was amplified under
the following conditions: Initial heat activation for 30 sec at
95°C, followed by 40 cycles of 5 sec at 95°C for template
denaturation and 30 sec at 60°C for annealing and extension. GAPDH
was used as an internal reference gene. Data were analyzed using
the 2−ΔΔCq method (21). The primers of the genes
investigated in the present study are listed in Table I.
 | Table IPrimer sequences used in reverse
transcription-quantitative polymerase chain reaction analysis. |
Table I
Primer sequences used in reverse
transcription-quantitative polymerase chain reaction analysis.
| Target gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| LC3 |
TCACGGACGGAAGCCAACACA |
AATCCTTCCCGACCGCACCAT |
| Beclin-1 |
ACAGCGGACAATTTGGCACGAT |
TGGAGCAACAACACTGTCTGGC |
| Caspase-3 |
GCACACGGGACTTGGAAAGCAT |
AGCGATGACTCAGCACCTCCAT |
| GAPDH |
CCAGCAAGAGCACAAGAGGAAGAG |
GGTCTACATGGCAACTGTGAGGAG |
Western blot analysis
Total proteins were extracted from NP cells using
radioimmunoprecipitation assay (RIPA) lysis buffer (Thermo Fisher
Scientific, Inc.). The concentration of protein was quantified
using the bicinchoninic acid method (Beyotime Institute of
Biotechnology). Protein samples (50 µg) were separated using
10% SDS-PAGE and transferred onto polyvinylidene difluoride
membranes (EMD Millipore, Billerica, MA, USA). The membranes were
blocked with 5% milk in TBST for 1 h at 37°C to block nonspecific
protein binding. The membranes were then incubated at 4°C overnight
with primary antibodies, following which the membranes were washed
with TBST and incubated at 37°C for 1 h with HRP-conjugated
secondary antibodies. Immunolabeling was detected using ECL
reagents (Thermo Fisher Scientific, Inc.). The optical density of
the bands was measured using Image J software (National Institutes
of Health).
Transmission electron microscopy
(TEM)
Following stretching, the NP cells were isolated
with trypsin and fixed in 2% glutaraldehyde at 4°C for 2 days. The
cells were then treated with 1% osmium tetroxide (Sigma-Aldrich;
Merck KGaA) for 30 min at room temperature, dehydrated in an
ethanol gradient and embedded with Epon 812 (Shell Chemical Co.,
Houston, TX, USA). The fixed cells were then sliced into ultrathin
sections (60 nm) and observed under a Tecnai-10 TEM (Philips
Healthcare, Amsterdam, The Netherlands).
Apoptotic incidence measurement
The apoptotic incidence was detected using the
Annexin V-FITC apoptosis detection kit I (BD Biosciences, San Jose,
CA, USA) according to the manufacturer's instructions. Following
stretching, the NP cells were isolated with trypsin and washed
twice with PBS. The NP cells were then resuspended in 500 µl
binding buffer at a concentration of 1×106 cells/ml. PE
Annexin V solution (5 µl) and 7-amino-actinomycin D (5
µl) were added to the cells at room temperature for 15 min
in the dark. Finally, the cells were analyzed on a flow cytometer
(Beckman Coulter, Inc., Brea, CA, USA).
Autophagy incidence measurement
Following stretching, the NP cells were isolated
with trypsin and washed twice with PBS, resuspended in PBS with
0.05 mM monodansylcadaverine (Merck KGaA) at a concentration of
1×106 cells/ml, and incubated at 37°C and 5%
CO2 for 20 min in the dark. Following incubation, the
cells were washed three times with PBS. The mean fluorescence
intensity (MFI) was analyzed on a flow cytometer (Beckman Coulter,
Inc.).
Reactive oxygen species (ROS)
measurement
Following stretching, the NP cells were isolated
with trypsin and washed twice with PBS. The cells were then
resuspended using serum-free DMEM/F12 medium with 25 µM
2',7'-dichlorodi-hydrofluorescein diacetate (Merck KGaA) at a
concentration of 1×106 cells/ml, and incubated at 37°C
and 5% CO2 for 20 min. Following incubation, the cells
were washed three times with serum-free DMEM/F12 medium. The MFI
was analyzed on a flow cytometer (Beckman Coulter, Inc.).
Statistical analysis
Data are presented as the mean ± standard deviation.
All experiments were repeated at least three times. For comparisons
between two independent groups, Student's t-test was used. For
comparisons among three or more groups, one-way ANOVA and least
significant difference multiple comparisons were used. The RT-qPCR
data were analyzed using Kruskal-Wallis nonparametric analysis and
Mann-Whitney U post hoc tests. Data were analyzed and displayed
using SPSS version 22.0 (International Business Machines Corp.,
Armonk, NY, USA) and GraphPad Prism 6 software (GraphPad Software
Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
Morphology of NP cells following CMT
application
Following cyclic tension loading, the morphology of
NP cells was observed using a phase contrast microscope. The NP
cells that adhered to the outer part of the membrane became clearly
spindle-shaped and exhibited a circular arrangement perpendicular
to the axial tension (Fig. 1).
Only a few cells were detached from the membrane surface.
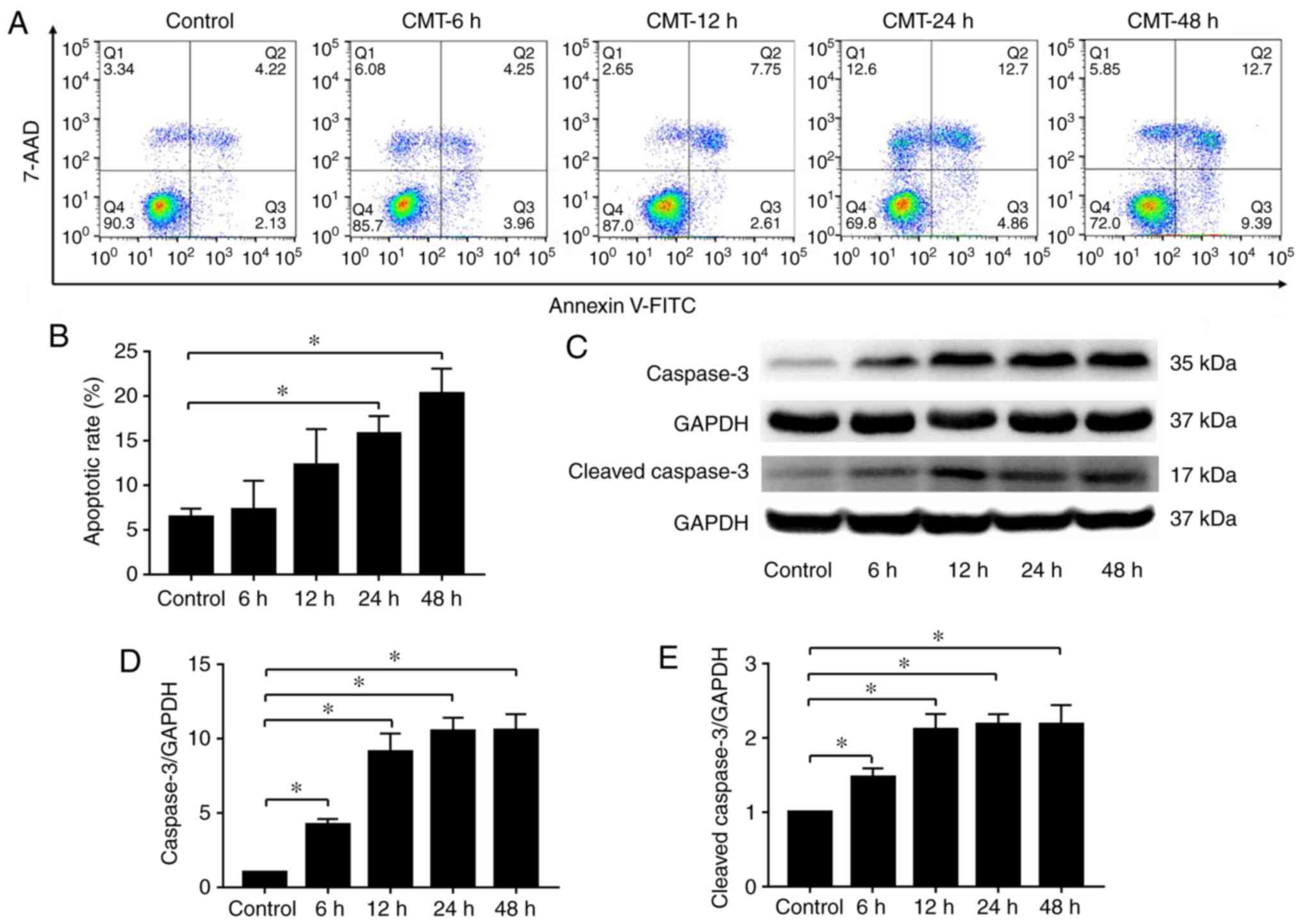
Effect of CMT on apoptosis of rat NP
cells
Following 20% CMT at a frequency of 1 Hz for 6, 12,
24 and 48 h, the apoptotic rate of the NPs was detected by flow
cytometry with Annexin V and 7AAD double-labeling. The results
demonstrated a time-dependent increase in the apoptotic rate of rat
NP cells subjected to CMT (Fig.
2A), and significant differences in the apoptotic rate were
observed after 24 h (Fig. 2B).
Consistent with the results of flow cytometry, the western blot
results showed that the levels of caspase-3 and cleaved caspase-3
were also increased in the NP cells subjected to CMT (Fig. 2C-E).
Effect of CMT on the autophagy of rat NP
cells
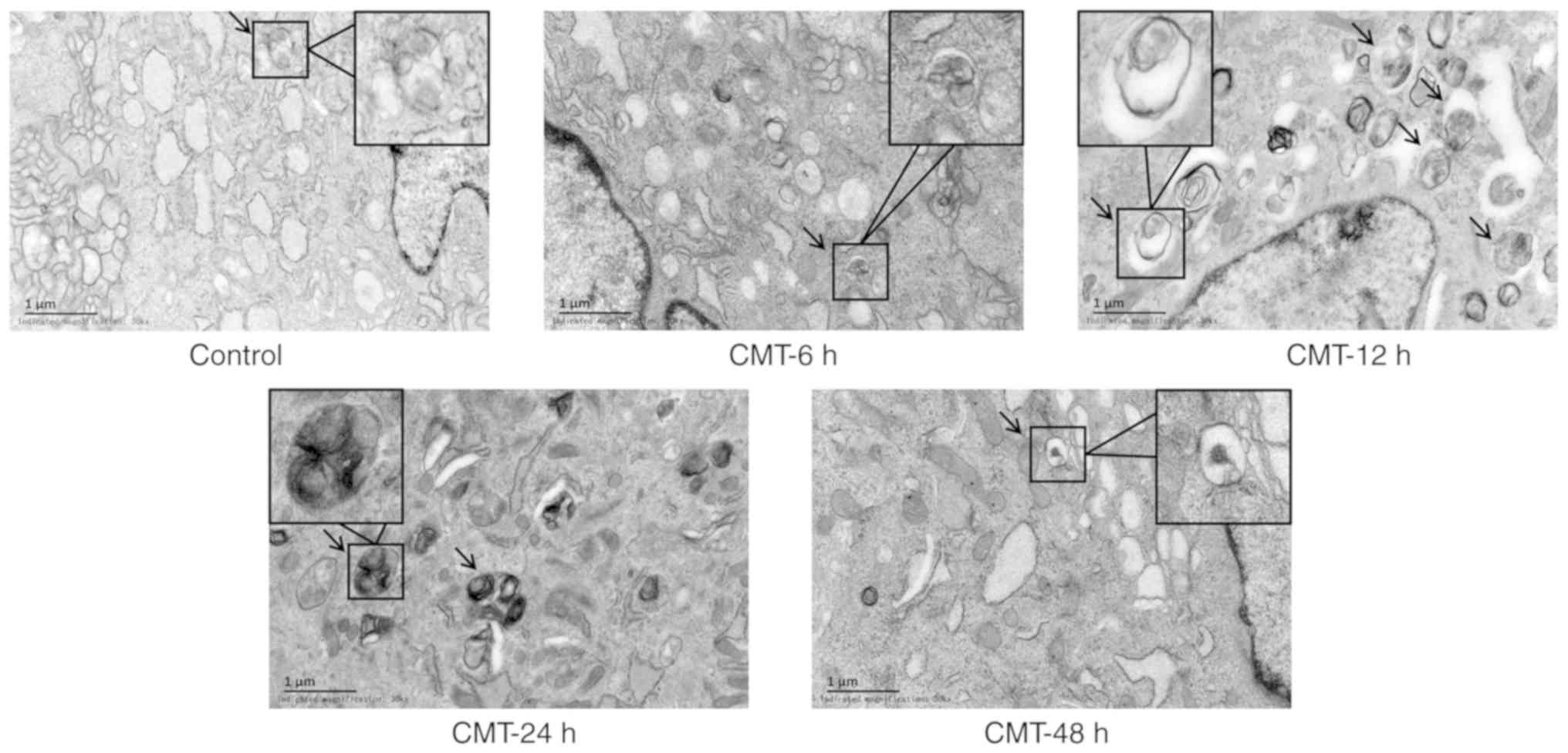
Examining the ultrastructure of cells by TEM is the
gold standard for the detection of autophagy (14). The ultrastructure of the NP cells
was observed under TEM. The autophagosomes, manifested as
double-membrane vacuolar structures containing organelles and parts
of cytoplasm, were observed in NP cells. The number of
autophagosomes in the CMT groups was higher than the number in the
static control groups (Fig. 3).
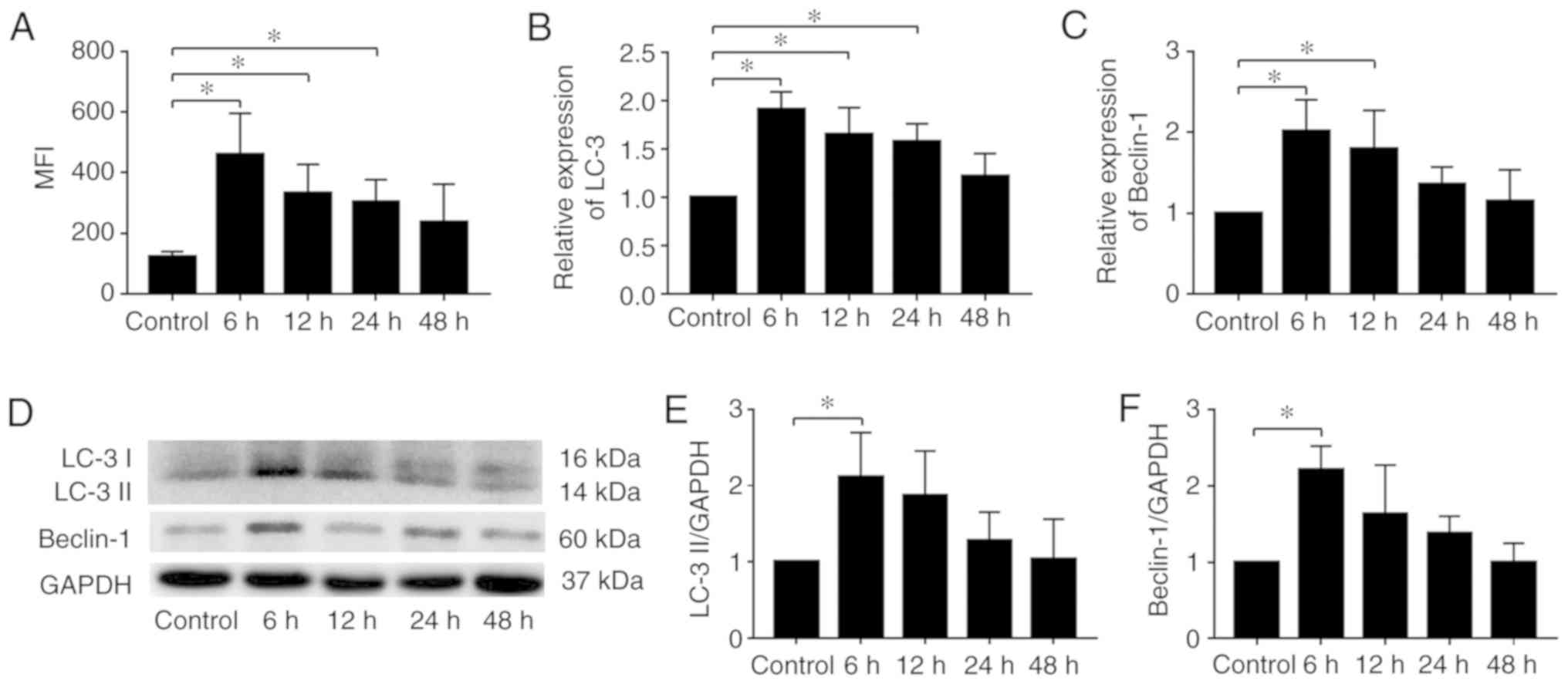
MDC staining and a flow cytometry quantitative assay were used to
quantify the expression of autophagy. The increase in MFI in each
CMT group was measured. There was a time-dependent decrease in the
MFI of NP cells subjected to 20% CMT for 6, 12, 24 and 48 h
(Fig. 4A). The mRNA expression
levels of LC3 and Beclin-1 showed a similar trend (Fig. 4B and C). The protein expression of
LC3 II and Beclin-1 was also increased significantly in the NP
cells subjected to CMT for 6 h (Fig.
4D-F), and exhibited a time-dependent decrease after 6 h. These
results indicated the upregulation of autophagy in NP cells
subjected to CMT. Furthermore, the protein expression of LC3 II was
measured at 2, 4 and 6 h to determine when autophagy was the most
marked. The results showed that autophagy increased within 6 h and
reached a maximum level between 4 and 6 h (Fig. S1).
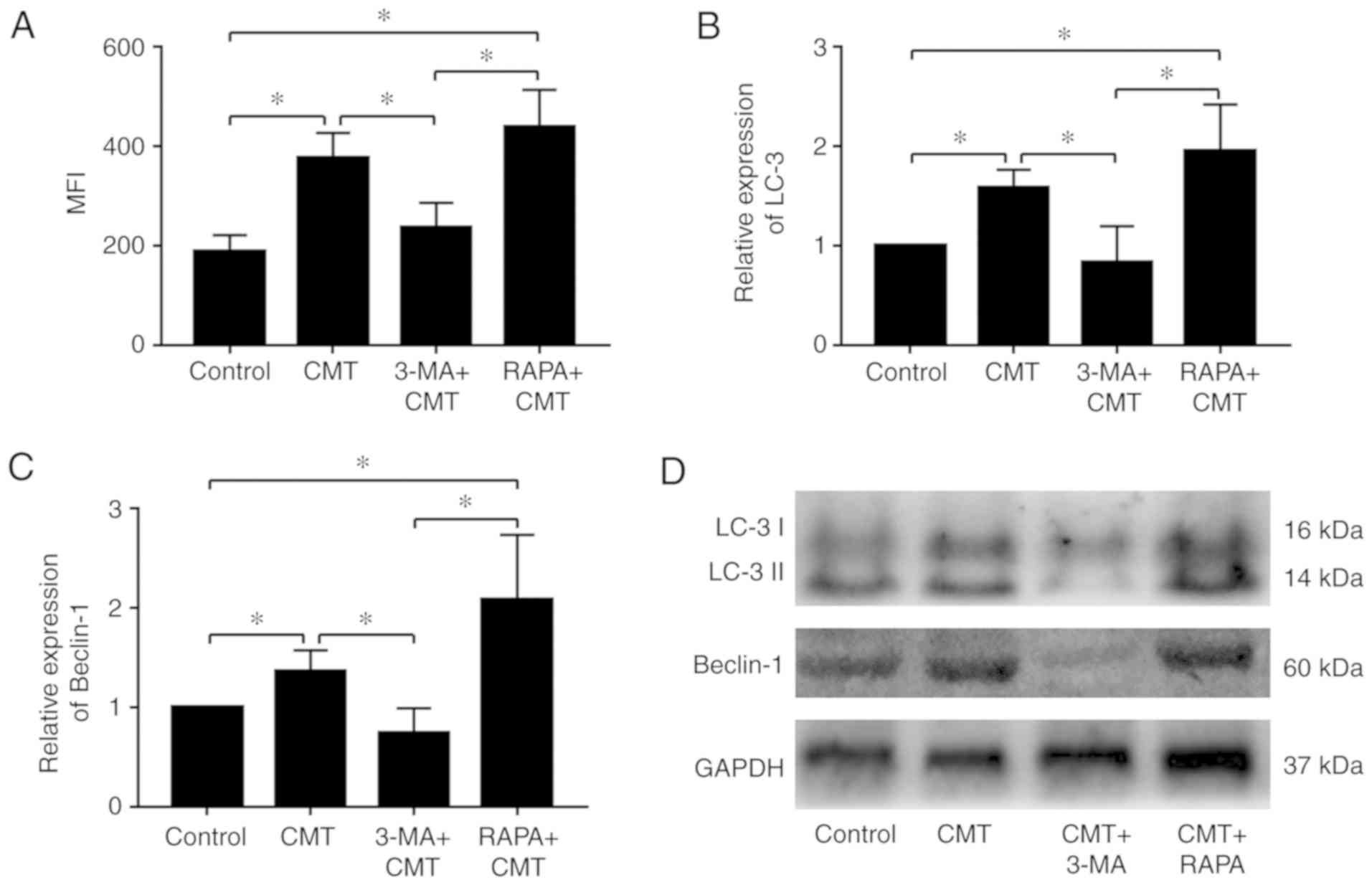
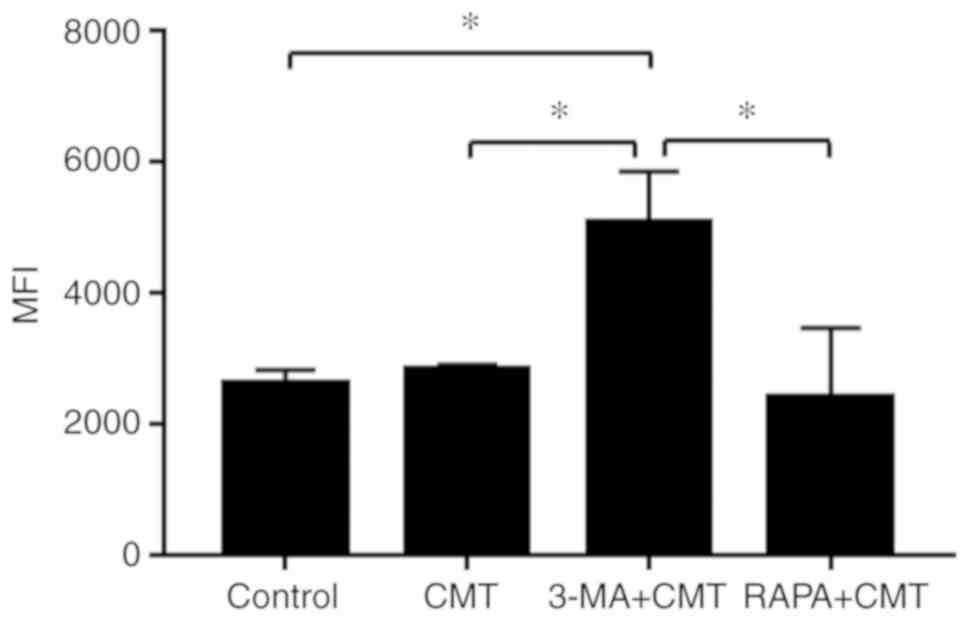
Autophagy is modulated by rapamycin and
3-MA in NP cells subjected to CMT
The NP cells were administered with rapamycin (1
µM) or 3-MA (100 nM) and placed in an incubator for 1 h,
following which the NP cells were loaded with 20% CMT for 24 h. The
results of MDC staining and quantitative fluorescence measured by
flow cytometry indicated that the activities of autophagy in cells
following CMT loading were significantly reduced by 3-MA
(P<0.05; Fig. 5A). The
difference in autophagic activities between the CMT and the
combined CMT and rapamycin was not significant (P=0.39). The mRNA
expression levels of LC3 and Beclin-1 were decreased by 3-MA in NP
cells subjected to 20% CMT for 24 h, but did not differ
significantly following combined CMT and rapamycin treatment
(Fig. 5B). Consistent with the
results of the RT-qPCR analysis, the western blot analysis results
confirmed the downregulation of LC3 and Beclin-1 by 3-MA at the
protein level (Fig. 5C and
D).
ROS levels in NP cells subjected to 20%
CMT are increased by 3-MA
Our previous study indicated that CMT had minimal
effect on the production of ROS in NP cells (22). In the present study, the ROS
levels of NP cells pretreated with 3-MA or rapamycin following CMT
were measured at 24 h. The results showed that the level of ROS in
NP cells was increased significantly in the combined CMT and 3-MA
group (P<0.05), with no significant differences observed among
other groups (Fig. 6). In order
to further illustrate the effect of autophagy on ROS production,
ROS levels were measured at 6 h when autophagy was the highest
(Fig. S2). The results showed
that the level of ROS in NP cells increased significantly with 3-MA
treatment at 6 h (P<0.05).
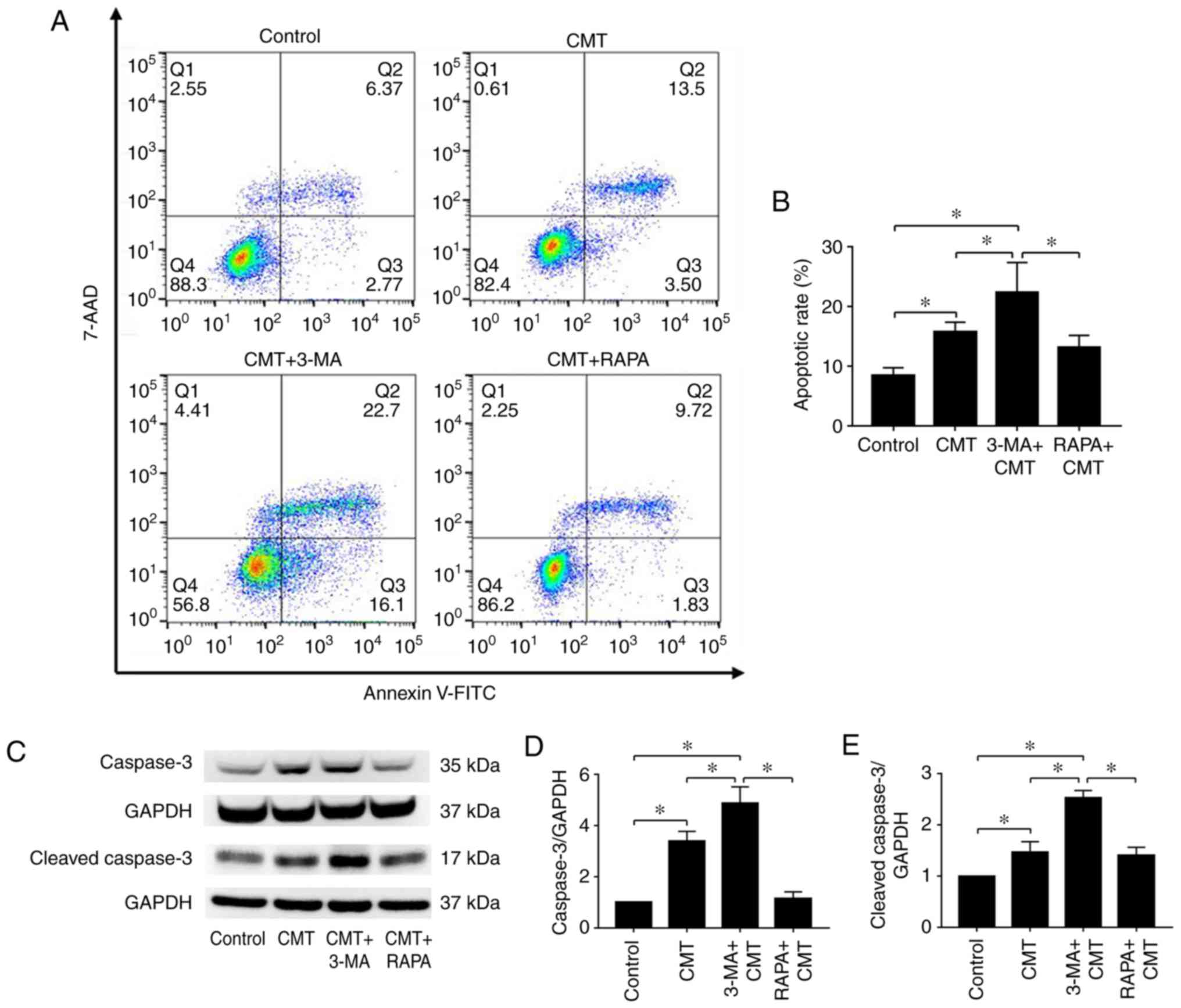
Apoptosis of NP cells loaded with 20% CMT
is induced by 3-MA
The NP cells were loaded with 20% CMT for 24 h
following incubation with 1 µM rapamycin or 100 nM 3-MA for
1 h. The apoptotic rates of NP cells were then measured (Fig. 7A). Compared with the CMT group,
the apoptotic rate of NP cells was significantly increased in the
combined CMT and 3-MA group (P<0.05), with no clear differences
observed in the combined CMT and rapamycin group (Fig. 7B). Changes in the protein
expression of caspase-3 and cleaved caspase-3 were similar
(Fig. 7C-E). According to the
results, autophagy was highest following application of CMT for 6
h. To verify the inhibitory effect of autophagy on apoptosis, the
apoptotic rates and expression of cleaved caspase-3 were measured
in NP cells treated with 3-MA and CMT at 6 h (Fig. S3). The apoptotic rate of NP cells
and the protein expression of cleaved caspase-3 were significantly
increased in the combined CMT and 3-MA group at 6 h
(P<0.05).
Discussion
IVDs are basic structures in the human body that are
important in spinal movement. Studies have demonstrated that the
initiation of IDD is associated with the degradation of
extracellular matrix and changes in the behavior of IVD cells,
including senescence, necrosis and apoptosis (11,23). NP cells, a major group of IVD
cells, are exposed to various mechanical stresses, including
tension (8). The loss of NP cells
has been closely correlated with the onset of IDD (9). Apoptosis, also known as type I PCD,
has been shown to be important in the decrease of NP cells
(24). Previous studies have
indicated that excessive stretching induced the apoptosis of rat AF
cells (19,25) and cartilage endplate-derived stem
cells (18). The present study
showed that 20% CMT, which is considered excessive stretch towards
NP cells, induced apoptosis in rat NP cells, with the incidence of
apoptosis increasing with the increasing duration of CMT. The
results indicated that apoptosis induced by prolonged excessive
mechanical stress in NP cells may be one of the factors causing a
decrease in the number of functional disc cells, consequently
accelerating the initiation and progression of IDD.
Autophagy is an adaptive response of cells to
survival when faced with different stimuli, including aging and
oxidative or mechanical stress. Autophagy is a conserved and
ubiquitous physiological mechanism that can degrade damaged
macromolecules and organelles to maintain cellular homeostasis
(26). The role of autophagy in
IDD has been widely investigated. Ye et al demonstrated that
autophagy existed in rat NP cells (27). Furthermore, it has been
demonstrated that autophagy is a protective mechanism against
apoptosis in podocytes and AF cells (28,29). However, the association between
autophagy and CMT-induced apoptosis in NP cells has remained
unclear.
In the present study, autophagy was observed in NP
cells and was upregulated by 20% CMT. The increasing level of
autophagy decreased in a time-dependent manner following the
application of CMT to NP cells for 6 h, and the incidence of
apoptosis in NP cells increased as the duration of CMT increased.
The results suggested that autophagy may be a protective mechanism
against apoptosis induced by CMT in NP cells. Therefore, the
association between autophagy and stretch-induced apoptosis in NP
cells was investigated. Rapamycin and 3-MA were used to regulate
autophagy in the NP cells. The results showed that autophagy was
significantly inhibited by 3-MA in NP cells subjected to CMT,
whereas rapamycin had minimal effect. The group treated with 3-MA,
an autophagy inhibitor, exhibited a significant decrease in the
incidence of autophagy following 24 h of CMT treatment, whereas an
increase in the incidence of apoptosis was noted. Rapamycin, an
autophagy promoter, had no significant effect on autophagy. In
conclusion, the inhibition of autophagy causes an increase in the
incidence of apoptosis in NP cells subjected to CMT. The results
suggested that autophagy protects against CMT-induced apoptosis in
NP cells. Furthermore, the underlying mechanism of autophagy in the
protection against CMT-induced apoptosis of NP cells warrants
further investigation.
Oxidative stress caused by excessive ROS generation
is also an essential trigger of NP cell apoptosis (30). It has been reported that ROS can
trigger autophagy in cancer cells as signaling molecules (16). In turn, autophagy may contribute
to reducing the generation of intracellular ROS and oxidative
damage by clearing oxidized macromolecules and organelles (20). Our previous study suggested that
CMT had little effect on the production of ROS in NP cells.
However, NP cells grow in a low oxygen environment in normal IVD,
and cultured NP cells in vitro grow in a high oxygen
environment (31,32). ROS was overgenerated by high
oxygen stimuli following isolation of the NP cells from the tissue
and culture in vitro (33). This may be the reason for CMT
having little effect on the production of ROS in NP cells in our
previous study. In contrast with our previous study, the present
study found that the ROS level was significantly increased in the
group in which autophagy was inhibited by 3-MA; apoptosis in NP
cells was also enhanced. In combination, the results of the present
study suggested that ROS serve an important role in CMT-induced
apoptosis, and that autophagy protects against apoptosis by
reducing the generation of intracellular ROS and oxidative damage
in NP cells. Details of the functions and mechanisms underlying the
associations among ROS, autophagy and CMT-induced apoptosis require
further investigation.
In conclusion, the present study suggested that the
effect of CMT on NP cell apoptosis is duration-dependent. In
addition, autophagy protected against CMT-induced apoptosis in NP
cells in vitro, with ROS potentially having an important
role in this process. These results suggest that maintaining the
autophagy level of disc cells is conducive to alleviating
apop-tosis caused by mechanical stress and may assist in delaying
the process of IDD. However, the cellular biological response to
mechanical stress depends on different factors, including the
duration, frequency and magnitude of mechanical stress, in addition
to the disc cell type (22,34,35), and further experiments are
warranted to clarify these issues.
Supplementary Data
Abbreviations:
|
IVD
|
intervertebral disc
|
|
IDD
|
intervertebral disc degeneration
|
|
CMT
|
cyclic mechanical tension
|
|
NP
|
nucleus pulposus
|
|
ROS
|
reactive oxygen species
|
|
LBP
|
lower back pain
|
|
AF
|
annulus fibrosus
|
|
PCD
|
programmed cell death
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
MFI
|
mean fluorescence intensity
|
|
MDC
|
monodansylcadaverine
|
|
7-AAD
|
7-amino-actinomycin D
|
Acknowledgments
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant nos. 81672215, 81572186,
81702182 and 81874028).
Availability of data and material
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MY contributed to the conception and design of the
study, collection, analysis and interpretation of data and writing
of the manuscript. CF contributed to the conception and design of
the study, and writing and revision of the manuscript. YZ
contributed to the collection of data and provision of study
material. CL contributed to the collection of data and provision of
study material; BL contributed to the collection, analysis and
interpretation of data; QZ contributed to the collection, analysis
and interpretation of data; BH contributed to the conception and
design and final approval of the manuscript; YZ contributed to the
conception and design of the study, final approval of the version
to be published and financial and administrative support. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethics Committee of
Xinqiao Hospital. All protocols described in current study were in
accordance with the standards set forth in the eighth edition of
Guide for the Care and Use of Laboratory Animals published by the
National Academy of Sciences.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Haldeman S, Kopansky-Giles D, Hurwitz EL,
Hoy D, Mark Erwin W, Dagenais S, Kawchuk G, Strömqvist B and Walsh
N: Advancements in the management of spine disorders. Best Pract
Res Clin Rheumatol. 26:263–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meucci RD, Fassa AG and Faria NM:
Prevalence of chronic low back pain: Systematic review. Rev Saude
Publica. 49:2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cheung KM: The relationship between disc
degeneration, low back pain, and human pain genetics. Spine J.
10:958–960. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Roberts S, Evans H, Trivedi J and Menage
J: Histology and pathology of the human intervertebral disc. J Bone
Joint Surg Am. 88(Suppl 2): S10–S14. 2006.
|
|
5
|
Kadow T, Sowa G, Vo N and Kang JD:
Molecular basis of inter-vertebral disc degeneration and
herniations: What are the important translational questions? Clin
Orthop Relat Res. 473:1903–1912. 2015. View Article : Google Scholar
|
|
6
|
Kepler CK, Ponnappan RK, Tannoury CA,
Risbud MV and Anderson DG: The molecular basis of intervertebral
disc degeneration. Spine J. 13:318–330. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Setton LA and Chen J: Mechanobiology of
the intervertebral disc and relevance to disc degeneration. J Bone
Joint Surg Am. 88(Suppl 2): S52–S57. 2006.
|
|
8
|
Vergroesen PP, Kingma I, Emanuel KS,
Hoogendoorn RJ, Welting TJ, van Royen BJ, van Dieën JH and Smit TH:
Mechanics and biology in intervertebral disc degeneration: A
vicious circle. Osteoarthritis Cartilage. 23:1057–1070. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding F, Shao ZW and Xiong LM: Cell death
in intervertebral disc degeneration. Apoptosis. 18:777–785. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang F, Zhao X, Shen H and Zhang C:
Molecular mechanisms of cell death in intervertebral disc
degeneration (Review). Int J Mol Med. 37:1439–1448. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao CQ, Jiang LS and Dai LY: Programmed
cell death in inter-vertebral disc degeneration. Apoptosis.
11:2079–2088. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ariga K, Yonenobu K, Nakase T, Hosono N,
Okuda S, Meng W, Tamura Y and Yoshikawa H: Mechanical
stress-induced apoptosis of endplate chondrocytes in organ-cultured
mouse intervertebral discs: An ex vivo study. Spine (Phila Pa
1976). 28:1528–1533. 2003. View Article : Google Scholar
|
|
13
|
Ma JF, Zang LN, Xi YM, Yang WJ and Zou D:
MiR-125a Rs12976445 polymorphism is associated with the apoptosis
status of nucleus pulposus cells and the risk of intervertebral
disc degeneration. Cell Physiol Biochem. 38:295–305. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hönscheid P, Datta K and Muders MH:
Autophagy: Detection, regulation and its role in cancer and therapy
response. Int J Radiat Biol. 90:628–635. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu HG, Yu YF, Zheng Q, Zhang W, Wang CD,
Zhao XY, Tong WX, Wang H, Liu P and Zhang XL: Autophagy protects
end plate chondrocytes from intermittent cyclic mechanical tension
induced calcification. Bone. 66:232–239. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Tan J, Miao Y, Lei P and Zhang Q:
ROS and autophagy: Interactions and molecular regulatory
mechanisms. Cell Mol Neurobiol. 35:615–621. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kang C, Xu Q, Martin TD, Li MZ, Demaria M,
Aron L, Lu T, Yankner BA, Campisi J and Elledge SJ: The DNA damage
response induces inflammation and senescence by inhibiting
autophagy of GATA4. Science. 349:aaa56122015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yuan C, Pu L, He Z and Wang J:
BNIP3/Bcl-2-mediated apoptosis induced by cyclic tensile stretch in
human cartilage endplate-derived stem cells. Exp Ther Med.
15:235–241. 2018.PubMed/NCBI
|
|
19
|
Zhang YH, Zhao CQ, Jiang LS and Dai LY:
Cyclic stretch-induced apoptosis in rat annulus fibrosus cells is
mediated in part by endoplasmic reticulum stress through nitric
oxide production. Eur Spine J. 20:1233–1243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ma KG, Shao ZW, Yang SH, Wang J, Wang BC,
Xiong LM, Wu Q and Chen SF: Autophagy is activated in
compression-induced cell degeneration and is mediated by reactive
oxygen species in nucleus pulposus cells exposed to compression.
Osteoarthritis Cartilage. 21:2030–2038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Feng C, Yang M, Zhang Y, Lan M, Huang B,
Liu H and Zhou Y: Cyclic mechanical tension reinforces DNA damage
and activates the p53-p21-Rb pathway to induce premature senescence
of nucleus pulposus cells. Int J Mol Med. 41:3316–3326.
2018.PubMed/NCBI
|
|
23
|
Vo NV, Hartman RA, Patil PR, Risbud MV,
Kletsas D, Iatridis JC, Hoyland JA, Le Maitre CL, Sowa GA and Kang
JD: Molecular mechanisms of biological aging in intervertebral
discs. J Orthop Res. 34:1289–1306. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gruber HE and Hanley EN Jr: Analysis of
aging and degeneration of the human intervertebral disc. Comparison
of surgical specimens with normal controls Spine (Phila Pa 1976).
23:751–757. 1998. View Article : Google Scholar
|
|
25
|
Rannou F, Lee TS, Zhou RH, Chin J, Lotz
JC, Mayoux-Benhamou MA, Barbet JP, Chevrot A and Shyy JY:
Intervertebral disc degeneration: The role of the mitochondrial
pathway in annulus fibrosus cell apoptosis induced by overload. Am
J Pathol. 164:915–924. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Levine B and Kroemer G: Autophagy in the
pathogenesis of disease. Cell. 132:27–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye W, Xu K, Huang D, Liang A, Peng Y, Zhu
W and Li C: Age-related increases of macroautophagy and
chaperone-mediated autophagy in rat nucleus pulposus. Connect
Tissue Res. 52:472–478. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shen C, Yan J, Jiang LS and Dai LY:
Autophagy in rat annulus fibrosus cells: Evidence and possible
implications. Arthritis Res Ther. 13:R1322011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dong C, Zheng H, Huang S, You N, Xu J, Ye
X, Zhu Q, Feng Y, You Q, Miao H, et al: Heme oxygenase-1 enhances
autophagy in podocytes as a protective mechanism against high
glucose-induced apoptosis. Exp Cell Res. 337:146–159. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sinha K, Das J, Pal PB and Sil PC:
Oxidative stress: The mitochondria-dependent and
mitochondria-independent pathways of apoptosis. Arch Toxicol.
87:1157–1180. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen JW, Ni BB, Zheng XF, Li B, Jiang SD
and Jiang LS: Hypoxia facilitates the survival of nucleus pulposus
cells in serum deprivation by down-regulating excessive autophagy
through restricting ROS generation. Int J Biochem Cell Biol.
59:1–10. 2015. View Article : Google Scholar
|
|
32
|
Wang F, Shi R, Cai F, Wang YT and Wu XT:
Stem cell approaches to intervertebral disc regeneration: Obstacles
from the disc microenvironment. Stem Cells Dev. 24:2479–2495. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Feng C, Zhang Y, Yang M, Lan M, Liu H,
Huang B and Zhou Y: Oxygen-sensing Nox4 generates genotoxic ROS to
induce premature senescence of nucleus pulposus cells through MAPK
and NF-κB pathways. Oxid Med Cell Longev. 2017:74264582017.
View Article : Google Scholar
|
|
34
|
Neidlinger-Wilke C, Galbusera F, Pratsinis
H, Mavrogonatou E, Mietsch A, Kletsas D and Wilke HJ: Mechanical
loading of the intervertebral disc: From the macroscopic to the
cellular level. Eur Spine J. 23(Suppl 3): S333–S343. 2014.
View Article : Google Scholar
|
|
35
|
Daly C, Ghosh P, Jenkin G, Oehme D and
Goldschlager T: A review of animal models of intervertebral disc
degeneration: Pathophysiology, regeneration, and translation to the
clinic. Biomed Res Int. 2016:59521652016. View Article : Google Scholar : PubMed/NCBI
|