Introduction
Breast cancer is the most common type of cancer
diagnosed in women and the leading cause of female cancer-related
mortality worldwide (1). Breast
cancer is currently considered a heterogeneous disease that
includes multiple subtypes that differ in origin, dynamics,
response to treatments, risk of recurrence and survival (2–5).
Surgery is the primary therapeutic option for
treating breast cancer and other solid neoplasms. However, surgical
treatment alone is often insufficient to eradicate the disease
since most patients develop distant tumors that were present as
undetectable micrometastases at the time of diagnosis, keeping a
higher risk of recurrence after tumor removal. Furthermore,
although surgery results in a considerable increase in overall
survival for most patients, evidence suggests that tumor removal
may unfavorably alter the natural history of the disease. Based on
preclinical models and the analysis of large series of patients,
several authors have postulated a link between surgery and
recurrence (6–9). This hypothesis is supported by the
existence of a nonproliferative, dormant state of distal
micrometastatic foci that can be disrupted after the surgical
depletion of the primary tumor (10–12).
Surgery-driven escape from dormancy is proposed to be a systemic
process mediated by soluble secreted proteins, such as growth
factors, chemokines or angiogenic factors, that also play a
critical role in wound healing and tissue regeneration following
surgery (13,14).
Changes in the concentration of serum proteins have
traditionally been measured by techniques based on highly sensitive
and specific antibody-antigen recognition, such as ELISA, RIA or
western blot analysis. Nevertheless, these techniques are suitable
for the quantification of only a single or a few analytes per
assay. The development of automated, high-throughput technologies
in the field of molecular biology has provided substantial advances
in the understanding of disease processes by enabling the
evaluation of a large number of samples in a single assay (15). Reverse phase protein microarrays
(RPPA) are high-throughput, multiplexed and miniaturized
immunoassays in which a small volume of protein samples is spotted
onto a capturing surface and probed with an antibody directed
against the analyte of interest (16–18).
Thus, RPPAs enable the mass detection of molecules and reduce cost,
time and sample volume without altering sensitivity or specificity.
These properties have made RPPAs a powerful tool in cancer research
(19).
The main aim of the present study was to detect
variations in serum protein levels that directly result from breast
surgery, focusing on those changes specifically induced by surgery
in patients with invasive breast cancer. We employed RPPAs to
examine the expression levels of 42 soluble proteins in serum
samples from healthy controls and breast cancer patients, both
before and after breast surgery.
Materials and methods
Sample preparation
Serum samples were obtained from 79 women who
underwent surgery at the Hospital Universitario Virgen de la
Victoria (Malaga, Spain) in our hospital between 1998 and 2005. All
subjects provided informed consent for study inclusion. Among the
patients, 56 were diagnosed with invasive breast cancer and 7 were
diagnosed with in situ breast carcinoma. Sixteen women who
developed benign breast fibroadenoma were included as healthy
controls. None of the patients received adjuvant chemotherapy
before surgery or immediate breast reconstruction after mastectomy.
Samples were collected 8 h before surgery, denoted t(0), and 24 h
after mastectomy/lumpectomy, denoted t(24). Blood was collected in 3 ml
serum-separating tubes (SST, Becton-Dickinson, Franklin Lakes, NJ,
USA) and processed within 1 h after collection. Samples were left
at room temperature for 30–40 min until clotted. Serum was obtained
by centrifugation at 4,000 rpm for 10 min at 4°C and stored in 200
μl aliquots at −80°C until use. The study was approved by
our hospital’s ethics committee.
Antibodies
A set of 45 polyclonal antibodies raised against 42
different serum soluble proteins was used to probe the RPPAs (SDI
Inc, Newark, DE, USA; Table II).
The human serum albumin level was determined on each spot using a
monoclonal α-HSA antibody (Sigma-Aldrich, St. Gallen, Switzerland)
and served as an intra-spot normalization control. Two different
fluorescent-labeled secondary antibodies were used: a donkey
α-mouse IgG-DyLight649 and a goat α-rabbit IgG-Cy3 (Jackson
Immunoresearch, Suffolk, UK).
 | Table IIList of antibodies used in the
study. |
Table II
List of antibodies used in the
study.
| Antibodies |
|---|
| αFP | ENG | IFNγ | IL18 | PDGF-B |
| CD31 | Factor XIIIa | IL1A | IL24 | THBS2 |
| CD44 | FasL (A) | IL1B | MMP1 | THBS3 |
| CEACAM | FasL (B) | IL5 | MMP11 | TNFα |
| CSF1 | HER2 | IL6 | MMP3 | VCAM1(A) |
| CSF2 | HER3 | IL6ST | MMP7 | VCAM1(B) |
| CTS D | ICAM5 | IL7 | MMP9 | VEGF-A |
| E-Cad (A) | IFNα1 | IL12A | MUC16 | VEGF-B |
| E-Cad (B) | IFNβ1 | IL16 | OPN | VWF |
Microarray generation
Serum samples were diluted in Denaturing Printing
Buffer (DPB) at a final concentration of 8% glycerol, 2% SDS, 50 mM
Tris-HCl, pH 6.8 and 2% β-ME, as previously described (20), loaded onto 384-well plates and
denatured by boiling at 95°C for 10 min. Ten picoliters from each
sample were spotted onto low-fluorescence Immobilon-FL membranes
(Merck Millipore, Darmstadt, Germany) using a BioOdyssey
Calligrapher MiniArrayer (Bio-Rad, Hercules, CA, USA). Spots were
printed in duplicate on each membrane following a predefined 24×24
matrix pattern and dried for 2 h inside the printer. Subsequently,
the membranes were stored at 4°C in a desiccated chamber with NaCl.
Prior to use, membranes were stained with Ponceau red solution
(AppliChem, Darmstadt, Germany) to ensure that the spots were
printed successfully. The membranes were then washed once with
double-distilled water and twice with TBS-T (50 mM Tris-HCl, pH
7.5, 150 mM NaCl, 0.1% Tween-20) and blocked with blocking buffer
(3% bovine serum albumin in TBS-T) for 30 min at room temperature.
Next, the membranes were incubated for 1 h at room temperature in
blocking buffer containing both the specific primary antibody and
the HSA antibody used for normalization. After three washes in
TBS-T, membranes were incubated for 30 min at room temperature in
blocking buffer containing both fluorescent-labeled secondary
antibodies. The membranes were washed three times in TBS-T and once
in double-distilled water and then air-dried in darkness. Finally,
fluorescence levels were quantified in a LuxScan 10K/A fluorescence
scanner (CapitalBio, Beijing, China) at 570 nm for the Cy3-labeled
specific protein of interest and 670 nm for the DyLight649-labeled
HSA.
To perform robust inter-array comparisons, we
referenced the raw fluorescence data from the protein of interest
(green) to the level of endogenous human albumin (red) in each
spot. This approach also enabled us to discriminate between
specific variations and fluctuations related to surgery-associated
protein leakage and stress, as previously described (21).
Data analysis
The complete analysis of microarray data was
performed using the R programming language (http://www.r-project.org), which has become a de facto
standard in the field. Foreground and background intensities from
scanned images were extracted using data input functions for
two-color microarrays in the limma package (22). The background intensities were
subtracted from the foreground intensities for each spot on the
arrays, and then a quantile normalization procedure was implemented
to make intensities consistent between arrays. Quantile
normalization was proposed by Yang and Thorne (23) for two-color cDNA arrays, with the
aim of ensuring that the intensities have the same empirical
distribution across arrays and across channels. The values of
replicate spots within each array were then replaced with their
average values.
Multiple linear models were fitted to the expression
data (in log-ratio scale) for each antibody. Thus, the coefficients
of the fitted models describe the differences between the sources
hybridized to the arrays. Assessment of differential expression was
then completed by performing hypothesis tests and adjusting the
p-values for multiple tests. The basic statistical method used to
determine significance was the moderated t statistic, which is
computed for each probe and for each contrast using a simple
Bayesian model that borrows information from the ensemble of
antibodies to aid with inference for each individual antibody
(24). The method of Benjamini and
Hochberg (25) was used to control
the false discovery rate and adjust for q-values such that all
antibodies with a q-value below a threshold (typically 0.05) were
selected as differentially expressed.
Results
To measure the impact of surgery on the serum
protein profile, we designed a longitudinal study with serum
samples obtained from 63 breast cancer patients who underwent
breast surgery at the Hospital Universitario Virgen de la Victoria
in Malaga. To provide a control set, we collected samples from 16
healthy women who underwent operations for fibroadenoma. The
clinical characteristics of patients and controls are detailed in
Table I. Samples were obtained
prior to surgery, denoted t(0), and 24 h after the operation,
denoted t(24), to prevent
potential interference from uncontrolled factors, such as diet,
post-surgical infections or adjuvant treatment. Variations in serum
protein levels were quantified using reverse phase protein arrays
(RPPAs) and a set of 45 antibodies raised against 42 proteins
involved in angiogenesis, proliferation, apoptosis, inflammation or
wound healing (Table II). A
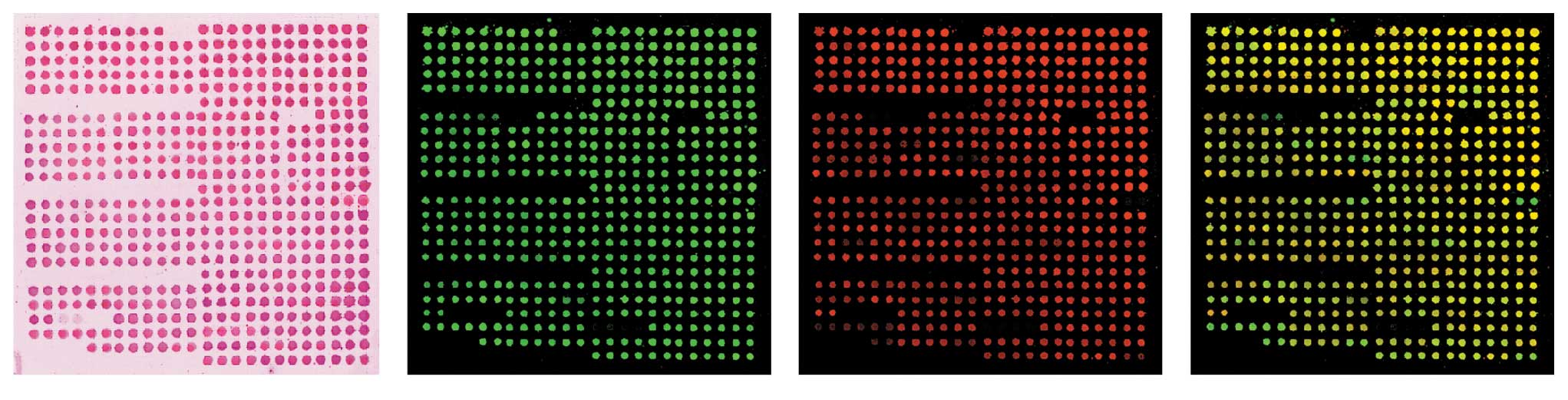
representative image of a hybridized array is shown in Fig. 1.
 | Table ICharacteristics of the study
population. |
Table I
Characteristics of the study
population.
| Invasive | In situ | Control |
|---|
| Mean age, years
(range) | 58 (27–87) | 54 (45–74) | 36 (18–55) |
| Hormonal
status | | | |
|
Premenopausal | 22 | 3 | 14 |
|
Postmenopausal | 24 | 4 | 2 |
| Surgery | | | |
| Mastectomy | 30 | 1 | - |
| Breast
conserving | 26 | 6 | 16 |
| Tumor size | | | |
| T1 (<2
cm) | 20 | | |
| T2 (2–5 cm) | 32 | | |
| T3 (>5
cm) | 2 | | |
| NA | 2 | | |
| Tumor grade | | | |
| I | 10 | | |
| II | 31 | | |
| III | 13 | | |
| NA | 2 | | |
| ER expression | | | |
| Negative | 16 | | |
| Positive | 40 | | |
| PgR expression | | | |
| Negative | 16 | | |
| Positive | 40 | | |
| HER2 status | | | |
| Negative | 35 | | |
| Positive | 13 | | |
| NA | 8 | | |
| Nodal status | | | |
| Negative | 26 | | |
| Positive | 25 | | |
| NA | 5 | | |
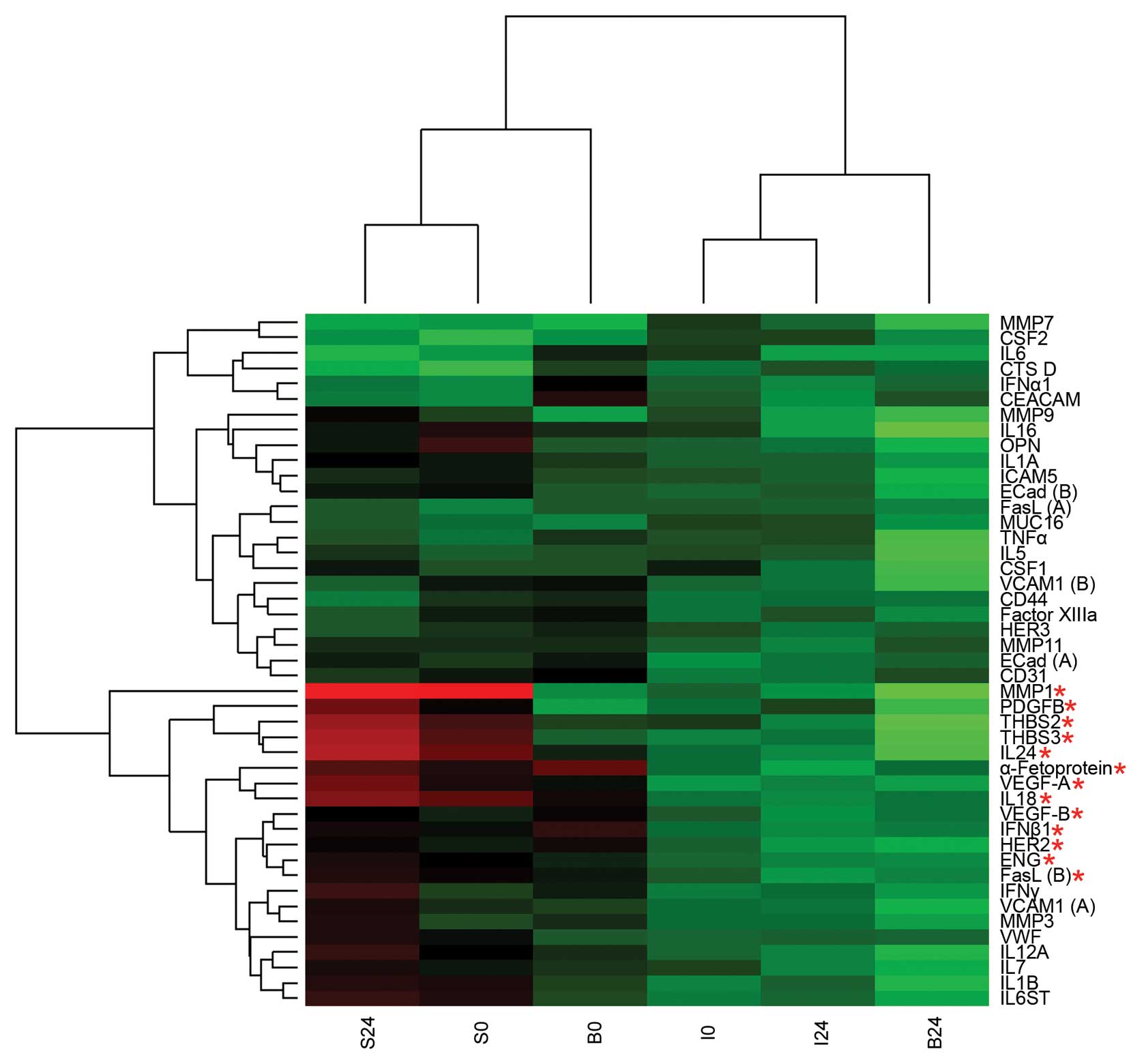
Mean levels of each antigen were calculated and
represented as a heat map (Fig.
2). Surgery-induced changes in the protein profiles of invasive
breast cancer patients (I), in situ breast cancer patients
(S) and fibroadenoma control subjects (B) were examined, with a
particular focus on common changes and specific differences that
occurred among the different groups of patients. Several cytokines
(CSF1, IL6, IL7 and IL16) and angiogenesis-related factors (THBS2
and VEGF-B) were consistently increased after surgery in the
overall population (Table III),
suggesting that these proteins are involved in a common response to
surgical injury in both breast cancer patients and healthy women.
However, we identified several proteins that were differentially
expressed in B samples before and after surgery (Fig. 2, red asterisks), most of which were
angiogenic factors; the concentrations of these proteins were
higher in samples collected after the surgical removal of the
primary tumor than in preoperative samples. Markedly, we discovered
that S and I samples were grouped based on the expression of this
same set of proteins. S samples clustered with preoperative B
samples whereas I samples clustered with postoperative B samples,
regardless of surgery.
 | Table IIIChanges after surgery in the overall
population. |
Table III
Changes after surgery in the overall
population.
| Soluble factor | Variation after
surgery (%) |
|---|
| IL16 | 11.9* |
| IL6 | 10.1* |
| CSF1 | 8.4 |
| THBS2 | 7.9 |
| HER2 | 7.2 |
| VEGF-B | 6.3 |
| IL7 | 6.2 |
| FasL | 6.2 |
Since most relapses are caused by invasive tumors,
we focused on specific differences between I and B or between I and
nI (noninvasive disease, which includes both in situ
carcinoma and fibroadenoma samples). Although we were unable to
detect dramatic differences in any single factor after comparing
groups, we observed significant variations in several proteins that
defined a specific pattern of response to surgery in invasive
breast cancer patients (Table IV).
We determined that, compared with noninvasive samples or healthy
controls, I samples were characterized by higher preoperative
concentrations of αFP, INFβ1, VEGF-A, IL18, soluble E-cadherin,
CD31, factor XIIIa, VEGF-A and IL18 and lower postoperative
concentrations of TNFα and IL5. Next, we examined the velocity of
accumulation of the 42 analytes. This rate of change over time has
been used to identify a signature of serum proteins associated with
breast cancer relapse (26) and
provides information about surgery-induced analyte dynamics
regardless of the initial concentration of each protein. Following
this approach, we compared the velocities of each analyte between I
and B as well as between I and nI, and we detected significant
differences in the velocities of VEGF-A and IL-16 (Table V). We also observed a decrease in
the velocity of endoglin accumulation in I compared with B and a
decrease in the velocity of IL24 accumulation in I compared with
nI.
 | Table IVSurgery-induced variations among
groups. |
Table IV
Surgery-induced variations among
groups.
| I vs. B (%)
| I vs. nI (%)
|
|---|
| Soluble factor | t(0) | t(24) | t(0) | t(24) |
|---|
| MMP7 | −10.6 | −8.2 | −9.7* | −7.5 |
| αFP | 21.8 | - | 19.3 | - |
| IFNβ1 | 15.8 | - | 14.1 | - |
| VEGF-A | 14.2 | - | 15.1* | - |
| IL18 | 12.9 | - | 15.5* | 9.6 |
| E-Cad (A) | 12.1 | - | 10.9 | - |
| CD31 | 12.1 | - | 11.5 | - |
| Factor XIIIa | 10.7 | - | 10.1 | - |
| IL24 | - | - | 12.3 | - |
| CSF2 | - | - | −7.9 | −6.9 |
| CD44 | - | - | 7.1 | - |
| TNFα | - | −12.7 | - | −9.2 |
| IL5 | - | −11.4 | - | −7.0 |
| PDGF-B | - | −11.8 | - | - |
 | Table VSurgery-induced changes in analyte
velocities. |
Table V
Surgery-induced changes in analyte
velocities.
| Soluble factor | logFC I vs. B | logFC I vs. nI |
|---|
| IFNβ1 | 20.77 | 14.56 |
| VEGF-A | −10.60 | −9.65 |
| IL16 | 26.51 | 17.93 |
| IL24 | −4.56 | - |
| ENG | - | −11.25 |
Discussion
Wound healing is a highly dynamic process that is
classically defined by three overlapping stages: inflammation,
tissue formation and tissue remodeling (27). These stages involve many different
cells, soluble factors and extracellular matrix molecules that
possess critical roles in tissue repair, formation of new blood
vessels and maintenance of homeostasis. Furthermore, these factors
act as chemoattractants to recruit white blood cells from the
bloodstream and promote a systemic response. Similarly, surgical
lesions activate the release of inflammatory and angiogenic
mediators that trigger the wound healing response. With regard to
cancer, the impact of surgery on serum factor dynamics is of
particular interest due to its involvement in the escape from
dormancy and cancer recurrence. In this study, we have gained
insight into the effect of breast surgery on the expression of 42
serum proteins and focused on the differential behavior that these
factors display between breast cancer patients and healthy controls
during the first 24 h following surgery.
In healthy women, the concentration of most of the
analyzed proteins increased after surgery. However, only a few of
them were commonly elevated in both controls and patients. Some of
these changes were expected, such as increased IL6 and CSF-1, due
to their established role in postoperative inflammation and
angiogenesis (28,29). Other proteins that exhibited
interesting changes in expression were THBS2, IL7 and IL16.
Although THBS2 was first described as an inhibitor of both
angiogenesis and tumor metastasis (30,31),
it is necessary for the proper regeneration of connective tissue
during wound healing (32) and its
serum levels rise in mice during the first 10 days after wound
induction (33). IL7 is involved
in skin repair (34) and enhances
endothelial cell growth and migration (35). IL16 is a potent chemoattractant for
white blood cells (36) and is
indirectly associated with neovascularization and wound healing
(37). These proteins have also
been shown to participate in cancer growth and metastasis (38–40).
We also examined the effect of surgery on the
protein profile of healthy women, invasive breast cancer patients
and in situ carcinoma breast cancer patients. Although
higher preoperative levels of most of the proteins were detected in
I rather than in S or B samples, a small increase or even decrease
in the expression of some of these proteins was detected after
surgery when compared with B. This observation suggests that the
sustained production of several factors by the tumor and the
surrounding stroma could lead to a systemic desensitization that
alters the response to surgery in cancer patients compared with
healthy women.
Angiogenesis is an essential process in wound
healing, however, it is also a hallmark of cancer (41). Vascularization is highly associated
with aggressive disease, invasiveness and worse outcome; invasive
tumors are often highly vascularized while in situ
carcinomas are poorly vascularized. In agreement with these
reports, our data indicate that the concentrations of several
angiogenesis-related proteins are lower in S than in I. Strikingly,
S was more similar to a preoperative B profile, while I was more
similar to B after breast surgery, once the angiogenic cascade is
triggered.
Several authors have used different proteomic
approaches to identify cancer-related serum biomarkers. Mass
spectrometry-based assays and antibody arrays have been used to
detect metastasis-related signatures in the serum samples of breast
cancer patients (26,42–44);
nevertheless, few of these studies have considered the effect of
surgery on the serum proteome. Based on a MALDI-TOF analysis,
Pietrowska et al examined therapy-induced changes in the
serum profiles of breast cancer patients (44). In their study, serum samples from
70 early breast cancer patients who underwent surgery and then
received adjuvant chemotherapy, radiotherapy or chemo-radiotherapy
were analyzed before the surgery (A), 7–14 days after the surgery
but before starting the adjuvant treatment (B) and 1 year after the
surgery, once the treatment was finished (C). Although several
variations between B and C were described, no clear differences
between A and B were detected. Thus, the authors suggested that
tumor resection had either minimal or no short-term influence on
the serum profile dynamics of breast cancer patients. Using an
antibody microarray platform, Carlsson et al recently
identified a signature of distant metastasis in serum samples from
64 breast cancer patients that were collected both before surgery
and 3–6 months after the removal of the primary tumor (26). These authors proposed the velocity
of accumulation to be a more sensitive and informative predictor
than analyte concentration at a given time. By analyzing this
parameter, the authors reported a 21-protein signature associated
with distant metastasis; strikingly, the velocity of accumulation
of IL16 was identified as an accurate predictor within this
signature, exhibiting higher values in metastatic breast cancer
samples compared with non-metastatic breast cancer samples. The
number of patients with early recurrence in our study cohort was,
however, insufficient to obtain any significant conclusion.
Instead, we detected higher velocities of IL16 accumulation during
the first 24 h in I samples compared with both B and nI samples,
although surgical injury increased serum IL16 levels in a
cancer-independent manner. Based on these data, we suggest that
IL16 may be involved in the surgery-driven escape from dormancy:
not only is IL16 behavior modified as a consequence of breast
surgery, but also higher IL16 velocities are observed in invasive
breast cancer and, as reported by Carlsson et al, IL16 may
be associated with the development of distal metastases.
Although surgical resection is the first line of
treatment for solid tumors, little is known about the direct
effects of surgery on the serum proteome, and even less is known
about how these effects may impact disseminated cancer cells. We
conducted a molecular dissection of the changes in several serum
proteins during the first postoperative 24 h. Inflammation and
tissue regeneration, the two main processes occurring during this
period of time, are stimulated by the production of soluble
mediators that ensure a systemic response to local injury. However,
we also identified specific variations in the serum protein
profiles of invasive breast cancer patients. These observed changes
in serum profiles may be strongly related to surgery-induced cancer
relapse and, in agreement with previous studies, an interruption of
dormancy in disseminated cancer foci.
Abbreviations:
|
αFP
|
α-fetoprotein;
|
|
CTS D
|
cathepsin D;
|
|
E-cad
|
E-cadherin;
|
|
ENG
|
endoglin;
|
|
FasL
|
Fas ligand;
|
|
HSA
|
human serum albumin;
|
|
IFN
|
interferon;
|
|
OPN
|
osteopontin;
|
|
RRPA
|
reverse phase protein array;
|
|
THBS
|
thrombospondin;
|
|
VWF
|
von Willebrand factor
|
Acknowledgements
The authors acknowledge support
through grants from the Junta de Andalucia (0199/2006 and
TIC-4026), the Fundacion Mutua Madrileña and the Spanish MINECO
(TIN2010-16556).
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Perou CM, Sørlie T, Eisen MB, van De Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Anderson WF and Matsuno R: Breast cancer
heterogeneity: a mixture of at least two main types? J Natl Cancer
Inst. 98:948–951. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Børresen-Dale AL, Sørlie T and Kristensen
VN: On the molecular biology of breast cancer. Mol Oncol.
4:171–173. 2010.
|
|
5.
|
Sørlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van De Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001.PubMed/NCBI
|
|
6.
|
Baum M, Demicheli R, Hrushesky W and
Retsky MW: Does surgery unfavourably perturb the ‘natural history’
of early breast cancer by accelerating the appearance of distant
metastases? Eur J Cancer. 41:508–515. 2005.PubMed/NCBI
|
|
7.
|
Demicheli R, Retsky MW, Hrushesky WJM,
Baum M and Gukas ID: The effects of surgery on tumor growth: a
century of investigations. Ann Oncol. 19:1821–1828. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Fehm T, Mueller V, Marches R, Klein G,
Gueckel B, Neubauer H, Solomayer E and Becker S: Tumor cell
dormancy: implications for the biology and treatment of breast
cancer. Acta Pathol Microbiol Immunol Scand. 116:742–753. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Tseng WW, Fadaki N and Leong SP:
Metastatic tumor dormancy in cutaneous melanoma: Does surgery
induce escape? Cancers. 3:730–746. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Retsky MW, Demicheli R, Hrushesky W, Baum
M and Gukas I: Surgery triggers outgrowth of latent distant disease
in breast cancer: An inconvenient truth? Cancers. 2:305–337. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Uhr JW and Pantel K: Controversies in
clinical cancer dormancy. Proc Natl Acad Sci USA. 108:1–5.
2011.
|
|
12.
|
Demicheli R, Retsky MW, Hrushesky WJM and
Baum M: Tumor dormancy and surgery-driven interruption of dormancy
in breast cancer: learning from failures. Nat Clin Pract Oncol.
4:699–710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Aguirre-Ghiso JA: Models, mechanisms and
clinical evidence for cancer dormancy. Nat Rev Cancer. 7:834–846.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Coffey JC, Wang JH, Smith MJF,
Bouchier-Hayes D, Cotter TG and Redmond HP: Excisional surgery for
cancer cure: therapy at a cost. Lancet Oncol. 4:760–768. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Spurrier B, Honkanen P, Holway A, Kumamoto
K, Terashima M, Takenoshita S, Wakabayashi G, Austin J and
Nishizuka S: Protein and lysate array technologies in cancer
research. Biotechnol Adv. 26:361–369. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Mueller C, La Liotta and Espina V: Reverse
phase protein microarrays advance to use in clinical trials. Mol
Oncol. 4:1–21. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Sheehan KM, Calvert VS, Kay EW, Lu Y,
Fishman D, Espina V, Aquino J, Speer R, Araujo R, Mills GB, et al:
Use of reverse phase protein microarrays and reference standard
development for molecular network analysis of metastatic ovarian
carcinoma. Mol Cell Proteomics. 4:346–355. 2005. View Article : Google Scholar
|
|
18.
|
Tibes R, Qiu Y, Lu Y, Hennessy B, Andreeff
M, Mills GB and Kornblau SM: Reverse phase protein array:
validation of a novel proteomic technology and utility for analysis
of primary leukemia specimens and hematopoietic stem cells. Mol
Cancer Ther. 5:2512–2521. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Brennan DJ, O’Connor DP, Rexhepaj E,
Ponten F and Gallagher WM: Antibody-based proteomics: fast-tracking
molecular diagnostics in oncology. Nat Rev Cancer. 10:605–617.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Grote T, Siwak DR, Fritsche HA, Joy C,
Mills GB, Simeone D, Whitcomb DC and Logsdon CD: Validation of
reverse phase protein array for practical screening of potential
biomarkers in serum and plasma: accurate detection of CA19-9 levels
in pancreatic cancer. Proteomics. 8:3051–3060. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Holdaway IM, Lethaby AE, Mason BH, Singh
V, Harman JE, MacCormick M and Civil ID: Effect of breast surgery
on serum levels of insulin-like growth factors (IGF-I, IGF-II, and
IGF binding protein-3) in women with benign and malignant breast
lesions. Ann Surg Oncol. 8:25–31. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irizarry
R and Huber W: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
23.
|
Yang YH and Thorne NP: Normalization for
two-color cDNA microarray data. Institute of Mathematical
Statistics; 2003
|
|
24.
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Gen Mol Biol. 3:Article3, Feb 12, 2004 (Epub
ahead of print).
|
|
25.
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J Roy Stat Soc Ser B (Stat Method). 57:289–300.
1995.
|
|
26.
|
Carlsson A, Wingren C, Kristensson M, Rose
C, Fernö M, Olsson H, Jernström H, Ek S, Gustavsson E, Ingvar C, et
al: Molecular serum portraits in patients with primary breast
cancer predict the development of distant metastases. Proc Natl
Acad Sci USA. 108:14252–14257. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Gurtner GC, Werner S, Barrandon Y and
Longaker MT: Wound repair and regeneration. Nature. 453:314–321.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Hamilton JA: Colony-stimulating factors in
inflammation and autoimmunity. Nat Rev Immunol. 8:533–544. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Lin Z-Q, Kondo T, Ishida Y, Takayasu T and
Mukaida N: Essential involvement of IL-6 in the skin wound-healing
process as evidenced by delayed wound healing in IL-6-deficient
mice. J Leukoc Biol. 73:713–721. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Oshika Y, Masuda K, Tokunaga T, Hatanaka
H, Kamiya T, Abe Y, Ozeki Y, Kijima H, Yamazaki H, Tamaoki N, et
al: Thrombospondin 2 gene expression is correlated with decreased
vascularity in non-small cell lung cancer. Clin Cancer Res.
4:1785–1788. 1998.PubMed/NCBI
|
|
31.
|
Tokunaga T, Nakamura M, Oshika Y, Abe Y,
Ozeki Y, Fukushima Y, Hatanaka H, Sadahiro S, Kijima H, Tsuchida T,
et al: Thrombospondin 2 expression is correlated with inhibition of
angiogenesis and metastasis of colon cancer. Br J Cancer.
79:354–359. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Maclauchlan S, Skokos EA, Agah A, Zeng J,
Tian W, Davidson JM, Bornstein P and Kyriakides TR: Enhanced
angiogenesis and reduced contraction in thrombospondin-2-null
wounds is associated with increased levels of matrix
metalloproteinases-2 and -9, and soluble VEGF. J Histochem
Cytochem. 57:301–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Kyriakides TR, Tam JW and Bornstein P:
Accelerated wound healing in mice with a disruption of the
thrombospondin 2 gene. J Invest Dermatol. 113:782–787. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Jameson J and Havran WL: Skin gammadelta
T-cell functions in homeostasis and wound healing. Immunol Rev.
215:114–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Al-Rawi MAA, Watkins G, Mansel RE and
Jiang WG: The effects of interleukin-7 on the lymphangiogenic
properties of human endothelial cells. Int J Oncol. 27:721–730.
2005.PubMed/NCBI
|
|
36.
|
Cruikshank W and Little F: lnterleukin-16:
the ins and outs of regulating T-cell activation. Crit Rev Immunol.
28:467–483. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Stabile E, Kinnaird T, la Sala A, Hanson
SK, Watkins C, Campia U, Shou M, Zbinden S, Fuchs S, Kornfeld H, et
al: CD8+ T lymphocytes regulate the arteriogenic
response to ischemia by infiltrating the site of collateral vessel
development and recruiting CD4+ mononuclear cells
through the expression of interleukin-16. Circulation. 113:118–124.
2006.
|
|
38.
|
Al-Rawi MAA, Rmali K, Watkins G, Mansel RE
and Jiang WG: Aberrant expression of interleukin-7 (IL-7) and its
signalling complex in human breast cancer. Eur J Cancer.
40:494–502. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Germano G, Allavena P and Mantovani A:
Cytokines as a key component of cancer-related inflammation.
Cytokine. 43:374–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Studebaker AW, Storci G, Werbeck JL,
Sansone P, Sasser AK, Tavolari S, Huang T, Chan MWY, Marini FC,
Rosol TJ, et al: Fibroblasts isolated from common sites of breast
cancer metastasis enhance cancer cell growth rates and invasiveness
in an interleukin-6-dependent manner. Cancer Res. 68:9087–9095.
2008. View Article : Google Scholar
|
|
41.
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar
|
|
42.
|
Carlsson A, Wingren C, Ingvarsson J,
Ellmark P, Borrebaeck CAK, Baldertorp B, Fernö M and Olsson H:
Serum proteome profiling of metastatic breast cancer using
recombinant antibody microarrays. Eur J Cancer. 44:472–480. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Gast M-CW, Zapatka M, van Tinteren H,
Bontenbal M, Span PN, Tjan-Heijnen VCG, Knol JC, Jimenez CR,
Schellens JHM and Beijnen JH: Postoperative serum proteomic
profiles may predict recurrence-free survival in high-risk primary
breast cancer. J Cancer Res Clin Oncol. 137:1773–1783. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Pietrowska M, Polanska J, Marczak L,
Behrendt K, Nowicka E, Stobiecki M, Polanski A, Tarnawski R and
Widlak P: Mass spectrometry-based analysis of therapy-related
changes in serum proteome patterns of patients with early-stage
breast cancer. J Transl Med. 8:662010. View Article : Google Scholar : PubMed/NCBI
|