Introduction
Esophageal squamous cell carcinoma (ESCC) is known
to have one of the worst prognoses of any cancers worldwide, and
the overall 5-year survival rate of ESCC is currently 25–30%
(1,2). Recent advances in the
multidisciplinary diagnostic and therapeutic approaches have
improved the prognosis of the patients with ESCC (3,4), but
the prognosis of these patients is still poor compared to that of
the patients with breast and colon cancer (5,6). The
elucidation of the molecular features of ESCC cells could
contribute to improvements in the diagnosis and treatment of
patients with ESCC.
MicroRNA (miRNA) is regarded to be one of the most
practical biological approaches to investigate the causative
mechanisms of various diseases. It has been suggested that miRNA
can suppress the expression of target genes by repressing mRNA
translation or cleaving the target mRNA, and miRNAs are thought to
regulate a wide range of biological phenomena, such as cell growth,
proliferation, differentiation, and death (7–10).
Therefore, it has been supposed that aberrations in the expression
of miRNAs might affect the development of various malignancies by
altering the expression of oncogenes and tumor suppressor genes
(11–14), and several studies have showed that
identifying a unique subset of miRNAs that are differentially
expressed in tumors might contribute to improvements in the
diagnosis and treatment of various types of cancers (15,16).
A few genome-wide miRNA expression studies of ESCC have been
published, including our previous report, and some promising miRNAs
for the identification and/or treatment of ESCC have been
identified (17–20), but the biological behavior of
miRNAs in ESCC cells remains unclear. Therefore, further
investigations are needed to clarify the role of miRNAs in the
development of ESCC.
We previously identified a novel subset of 15
downregulated miRNAs by our comprehensive miRNA profiling study of
ESCC specimens. In this study, we analyzed the effects of miR-203,
which was one of the 15 downregulated miRNAs, on ESCC cell lines
using proliferation, migration, and invasion assays. Of the
candidate target genes of miR-203, we selected LIM and SH3 protein
1 (LASP1) for a further analysis, because it was reported that
LASP1 is involved in breast and colorectal cancer metastasis
(21,22), and verified that LASP1 was directly
regulated by miR-203 in ESCC. Furthermore, the expression of
miR-203 was found to be inversely correlated with the LASP1
expression in ESCC specimens. The finding of the association
between miR-203 and LASP1 might assist in understanding the
molecular mechanism of the progression of ESCC.
Materials and methods
Clinical ESCC specimens and ESCC cell
culture
Eighteen pairs of primary esophageal squamous cell
carcinoma and corresponding normal esophageal epithelia tissue
sections were obtained from patients who had undergone surgery at
Chiba University Hospital from 2004 to 2005. All tissue specimens
were obtained from previously untreated patients undergoing primary
surgical treatment. Normal tissues were obtained far from the
center of the cancer from surgical specimens. No cancer cells were
detected in neighboring formalin-fixed paraffin-embedded (FFPE)
specimens. Written consent for tissue donation for research
purposes was obtained from each patient before tissue collection.
The protocol was approved by the Institutional Review Board of
Chiba University. The specimens were snap-frozen in liquid nitrogen
and stored at −80°C.
TE2 cells, a squamous cancer cell line with
wild-type p53 derived from human ESCC, were provided by the
Department of Surgery, Tohoku University School of Medicine. T.Tn
cells, from a human ESCC cell line known to contain an oncogenic
p53 mutation, were provided by the Japanese Cancer Research
Resources Bank. These cell lines have been tested and authenticated
as described in the literature (23,24).
Both cell lines were cultured in Dulbecco’s modified Eagle’s medium
nutrient mixture (DMEM; Life Technologies, Grand Island, NY)
supplemented with 10% fetal bovine serum (FBS) in a humidified
atmosphere containing 5% CO2 at 37°C.
RNA isolation
Tissues and cells were treated with the TRIzol
reagent (Invitrogen, Carlsbad, CA, USA), according to the
manufacturer’s instructions, for total-RNA extraction. The
extracted RNA was quantified by absorbance at 260 nm, and its
purity was evaluated by examining the 260/280 ratio of absorbance
using a SmartSpec™ Plus spectrophotometer (Bio-Rad
Laboratories, Hercules, CA, USA).
Mature miRNA transfection and small
interfering RNA (siRNA) treatment
Mature miRNA molecules, pre-miR™ miRNA
precursors (hsa-miR-203; Pre-miR ID: PM10152 and negative control
miRNA; P/N: AM17110) (Applied Biosystems, Foster City, CA, USA)
were incubated with Opti-MEM (Invitrogen) and the
siPORT™ NeoFX™ transfection reagent
(Invitrogen).
The small interfering RNA (Silencer®
Select Pre-designed siRNA; LASP1 Cat. no. 4392420) and negative
control siRNA (Silencer® Select Negative Control no. 1
siRNA Cat. no. 4390843) (Ambion) were incubated with Opti-MEM
(Invitrogen) and the Lipofectamine™ RNAiMax Reagent
(Invitrogen).
As recommended by the manufacturer, we first tested
the transfection efficacy of miRNA into the ESCC cell lines based
on the downregulation of protein tyrosine kinase 9 (PTK9) mRNA
after transfection with miR-1.
Cell proliferation, migration, and
invasion assays in ESCC cell lines
Cells were reverse transfected with 10 nM miRNA or
si-LASP1 and plated in 96-well plates at 3x103 cells per
well in 100 μl of medium containing 10% FBS. After 72 h,
cell proliferation was assessed with the Cell Counting Kit-8
(Dojindo, Kumamoto, Japan). Triplicate wells were measured for cell
viability in each treatment group.
Cell motility was determined using a micropore
chamber assay; 8x104 cells were seeded onto the top chamber of a
24-well micropore polycarbonate membrane filter with 8 μm
pores (BD Biosciences, Bedford, MA, USA) and the bottom chamber was
filled with DMEM containing 10% FBS as a chemoattractant. After 24
h of incubation, the membranes were fixed and stained by the
DiffQuik reagent (International Reagents, Kobe, Japan) and the
cells on the upper surface were carefully removed with a cotton
swab. Cell migration was quantified by counting the average number
of migrated cells in 4 random high-powered fields per filter. For
the assessment of cell invasion, the migration assay was repeated
with a 24-well Matrigel-coated micropore membrane filter with 8
μm pores (BD Biosciences). Cell invasion was quantified by
counting the number of cells that had migrated through the
membrane.
Screening for miR-203 target genes by a
microarray analysis
A genome-wide screen was performed for identify the
genes silenced by miR-203 in ESCC cells (TE2 and T.Tn). GeneChip
U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA, USA) were used
for the expression profiling of miR-203 transfectants in comparison
with miRNA-negative control transfectants in the ESCC cell lines.
Starting with 250 ng of total-RNA, the GeneChip U133 Plus 2.0 Array
was conducted in accordance with manufacturer’s protocol, and using
the GeneChip 3′IVT Express Kit (Affymetrix) and GeneChip
Hybridization Wash and Stain Kit (Affymetrix). The raw expression
signal obtained from the GeneChip Instrument System (Affymetrix)
was normalized by scaling to the target signal of 100.
The predicted target genes and their conserved sites
where the seed region of each miRNA binds were investigated using
the TargetScan database (release 5.1, http://www.targetscan.org/). The sequence of the
predicted mature microRNA was confirmed using the miRBase software
program (release 17.0, http://microrna.sanger.ac.uk/).
Quantitative reverse-transcription-PCR
(qRT-PCR)
First-strand cDNA was synthesized from 1.0 μg
of total-RNA using a High Capacity cDNA Reverse Transcription kit
(Applied Biosystems). Gene-specific PCR products were assayed
continuously using a 7300 real-time PCR system (Applied Biosystems)
according to the manufacturer’s protocol. TaqMan® probes
and primers for LASP1 (P/N: Hs01078815_m1) and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (P/N:
Hs02758991_g1) as an internal control were obtained from Applied
Biosystems (Assay-On-Demand Gene Expression Products). The
expression levels of miR-203 (assay ID: 000507) were analyzed by
TaqMan quantitative real-time PCR (TaqMan MicroRNA Assay, Applied
Biosystems) and normalized to the level of RNU6B (assay ID:
001093). All reactions were performed in triplicate, and included a
negative control that lacked cDNA.
Western blot analysis
The cells were harvested and lysed 72 h after
transfection. Protein extracts (40 μg) were separated by
sodium dodecyl sulfate-polyacrylamide gel electrophoresis and
transferred to polyvinylidene difluoride membranes. Membranes were
blocked in PBS containing 5% non-fat dried milk and 0.1% Tween-20.
The anti-human LASP1 rabbit polyclonal IgG was used at a dilution
of 1:100 and the anti-rabbit IgG-peroxidase antibody produced in a
goat (Sigma-Aldrich, St. Louis, MO, USA) was used at a dilution of
1:5,000. The anti-β-actin polyclonal antibody (Santa Cruz
Biotechnology) at a dilution of 1:200 and anti-goat IgG-peroxidase
antibody produced in rabbits (Sigma-Aldrich) at a dilution of
1:5,000 were used as controls. After incubation with a primary
antibody for 2 h at room temperature, the membranes were incubated
for 1 h with the secondary antibody, also at room temperature. The
analysis was performed using ECL Western blotting detection
reagents (Amersham Pharmacia Biotech, Piscataway, NJ, USA). The
expression levels of these proteins were evaluated by ImageJ
software (version 1.44; http://rsbweb.nih.gov/ij/index.html).
Target site inhibition assays
The miScript Target Protector for the miR-203
binding site in the 3′UTR of LASP1 mRNA was obtained from Qiagen
(target binding site sequence provided:
5′-GAAAUUUGCCUCUGUCCAAACAUUUCAUCC-3′). The miScript Target
Protector is single-stranded, modified RNA that specially
interferes with the interaction of a miRNA with a single target,
while leaving the regulation of other targets of the same miRNA
unaffected. Syn-hsa-miR-203 miScript miRNA Mimic (Qiagen, miScript
miRNA Mimic for: GUGAAAUGU UUAGGACCACUAG) and miScript Target
Protector were co-transfected into ESCC cell lines (TE2 and T.Tn)
according to the manufacturer’s protocol. Syn-hsa-miR-203 miScript
miRNA Mimic and Negative control miScript Target Protector designed
not to bind the mRNA of mammals were co-transfected into ESCC cell
lines as a negative control. After 48 h of transfection, the
total-RNA was isolated and the mRNA expression levels of LASP1 were
measured by TaqMan quantitative real-time PCR.
Statistical analysis
The relationship between two variables and numerical
values obtained by real-time RT-PCR were analyzed using the
Mann-Whitney U test or paired t-test. The relationships between
miR-203 expression and LASP1 expression were analyzed using the
Spearman rank correlation. To evaluate the significance of the
expression of miR-203 as a prognostic factor, we performed an
analysis using the Kaplan-Meier method and the log-rank test. The
Expert StatView software program (version 5, SAS Institute Inc.,
Cary, NC, USA) was used for the analysis, with statistical
significance defined as P<0.05.
Results
The effects of miRNA transfection on ESCC
cell lines
The expression levels of miR-203 were extremely low
in ESCC cell lines (data not shown), suggesting that there was no
effect of the corresponding endogenous miR-203 on the viability of
these cell lines. In a gain-of-function analysis, two cell lines
(TE2 and T.Tn) were transfected with miR-203.
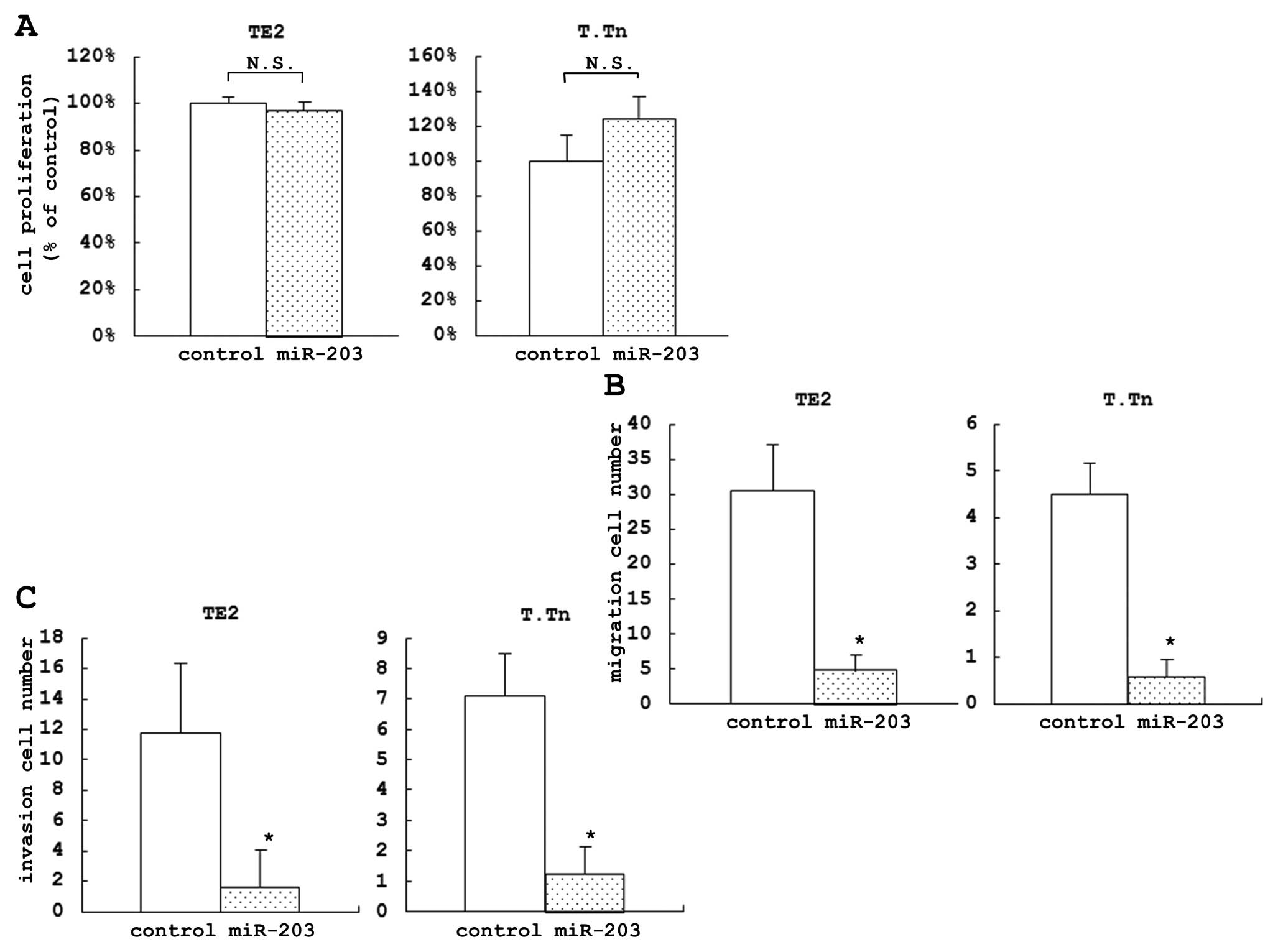
The proliferation assay, which was used to evaluate
the proportion of cell growth inhibition of the cells transfected
with miR-203 compared to the control cells at 72 h after the
transfection, indicated that there was no significant differences
between the cells transfected with miR-203 and the control in both
the TE2 and T.Tn cell lines (Fig.
1A). The proliferation assay did not show any significant
difference in the cell growth inhibition at 24, 48 and 96 h (data
not shown).
In the migration assay, we evaluated the proportion
of the inhibition of cell motility of the cells transfected with
miR-203 compared to the control cells at 72 h. After the
transfection, we found a significant difference between these cell
lines (84.2% in TE2 and 87.0% in T.Tn) (Fig. 1B). The invasion assay was used to
evaluate the proportion of the inhibition of cell invasion of the
cells transfected with miR-203 compared to the control cells. Using
this assay, we also found a significant difference between the
miRNA and control-transfected cells at 72 h after the transfection
(86.5% in TE2 and 82.4% in T.Tn) (Fig.
1C).
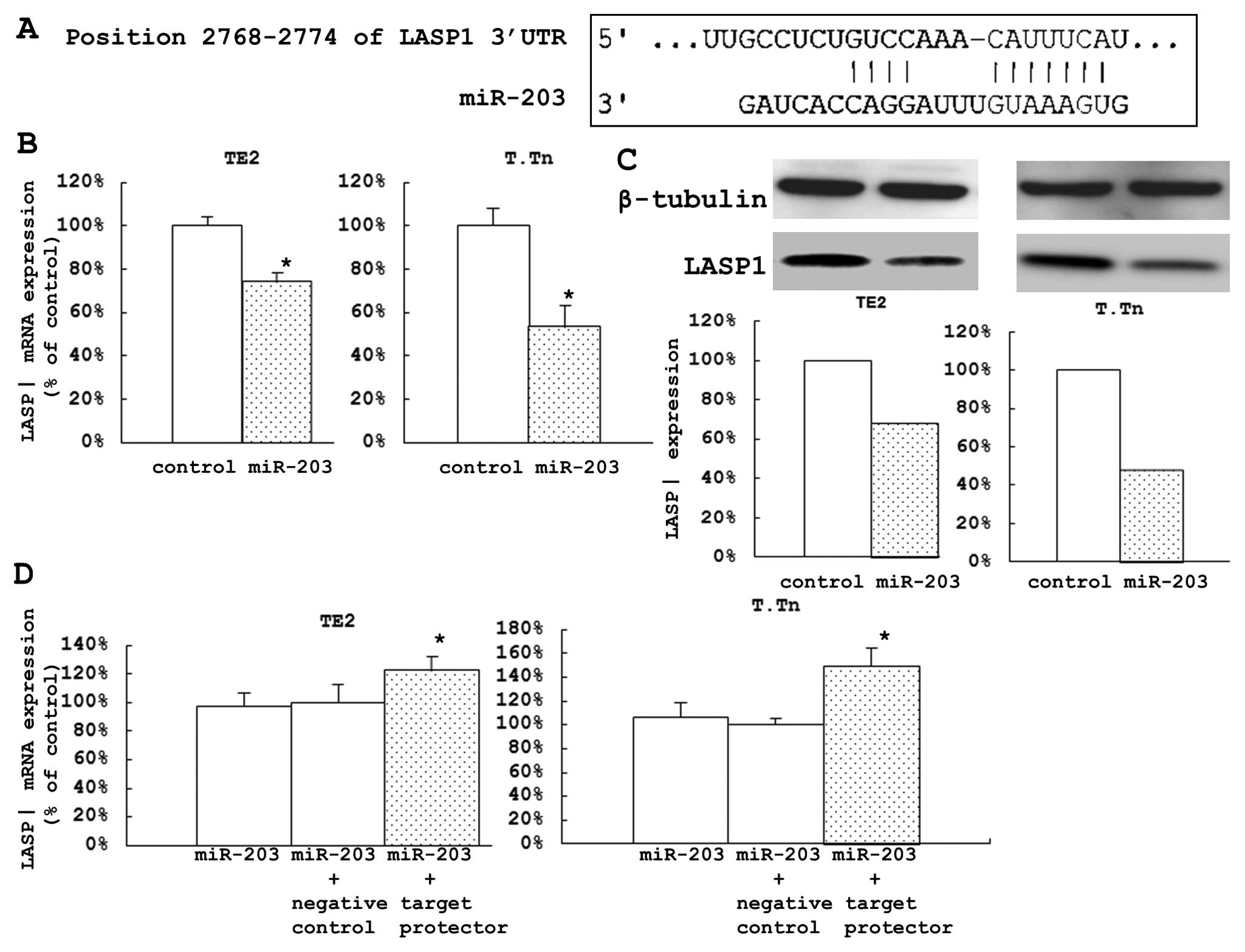
LASP1 as one of the direct target genes
of miR-203
An analysis of the genome-wide gene expression of
the target genes silenced by miR-203 was performed using a
microarray analysis of the ESCC cells transfected with the miR-203
and the control. A total of 22 genes were identified to be
downregulated, and we assessed the conserved sites in the 3′UTR
using the TargetScan database (Table
I). During the search of the TargetScan database, we found one
conserved site in the 3′UTR of LASP1 (Fig. 2A).
 | Table I.Gene downregulated in
miR-203-transfected ESCC cell lines compared to control cells,
which were found to have conserved sites in their 3′ untranslated
region using the TargetScan database. |
Table I.
Gene downregulated in
miR-203-transfected ESCC cell lines compared to control cells,
which were found to have conserved sites in their 3′ untranslated
region using the TargetScan database.
| No | Entrez gene ID | Gene symbol | Gene name | miR-203 target
site |
|---|
| 1 | 3927 | LASP1 | LIM and SH3 protein
1 | 1 |
| 2 | 90161 | HS6ST2 | Heparan sulfate
6-O-sulfotransferase 2 | 1 |
| 3 | 51496 | CTDSPL2 | CTD
(carboxy-terminal domain, RNA polymerase II, polypeptide A) small
phosphatase like 2 | 1 |
| 4 | 11333 | PDAP1 | PDGFA associated
protein 1 | 2 |
| 5 | 1352 | COX10 | COX10 homolog,
cytochrome c oxidase assembly protein, heme A:
farnesyltransferase | 1 |
| 6 | 3600 | IL15 | Interleukin 15 | 1 |
| 7 | 2186 | BPTF | Bromodomain PHD
finger transcription factor | 1 |
| 8 | 9037 | SEMA5A | Sema domain, seven
thrombospondin repeats (type 1 and type 1-like), transmembrane
domain (TM) and short cytoplasmic domain, (semaphorin) 5A | 2 |
| 9 | 27445 | PCLO | Piccolo
(presynaptic cytomatrix protein) | 1 |
| 10 | 54665 | RSBN1 | Round spermatid
basic protein 1 | 1 |
| 11 | 196528 | ARID2 | AT rich interactive
domain 2 (ARID, RFX-like) | 1 |
| 12 | 51714 | SELT | Selenoprotein
T | 1 |
| 13 | 3607 | FOXK2 | Forkhead box
K2 | 1 |
| 14 | 518 | ATP5G3 | ATP synthase, H+
transporting, mitochondrial Fo complex, subunit C3 (subunit 9) | 1 |
| 15 | 55573 | CDV3 | CDV3 homolog | 1 |
| 16 | 114882 | OSBPL8 | Oxysterol binding
protein-like 8 | 1 |
| 17 | 57507 | ZNF608 | Zinc finger protein
608 | 1 |
| 18 | 54557 | SGTB | Small
glutamine-rich tetratricopeptide repeat (TPR)-containing, β | 1 |
| 19 | 160897 | GPR180 | G protein-coupled
receptor 180 | 1 |
| 20 | 91749 | KIAA1919 | KIAA1919 | 1 |
| 21 | 148867 | SLC30A7 | Solute carrier
family 30 (zinc transporter), member 7 | 1 |
| 22 | 55219 | TMEM57 | Tansmembrane
protein 57 | 1 |
The expression levels of LASP1 mRNA in the cells
transfected with miR-203 were significantly lower than those in the
control cells (25.9% of the control in TE2 cells and 46.6% in the
T.Tn cells) (Fig. 2B). After 72 h,
the protein expression levels were also reduced (Fig. 2C).
These two cell lines were also used to investigate
how miR-203 directly affects LASP1 expression. After 48 h of
co-transfection with Syn-hsa-miR-203 using a miScript miRNA Mimic
and miScript Target Protector/Negative control miScript Target
Protector, the expression levels of LASP1 mRNA in the transfected
cells were significantly higher than those in the control cells
(22.3% of the control in TE2 cells and 49.2% in T.Tn cells). These
results suggested that this specific target may be directly
controlled by miR-203 (Fig.
2D).
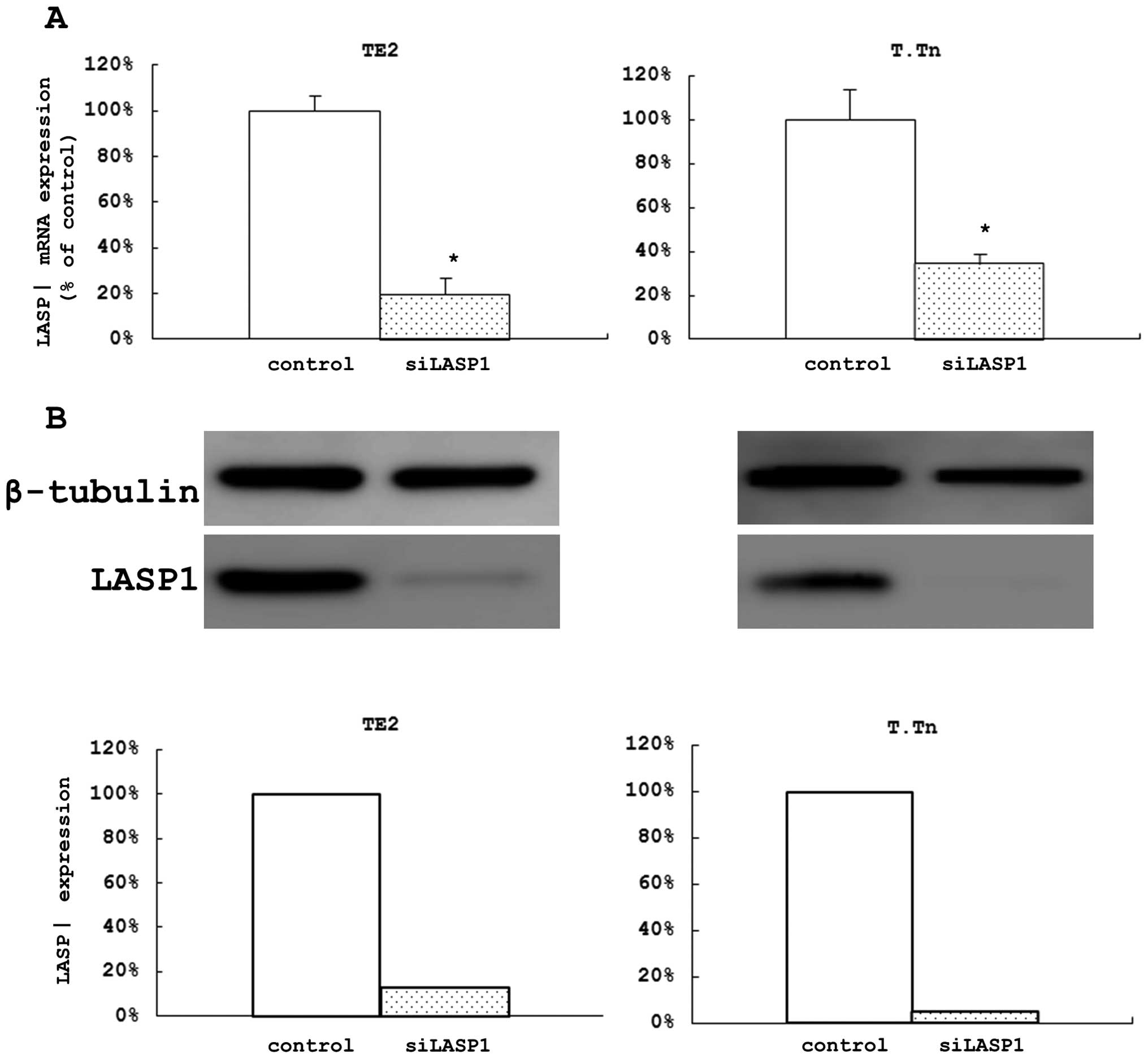
Evaluation of the effects of LASP1 on the
proliferation, migration, and invasion of ESCC cell lines using a
loss of function analysis
After 48 h of si-LASP1 transfection in the two cell
lines, we confirmed that the expression levels of LASP1 mRNA were
remarkably decreased (80.4% of the control in TE2 cells and 65.5%
in T.Tn cells) (Fig. 3A). We also
confirmed that the levels of the protein were remarkably decreased
after 72 h (Fig. 3B). In the
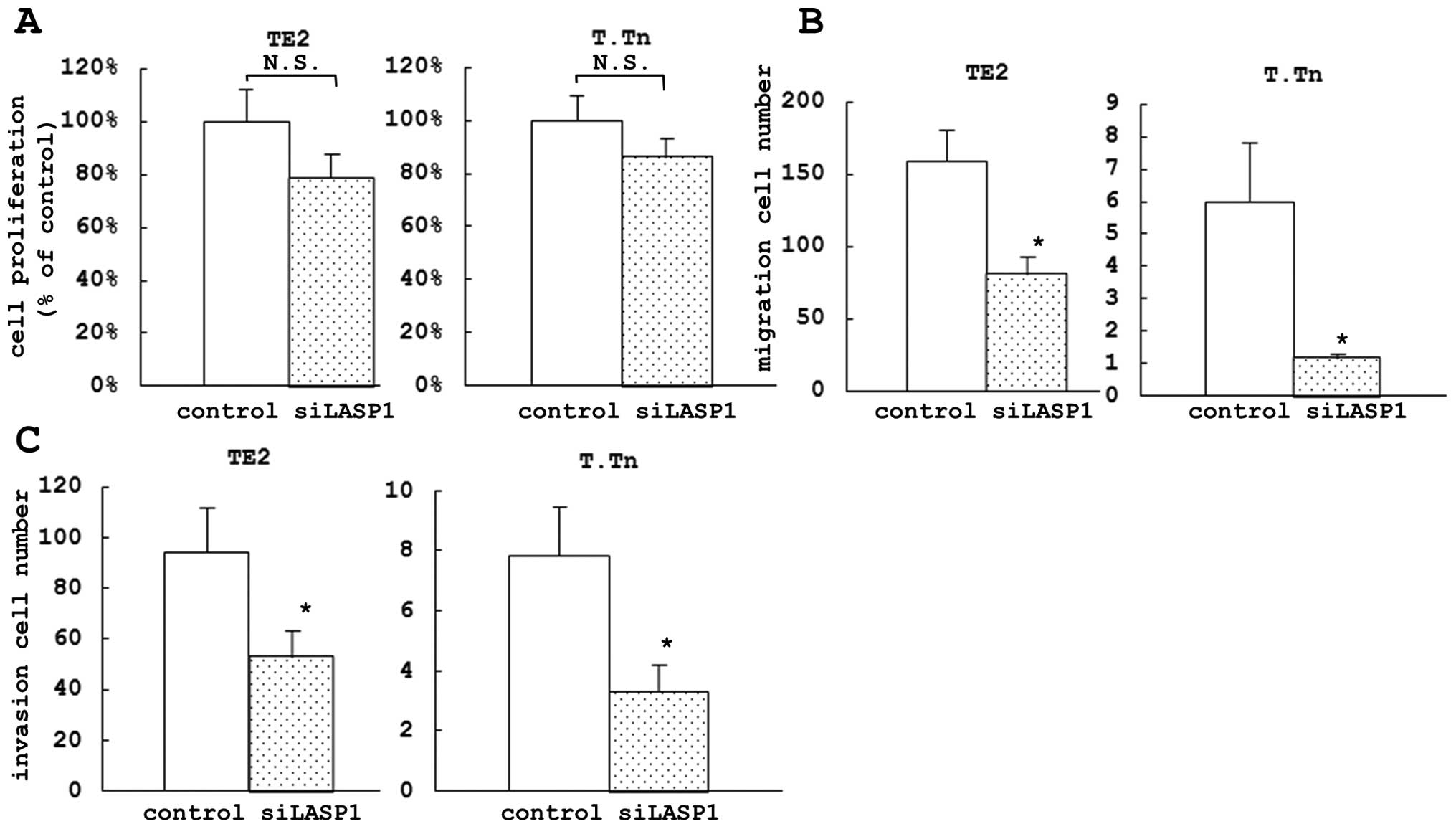
proliferation assay, we did not find any significant difference in
cell growth inhibition between the si-LASP1-transfected cells and
the control cells after 72 h (Fig.
4A) nor after 24, 48 or 96 h (data not shown), similar to the
effects observed following the miR-203 treatment.
In the migration assay, we found a significant
difference in the inhibition of cell motility between the
si-LASP1-transfected cells and the control cells after 72 h (49.0%
in TE2 and 80.6% in T.Tn) (Fig.
4B). Similarly, in the invasion assay, there was a significant
difference in the inhibition of cell invasion between the
si-LASP1-transfected cells and the control cells at 72 h after
transfection (43.8% in TE2 and 57.4% in T.Tn) (Fig. 4C).
The relationship between miR-203 and
LASP1 expression in ESCC clinical specimens
Total-RNA was isolated from matched non-cancerous
esophageal epithelia and ESCC tissues, from which the miRNA and
LASP1 mRNA levels were determined by TaqMan real-time PCR. In all
18 matched normal and cancer tissues, the expression levels of
miR-203 were significantly lower in cancer tissues compared to
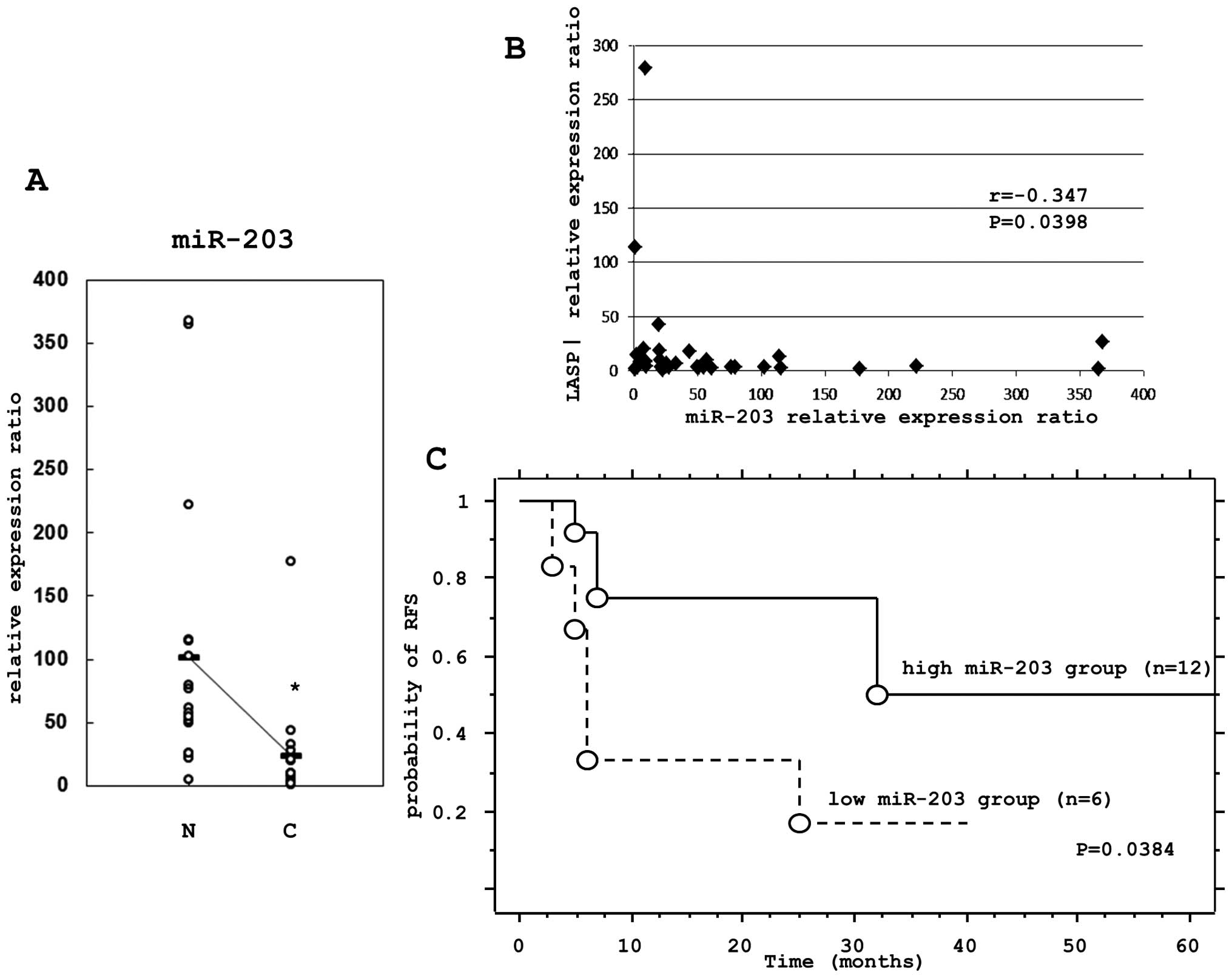
non-cancerous tissues (Fig. 5A).
The correlation of the expression levels of LASP1 and miR-203 were
tested using the Spearman rank correlation. There was a significant
inverse correlation between the LASP1 mRNA and miR-203 expression
levels (P=0.0398) (Fig. 5B).
The prognostic impact of miR-203 was further
examined using the Kaplan-Meier method and the log-rank test. There
was a significant correlation (P=0.0384) between patients with low
miR-203 expression and a poor relapse free survival (RFS) (Fig. 5C).
Discussion
We have conducted searches for tumor suppressive
miRNAs, such as miR-145, miR-133a, and miR-133b (20). In this study, we reported that
miR-203 might directly control the expression of LASP1 and
contribute to the inhibition of cell migration and invasion in
ESCC. Since Yi et al mentioned that miR-203 might control
epidermal differentiation by repressing p63 ‘stemness’ (25), several studies have focused on the
relationship between the expression of miR-203 and diverse
malignancies. For example, it was suggested that miR-203 might play
a crucial role in cell proliferation in head and neck squamous cell
carcinomas, hematopoietic malignancies, and hepatocellular
carcinoma (26–28). In ESCC, Feber et al
initially reported that the expression of miR-203 was downregulated
in Barrett esophagus and esophageal cancer compared to normal
tissues (17). Bueno et al,
Yuan et al and Bo et al revealed that miR-203
controlled cell proliferation in hematopoietic malignancies,
bladder cancer, and ESCC by targeting ABL1, bcl-w and p63 (27,29,30).
Moreover, other studies showed that miR-203 might be associated
with not only cell proliferation, but also cell migration, cell
invasion, and the epithelial-mesenchymal transition (EMT), by
targeting ZEB2, Bmi, survivin and Runx2 (31,32).
In our study using two ESCC cell lines, TE2 and
T.Tn, there was no relationship between miR-203 expression and cell
proliferation. However, we found that the migration and invasion of
both cell lines were significantly suppressed by miR-203. Our
results also showed that the cell proliferation controlled by
miR-203 in ESCC may depend on the individual cell line. In addition
to these analyses, miR-203 is also considered to be a suppressor of
cancer development, and Chen et al showed that higher
expression of miR-203 might be useful as an independent predictor
of a good prognosis (RFS and overall survival) of the patients with
hepatocellular carcinoma and cirrhosis after liver transplantation
(33). In our study, there was a
significant correlation between lower expression of miR-203 in ESCC
specimens and a poor RFS. Our result suggests that the expression
of miR-203 could be a novel prognostic marker for patients with
ESCC.
LASP1, known as one of the actin-binding proteins
(molecular weight: 40 kDa), is localized to multiple sites of
dynamic actin assembly, such as focal contacts, focal adhesions,
lamellipodia membrane ruffles, and pseudopodia, and is involved in
cell migration (21,34,35).
LASP1 was derived from a cDNA library of breast cancer metastases
(21). This protein was highly
overexpressed in 40% of metastatic human breast cancer tissues
(21). Zhao et al reported
that over-expression of LASP1 was also found in metastatic
colorectal cancer tissues, and its higher expression was closely
correlated with a poor prognosis of the patients with colorectal
cancer (22). Therefore, it was
suggested that LASP1 might have an important role in cancer cell
mobility and metastasis, but the precise function of LASP1 in the
various malignancies still remains unclear. No previous expression
and functional analysis of LASP1 in ESCC has been reported. In our
present study, there was no significant correlation between the
expression of LASP1 and cell proliferation. On the other hand, the
migration and invasion of ESCC cell lines were significantly
suppressed by si-LASP1. Chiyomaru et al reported that
miR-218, miR-1, and miR-133a regulated the expression of LASP1 in
urinary bladder cancer (36), and
Viticchiè G et al revealed that miR-203 regulated the
expression of LASP1 in prostate cancer (37). Our study suggested that miR-203 and
its target gene, LASP1, might be associated with the progression of
ESCC.
Castilla et al reported that miR-203 was
downregulated in the mesenchymal part of endometrial carcinosarcoma
(31), and Saini et al
mentioned that miR-203 regulated the EMT in prostate cancer
(32). In this study, miR-203,
which has been called an ‘antimetastatic miRNA’, was revealed to
target LASP1 in ESCC, thus further strengthening its connection
with cancer cell motility and metastasis.
The candidate target genes other than LASP1 were
identified by other studies. Sadanandam et al reported that
Semaphorin 5A (SEMA5A) was associated with tumor growth, invasion,
and metastasis in pancreatic cancer (38), and Choi et al reported that
overexpression of PDGFA associated protein 1 (PDAP1) was found in
human rectal carcinoma (39). Song
et al also reported that heparan sulfate
6-O-sulfotransferase 2 (HS6ST2) regulated the progression of
pancreatic cancer by potentiating Notch signaling (40). Thus, there is a possibility that
various pathways are affected by miR-203 in different cancers. We
hope that the further analyses of the mechanism underlying the
cancer progression regulated by miR-203 will contribute to the
development of new cancer treatments and provide a new marker for
the diagnosis of cancer in the future.
References
|
1.
|
Chen J, Pan J, Zheng X, et al: Number and
location of positive nodes, postoperative radiotherapy, and
survival after esophagectomy with three-field lymph node dissection
for thoracic esophageal squamous cell carcinoma. Int J Radiat Oncol
Biol Phys. 82:475–482. 2012. View Article : Google Scholar
|
|
2.
|
Yuequan J, Shifeng C and Bing Z:
Prognostic factors and family history for survival of esophageal
squamous cell carcinoma patients after surgery. Ann Thorac Surg.
90:908–913. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Altorki N, Kent M, Ferrara C and Port J:
Three-field lymph node dissection for squamous cell and
adenocarcinoma of the esophagus. Ann Surg. 236:177–183. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Medical Research Council Oesophageal
Cancer Working Group: Surgical resection with or without
preoperative chemotherapy in oesophageal cancer: a randomized
controlled trial. Lancet. 359:1727–1733. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Blamey RW, Hornmark-Stenstam B, Ball G, et
al: ONCOPOOL - a European database for 16,944 cases of breast
cancer. Eur J Cancer. 46:56–71. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Morris EJ, Sandin F, Lambert PC, et al: A
population-based comparison of the survival of patients with
colorectal cancer in England, Norway and Sweden between 1996 and
2004. Gut. 60:1087–1093. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Schickel R, Boyerinas B, Park SM and Peter
ME: MicroRNAs: key players in the immune system, differentiation,
tumori-genesis and cell death. Oncogene. 27:5959–5974. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
He L, Thomson JM, Hemann MT, et al: A
microRNA polycistron as a potential human oncogene. Nature.
435:828–833. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Calin GA, Trapasso F, Shimizu M, et al:
Familial cancer associated with a polymorphism in ARLTS1. N Engl J
Med. 352:1667–1676. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Feber A, Xi L, Luketich JD, et al:
MicroRNA expression profiles of esophageal cancer. J Thorac
Cardiovasc Surg. 135:255–260. 2008. View Article : Google Scholar
|
|
18.
|
Guo Y, Chen Z, Zhang L, et al: Distinctive
microRNA profiles relating to patient survival in esophageal
squamous cell carcinoma. Cancer Res. 68:26–33. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Hiyoshi Y, Kamohara H, Karashima R, et al:
MicroRNA-21 regulates the proliferation and invasion in esophageal
squamous cell carcinoma. Clin Cancer Res. 15:1915–1922. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Kano M, Seki N, Kikkawa N, et al: miR-145,
miR-133a and miR-133b: Tumor-suppressive miRNAs target FSCN1 in
esophageal squamous cell carcinoma. Int J Cancer. 127:2804–2814.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Tomasetto C, Regnier C, Moog-Lutz C, et
al: Identification of four novel human genes amplified and
overexpressed in breast carcinoma and localized to the q11-21.3
region of chromosome 17. Genomics. 28:367–376. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Zhao L, Wang H, Liu C, et al: Promotion of
colorectal cancer growth and metastasis by the LIM and SH3 domain
protein 1. Gut. 59:1226–1235. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Takahashi K, Kanazawa H, Chan CT, Hosono
T, Takahara M and Sato K: A case of esophageal carcinoma metastatic
to the mandible and characterization of 2 cell lines (T.T and T.
Tn) established from the oral tumor. Jpn J Oral Maxillofac Surg.
36:67–76. 1990. View Article : Google Scholar
|
|
24.
|
Nishihira T, Kasai M, Mori S, et al:
Characteristics of 2 cell lines (TE-1 and TE-2) derived from human
squamous cell carcinoma of the esophagus. Gann. 70:575–584.
1979.PubMed/NCBI
|
|
25.
|
Yi R, Poy MN, Stoffel M and Fuchs E: A
skin microRNA promotes differentiation by repressing ‘stemness’.
Nature. 452:225–229. 2008.
|
|
26.
|
Lena AM, Shalom-Feuerstein R, Rivetti di
Val Cervo P, et al: miR-203 represses ‘stemness’ by repressing
DeltaNp63. Cell Death Differ. 15:1187–1195. 2008.
|
|
27.
|
Bueno MJ, Pérez de Castro I, Gómez de
Cedrón M, et al: Genetic and epigenetic silencing of microRNA-203
enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell.
13:496–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Furuta M, Kozaki KI, Tanaka S, Arii S,
Imoto I and Inazawa J: miR-124 and miR-203 are epigenetically
silenced tumor-suppressive microRNAs in hepatocellular carcinoma.
Carcinogenesis. 31:766–776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Yuan Y, Zeng ZY, Liu XH, et al:
MicroRNA-203 inhibits cell proliferation by repressing ΔNp63
expression in human esophageal squamous cell carcinoma. BMC Cancer.
11:572011.
|
|
30.
|
Bo J, Yang G, Huo K, et al: microRNA-203
suppresses bladder cancer development by repressing bcl-w
expression. FEBS J. 278:786–792. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Castilla MÁ, Moreno-Bueno G, Romero-Pérez
L, et al: Micro-RNA signature of the epithelial-mesenchymal
transition in endometrial carcinosarcoma. J Pathol. 223:72–80.
2011.PubMed/NCBI
|
|
32.
|
Saini S, Majid S, Yamamura S, et al:
Regulatory role of mir-203 in prostate cancer progression and
metastasis. Clin Cancer Res. 17:5287–5298. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Chen HY, Han ZB, Fan JW, et al: miR-203
expression predicts outcome after liver transplantation for
hepatocellular carcinoma in cirrhotic liver. Med Oncol. Jul
24–2011.(Epub ahead of print).
|
|
34.
|
Chew CS, Chen X, Parente JA Jr, Tarrer S,
Okamoto C and Qin HY: Lasp-1 binds to non-muscle F-actin in vitro
and is localized within multiple sites of dynamic actin assembly in
vivo. J Cell Sci. 115:4787–4799. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Lin YH, Park ZY, Lin D, et al: Regulation
of cell migration and survival by focal adhesion targeting of
Lasp-1. J Cell Biol. 165:421–432. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Chiyomaru T, Enokida H, Kawakami K, et al:
Functional role of LASP1 in cell viability and its regulation by
microRNAs in bladder cancer. Urol Oncol. 30:434–443. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Viticchiè G, Lena AM, Latina A, et al:
miR-203 controls proliferation, migration and invasive potential of
prostate cancer cell lines. Cell Cycle. 10:1121–1131.
2011.PubMed/NCBI
|
|
38.
|
Sadanandam A, Varney ML, Singh S, et al:
High gene expression of semaphorin 5A in pancreatic cancer is
associated with tumor growth, invasion and metastasis. Int J
Cancer. 127:1373–1383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Choi SY, Jang JH and Kim KR: Analysis of
differentially expressed genes in human rectal carcinoma using
suppression subtractive hybridization. Clin Exp Med. 11:219–226.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Song K, Li Q, Peng YB, et al: Silencing of
hHS6ST2 inhibits progression of pancreatic cancer through
inhibition of Notch signalling. Biochem J. 436:271–282. 2011.
View Article : Google Scholar : PubMed/NCBI
|