Introduction
Ovarian cancer is the most lethal gynecologic
cancer, with high rate of mortality all over the world. Early
stages of ovarian cancer are generally asymptomatic and thus
diagnosis usually occurs after the disease has disseminated beyond
the ovaries (1). Therefore, 70% of
ovarian cancer patients are diagnosed with advanced-stage disease
and 5-year survival rates are less than 40%, with only modestly
improved survival over the past 40 year (2). Although the standard taxane/platinum
regimen achieves a complete response rate of 40 to 60% in advanced
ovarian cancer patients (2),
relapse occurs in over 70% of the patients, resulting in drug
resistance and finally leading to fatal disease (3).
Drug resistance in ovarian cancer normally develops
after the treatments to advanced stage cancer patients with
chemotherapies (3) and associates
with aberrant expression of some genes, such as tumor suppressor
genes (TSGs) and oncogenes. At least 16 candidate TSGs, 15
oncogenes, many other genes and more than 7 signaling pathways have
been implicated in aberrations in cell proliferation, apoptosis,
autophagy and changes in cell adhesion and motility in ovarian
cancer. All of these cellular processes contribute to cancer
development and metastasis (4).
Among all the genes and signaling pathways participated in the
development of ovarian cancer, genes [such as p53(5,6),
BRCA1(7,8), BRCA2(8) and ERBB2(9)] and pathways [such as p53 signaling
pathway (10) and mTOR signaling
pathway (11)] related to drug
resistance have been identified, suggesting that genes contributing
to advanced-stage ovarian cancer would also be noteworthy in drug
resistance.
The latest published research of ovarian cancer
reveals that CCL21 and SPARCL1 are noteworthy in
ovarian cancer because their promoters are hypermethylated and
silenced in the vast majority of the tumors (in a total of 489
high-grade serous ovarian adenocarcinomas), which is even more
notable than BRCA1 for which the promoter is hypermethylated
and silenced in only 56 of 489 (11.5%) tumors (12). BRCA1 is important in the
development of ovarian cancer and is involved in survival (13), metastasis (14), apoptosis (15) and drug resistance (16,17).
These findings suggest that CCL21 and SPARCL1 could
play important roles in advanced stage ovarian cancers (12).
In this study, based on the comprehensively
bioinformatics analysis through motif analysis, literature
co-occurrence, protein-protein interaction network, protein-small
molecule interaction network and microRNAs (miRNAs) enrichments, we
found that CCL21 and SPARCL1 directly or indirectly
interacted with many genes, proteins, small molecules and pathways
associated with drug resistance in ovarian cancer and other
cancers, suggesting that CCL21 and SPARCL1 might
contribute to drug resistance in ovarian cancer.
Materials and methods
The target genes (CCL21 and SPARCL1)
silenced in the vast majority of advanced-stage ovarian
adenocarcinomas (12) were
selected for bioinformatics analysis.
The motif analysis of proteins was performed with
SSDB Motif Search in Kyoto Encyclopedia of Genes and Genomes (KEGG)
online database (http://www.genome.jp/kegg/); the pathway searches were
performed with KEGG online database and GENEGO online database
(http://www.genego.com/). Protein domain
interactions were analyzed by DOMINE online database (18,19)
(http://domine.utdallas.edu/cgi-bin/Domine).
Literature Co-Occurrence was performed with Pubgene
online tool (20) (http://www.pubgene.org/index.cgi); the
gene/protein-gene/protein interaction network was generated with
GeneMANIA (21) (http://www.genemania.org/); the protein-small
molecules interaction network was generated with BiologicalNetworks
2 software (22) (downloaded from
http://biologicalnetworks.net/Software/index.php).
The miRNA target-gene prediction was performed by
miRWalk online tool (23)
(http://www.umm.uni-heidelberg.de/apps/zmf/mirwalk/),
for which 7 prediction programs (miRanda, miRDB, miRWalk, PICTAR4,
PICTAR5, RNA22 and Targetscan) were selected, and the same miRNA
predicted by at least 3 of these software was selected for
subsequent analysis. The pathway enrichment analysis of miRNAs was
performed with DIANA-mirPath web server (24) (http://diana.cslab.ece.ntua.gr/pathways/).
Results
The functions of CCL21 and SPARCL1 in
cancers
CCL21 is one of the chemokines which belong to the
small molecule chemoattractive cytokine family. Chemokines mediate
their chemical effect on target cells through G-protein-coupled
receptors, which are characterized structurally by 7 transmembrane
spanning domains and involved in the attraction and activation of
mononuclear and polymorphonuclear leukocytes (25). CCL21 participated in three
pathways, cytokine-cytokine receptor interaction, chemokine
signaling pathway and NF-κB signaling pathway, based on the
searches in KEGG database, and participated in apoptosis and
survival_Lymphotoxin-β receptor signaling pathway based on the
searches in GENEGO database. Besides, CCL21 plays roles in the
regulation of ERK pathway in human non-small cell lung cancer cells
(26). CCL21 is downregulated in
many cancers and associated with lymph node metastasis, poor
prognosis (27), apoptosis
(26), cell cycle (28) and tumor growth (29). SPARCL1 belongs to SPARC family that
contains ten protein members (30). SPARCL1 is widely expressed in
normal and cancer tissues, and it was initially identified as an
anti-adhesive extracellular matrix protein with anti-proliferative
effects mediated through cell-cell adhesion (31,32).
In addition, SPARCL1, which is downregulated in several tumor types
such as colorectal and gastric cancer, is associated with tumor
diagnosis, progression and prognosis (30,33,34).
Therefore, SPARCL1 is considered to be a TSG (35) and may have many further unexplored
functions in cancer development.
However, the studies on CCL21 and SPARCL1 associated
with drug resistance are rare. Only one study reports that SPARCL1
is an extracellular matrix remodeling gene and may contribute to
drug resistance in pediatric osteosarcoma (36). The research on CCL21 and SPARCL1 in
ovarian cancer is limited. It is reported that CCL21 potentiated
the cytotoxicity to ovarian cancer cells (37) and SPARCL1 is inactivated in ovarian
cancer (38). More recently, CCL21
and SPARCL1 are noteworthy in ovarian cancer because they are
silenced in the vast majority of high-grade serous ovarian
adenocarcinomas (12).
Function prediction through
motif-based approaches
Conserved protein sequence motifs are short
stretches of amino acid sequence patterns that potentially encode
the function of proteins (39).
Except in CfCCL21, IL8 domain (accession: PF00048) was a unique and
highly conserved motif in human CCL21 and its homologous proteins
(Table I) according to SSDB Motif
Search, indicating that IL8 domain might contribute to the function
of CCL21. IL8 domain originally came from IL8 protein which closely
related to drug resistance in many solid tumors and cancer cells.
The upregulated expression levels of IL6 and IL8 may contribute to
multidrug resistance in human breast cancer cells (40). Similarly, IL8 is overexpressed in
paclitaxel resistance SKOV3 cells, and therefore is considered to
be associated with paclitaxel resistance (41). These studies suggested that IL8
domain might closely relate to drug resistance, indicating that
CCL21 might associate with drug resistance.
 | Table IThe motifs of CCL21 and its
homologous proteins according to SSDB Motif Search. |
Table I
The motifs of CCL21 and its
homologous proteins according to SSDB Motif Search.
| Protein | KEGG ID | Motif
|
|---|
| IL8 | YqzE |
|---|
| HsCCL21 | hsa:6366 | * | - |
| AmCCL21-like | aml:100480794 | * | - |
| BtCCL21 | bta:511112 | * | - |
| CfCCL21 | cfa:448796 | * | * |
| EcCCL21-like | ecb:100060619 | * | - |
| EcCCL21-like | ecb:100063059 | * | - |
| MmCCL21 | mcc:574183 | * | - |
| MdCCL21-like | mdo:100028728 | * | - |
| MmCCL21-like | mmu:100041504 | * | - |
| MmCCL21-like | mmu:100041593 | * | - |
| MmCCL21B | mmu:100042493 | * | - |
| MmCCL21C-like | mmu:100862177 | * | - |
| MmCCL21A | mmu:18829 | * | - |
| MmCCL21C | mmu:65956 | * | - |
| OaCCL21-like | oaa:100092451 | * | - |
| PtCCL21 | ptr:746205 | * | - |
| RnCCL21 | rno:298006 | * | - |
| SsCCL21 | ssc:448797 | * | - |
| XtCCL21A-like | xtr:100490971 | * | - |
Four motifs comprising FOLN domain (accession:
PF09289), Kazal_1 domain (accession: PF00050), Kazal_2 domain
(accession: PF07648) and SPARC_Ca_bdg region (accession: PF10591)
were highly conserved in human SPARCL1 and its homologous proteins
in other species (Table II),
suggesting that these motifs might closely relate to the functions
of SPARCL1. Besides, efhand domain (accession: PF00036) was also
observed in most SPARCL1 proteins. It has been reported that serine
protease inhibitor Kazal-type 1, which contains Traw_N, Kazal_1 and
Kazal_2 domains, affects multiple aggressive properties in breast
cancer such as survival, invasiveness, and chemoresistance
(42). Similarly, serine
proteinase inhibitor Kazal-type 2, which contains only Kazal_1 and
Kazal_2 domains, is also found to play an important role in tumor
progression and response to the treatment in leukemia cell lines
(43). These studies indicated
that Kazal_1 and Kazal_2 domains might be associated with drug
resistance in cancers. A previous study revealed that protein
phosphatase with efhand domain may correlate with stress protective
responses, cell survival, growth, proliferation and drug resistance
(44). S100P with efhand domain is
detected in a spectrum of human tumor cell lines and tissues
derived from prostate, pancreas, breast, lung and colon, in which
it is connected with malignant phenotype, hormone independence and
resistance to chemotherapy (45).
These studies indicated that efhand domain might also associate
with drug resistance in cancers. In addition, on the basis of
DOMINE online analysis, FOLN domain interacted with Kazal_1 domain,
SPARC_Ca_bdg region interacted with FOLN and Kazal_1 domains and
efhand domain interacted with Kazal_1 domain, suggesting that all
these conserved motifs of SPARCL1 were closely related and
interacted with each other. Taken together, we concluded that FOLN,
Kazal_1, Kazal_2, SPARC_Ca_bdg, and efhand conserved in SPARCL1
were associated with drug resistance in cancers.
 | Table IIThe motifs of SPARCL1 and its
homologous proteins according to SSDB Motif Search. |
Table II
The motifs of SPARCL1 and its
homologous proteins according to SSDB Motif Search.
| Protein | KEGG ID | Motif
|
|---|
| EF_hand_3 | EF_hand_4 | EF_hand_5 | FOLN | Kazal_1 | Kazal_2 | SPARC_Ca_bdg | SSURE | efhand |
|---|
| HsSPARCL1 | hsa:8404 | - | - | - | * | * | * | * | - | * |
| HsSPARC | hsa:6678 | - | - | - | * | * | * | * | - | - |
| AcSPARC-like | acs:100554604 | - | - | - | * | * | * | * | - | * |
| AmSPARCL1 | aml:100476369 | - | - | - | * | * | * | * | - | * |
| BmSPARC
precursor | bmy:Bm1_31690 | * | - | - | * | * | * | * | - | * |
| BtSPARCL1 | bta:507537 | - | - | - | * | * | * | * | - | * |
| Cbost-1 | cbr:CBG22551 | * | * | - | * | * | * | * | - | * |
| Ceost-1 | cel:C44B12.2 | * | * | - | * | * | * | * | - | * |
| CfSPARCL1 | cfa:478470 | - | * | - | * | * | * | * | - | * |
| DrSPARCL1 | dre:567331 | - | - | - | * | * | * | * | - | * |
| EcSPARCL1 | ecb:100052928 | - | * | - | * | * | * | * | - | * |
| GgSPARCL1 | gga:422586 | - | - | - | * | * | * | * | - | * |
| MmSPARCL1 | mcc:701468 | - | - | - | * | * | * | * | - | * |
| MdSPARC-like | mdo:100024756 | - | - | - | * | * | * | * | - | - |
| MgSPARC-like
protein 1-like | mgp:100541910 | - | - | - | * | * | * | * | - | * |
| MmuSPARCL1 | mmu:13602 | - | * | - | * | * | * | * | - | * |
| OaSPARC-like
protein 1-like | oaa:100082443 | - | * | - | * | * | * | * | * | * |
| PaSPARCL1 | pon:100173668 | - | - | - | * | * | * | * | - | * |
| PtSPARCL1 | ptr:471247 | - | - | - | * | * | * | * | - | * |
| RnSPARCL1 | rno:25434 | - | * | - | * | * | * | * | - | * |
| SsSPARCL1 | ssc:100037275 | - | - | - | * | * | * | * | - | - |
| TgSPARC-like
protein 1 | tgu:100219989 | - | - | - | * | * | * | * | - | - |
| TsSPARC | tsp:Tsp_03863a | - | * | - | * | * | * | * | - | - |
| XlSPARC | xla:379277 | - | * | * | * | * | * | * | - | * |
| XtSPARC | xtr:394973 | - | * | * | * | * | * | * | - | * |
Function prediction and analysis based on
interaction networks
Function prediction and analysis based
on literature co-occurrence
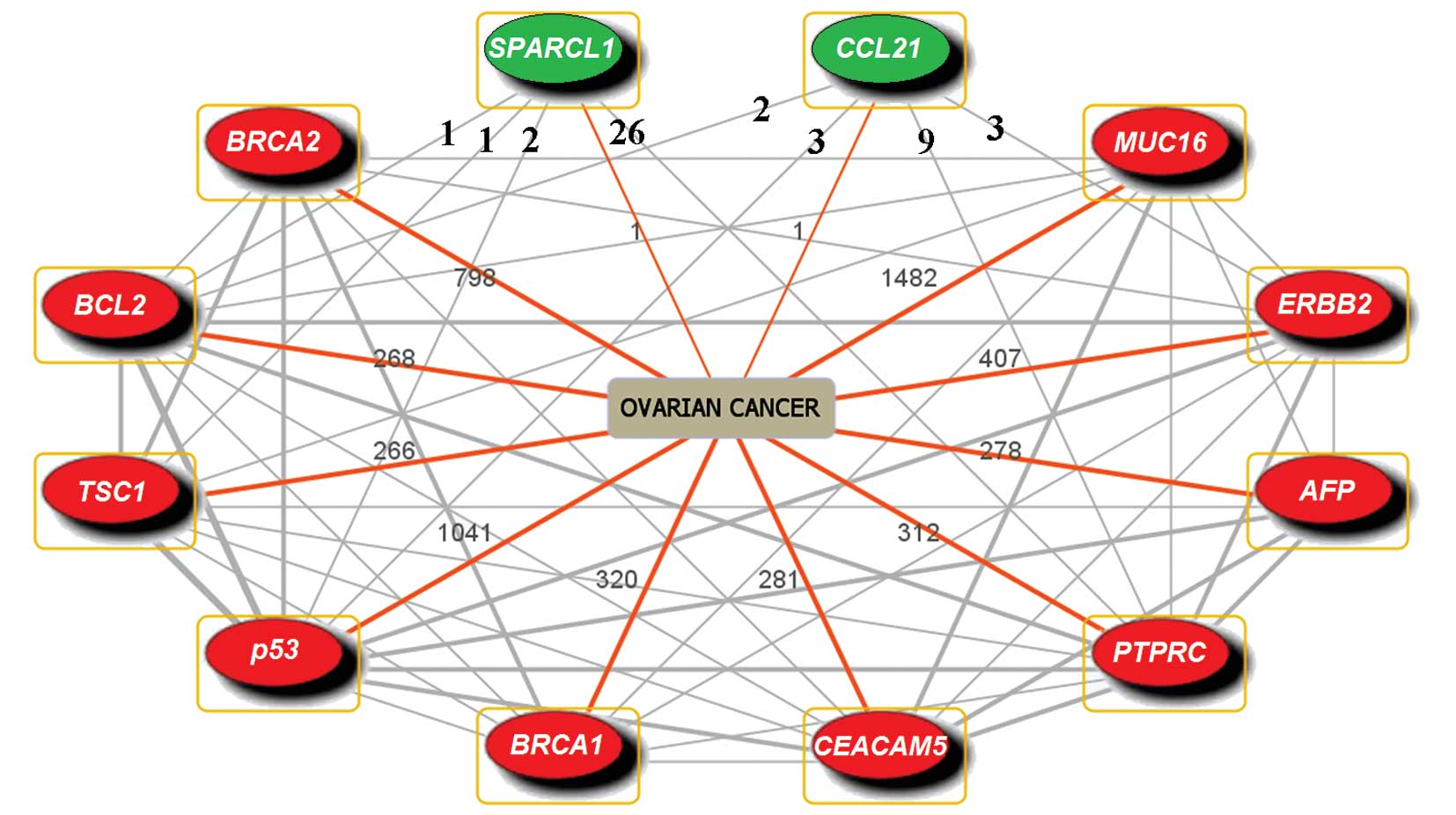
The involvement of CCL21 and SPARCL1 in cancer drug
resistance had not been reported on the basis of literature
co-occurrence, whereas there were 10 genes co-occurring with CCL21
and SPARCL1 in ovarian cancer (Fig.
1). Among those 10 genes, p53, BRCA1 and BRCA2 are well-known
TSGs, and downregulation of these 3 genes contributes to the
enhancement of drug resistance in ovarian cancer (5–8).
ERBB2 and BCL2 are oncogenes; ERBB2 takes part in drug resistance
in ovarian cancer (9), while BCL2
is reported to participate in drug resistance in other cancers
(46,47). Besides, AFP is a drug
resistance-related gene which plays a role in the expression of
P-glycoprotein (48); TSC1 is a
putative TSG participating in the signaling pathway of the
mammalian target of rapamycin (mTOR) associated with proliferation,
survival and drug resistance in leukemia cells (49); PTPRC, an apoptosis-related gene
near cis-regulatory elements (50), is regarded as underexpression in
breast cancer (51), suggesting
that this gene may relate to drug resistance.
The involvement of CCL21 and SPARCL1 in ovarian
cancer has rarely been studied. We observed that CCL21 had
co-occurrences with p53, BCL2, PTPRC and ERBB2; SPARCL1 had
co-occurrence with p53, BCL2, PTPRC and TSC1 (Fig. 1), suggesting that they might
interact directly or indirectly. Taken together, we found that 8 in
10 genes which had co-occurrences with CCL21 and SPARCL1 in
‘ovarian cancer’ were drug resistance-related genes in ovarian and
other cancers, suggesting that CCL21 and SPARCL1 might also be
involved in drug resistance.
Function prediction and analysis based
on gene/protein-gene/protein interactions.
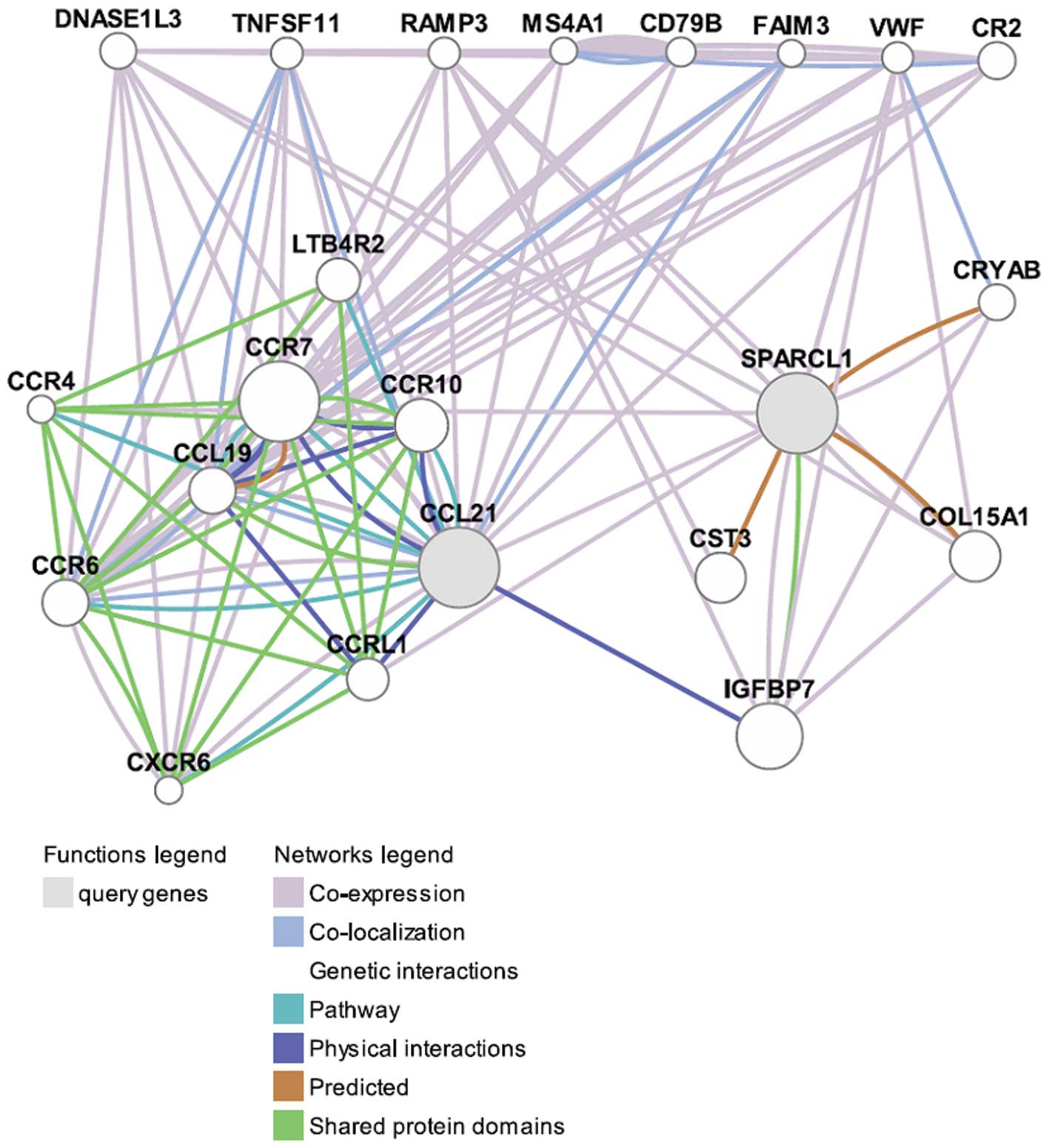
The functions of CCL21 and SPARCL1 were predicted
using GeneMANIA (as shown in Fig.
2). CCL21 was co-expressed, co-localized, physically
interacted, shared protein domains and pathways with a number of
proteins, especially with CCL19, CCR7 and CCR6, suggesting that
they were functionally related. In comparison, SPARCL1 had
considerably fewer interactions with other proteins.
Based on the annotated functions in accordance with
the GeneMANIA network (Table III),
CCL21, together with other proteins, played important roles in the
regulation of leukocytes, neutrophil chemotaxis, G-protein coupled
receptor activity and calcium ion. It has been proven that
leukocytes have close relationship with drug resistance, both in
vivo and in vitro. In a ‘blinded’ study of 21 patients
receiving combination cisplatin/carboplatin treatments, there was a
direct relationship between DNA damage in leukocytes and disease
response, and in leukocytes in vivo, persistence and
accumulation are prominent features of the cisplatin-DNA adduct
profile (52). Neutrophil
chemotaxis seemed to be associated with drug resistance in an
indirect way. For instance, celastrol is identified as an inhibitor
of neutrophil chemotaxis, and it induces synergistic apoptosis when
combined with conventional microtubule-targeting drugs and
manifested efficacy toward taxol-resistant cancer cells at the
cellular level (53). Similarly,
stress and drug-induced interleukin-8 (IL8) signaling has been
shown to confer chemotherapeutic resistance in cancer cells, while
IL8 is a proinflammatory CXC chemokine contributed to the promotion
of neutrophil chemotaxis and degranulation (54). G protein-coupled receptors
essentially regulate all cellular processes, including those that
are fundamental to cancer pathology, such as differentiation,
proliferation, migration, tissue invasion, survival and drug
resistance (55). Calcium content
increases in multidrug resistant (MDR) cells and the resistance
could be reversed by the calcium channel blocker verapamil,
suggesting that calcium ion may play a role in drug resistance
(56,57).
 | Table IIIThe annotated functions of CCL21 and
other proteins related to drug resistance in GeneMANIA network (as
shown in Fig. 2). |
Table III
The annotated functions of CCL21 and
other proteins related to drug resistance in GeneMANIA network (as
shown in Fig. 2).
| GO annotation | FDR (n/a)a | Genes/proteins in
the network |
|---|
| Regulation of
leukocyte chemotaxis, apoptosis, migration and activation | 6.22E-05 to
1.44E-02 | CCL21, CCL19, CCR7,
CCR6, TNFSF11 |
| Regulation of
neutrophil chemotaxis | 2.92E-04 to
1.44E-03 | CCL21, CCL19,
CCR7 |
| G-protein coupled
receptor activity | 3.18E-05 to
9.39E-03 | CCR7, CCR6, CCRL1,
CCR4 |
| Calcium ion
concentration, homeostasis, transportation and sequestering | 1.57E-04 to
6.21E-02 | CCL21, CCL19, CCR7,
CCR6, CCR4 |
| Positive regulation
of cell adhesion | 7.01E-03 | CCL21, CCR7,
TNFSF11 |
| Receptor-mediated
endocytosis | 1.80E-02 | CCL21, CCL19,
RAMP3 |
| Cell-substrate
adhesion | 3.70E-02 | CCL21, CCR7,
VWF |
There was no annotated function for SPARCL1 based on
GeneMANIA, but we could deduce its function through its
interactions with other proteins. As shown in Fig. 2, SPARCL1 was co-expressed with many
proteins such as RAMP3 and VWF. RAMP3 is associated with
receptor-mediated endocytosis which is involved in drug resistance.
Hsp47/CBP2 is a favorable candidate for targeted delivery of
anticancer drugs in human squamous cell carcinoma of the head and
neck, and the uptake of the targeted conjugate is inhibited in the
presence of an anti-Hsp47 antibody, suggesting the involvement of
active receptor mediated endocytosis in cell entry of the conjugate
(58). The VWF is related to
cell-substrate adhesion which is also involved in drug resistance.
It has been proven that cell-substrate adhesion contributes to drug
resistance via apoptosis in acute myeloid leukaemia, small cell
lung cancer cells, breast cancer and glioblastoma cells (59).
In addition, CCL21 and SPARCL1 were co-expressed and
interacted with each other indirectly through interacting with
other proteins. IGFBP7 was found to be the most important one for
CCL21 and SPARCL1 interactions. IGFBP7 had very strong physical
interactions with CCL21 and shared protein domains with SPARCL1,
indicating that CCL21, IGFBP7 and SPARCL1 might be functionally
related. IGFBP7 has been identified as one of these factors
responsible for the establishment and/or maintenance of
oncogene-induced senescence, and has been shown to be a TSG in a
variety of solid cancers (60).
Aberrant expression of IGFBP7 in adult leukemia is correlated with
chemotherapy resistance and shorter survival. Addition of IGFBP7 to
leukemic cell lines inhibits cell growth without induction of
apoptosis or senescence, suggesting a role of IGFBP7 in
contributing to drug resistance through reduced sensitivity to
cytostatic drugs (61).
Function prediction and analysis based
on protein-small molecules interactions
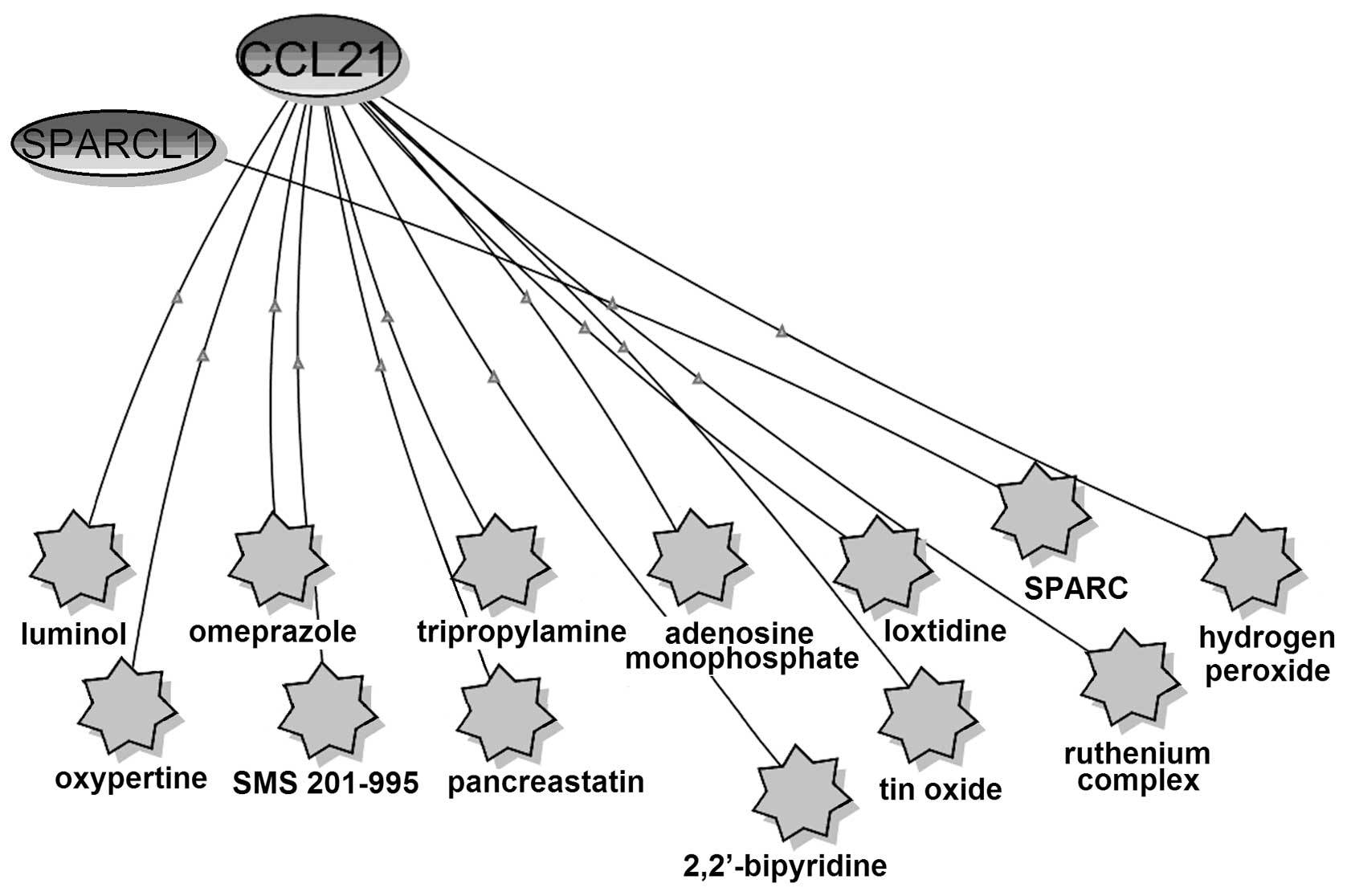
The relationship of CCL21, SPARCL1 and small
molecules were analyzed using BiologicalNetworks (Fig. 3). SPARCL1 had co-citation with only
one small molecule, SPARC, which is the peptides of SPARC protein
(SPARC113–130 and SPARC54–73) (62). SPARCL1 exhibits 62% identity with
the anti-adhesive extracellular matrix protein SPARC, over a region
of 232 aa spanning more than four-fifths of the SPARC coding
sequence (63). SPARCL1 shared
four domains (FOLN, Kazal_1, Kazal_2 and SPARC_Ca_bdg) with SPARC
based on the SSDB Motif Search results (Table II). Thus, SPARCL1 might have
similar functions with SPARC. SPARC is a candidate TSG and a
putative resistance-reversal gene and plays an important part in
drug resistance in ovarian cancer (64,65).
Therefore, we concluded that SPARCL1, which is considered as a TSG
(35), might also contribute to
drug resistance in ovarian cancer.
CCL21 had co-citations with 12 small molecules, and
half of them comprising omeprazole, SMS 201–995, adenosine
monophosphate, ruthenium complex, hydrogen peroxide and
2,2′-bipyridine are associated with drug resistance in cancers.
In vivo experiments show that oral pretreatment with
omeprazole induces a sensitivity of human solid tumors to
anticancer drugs (66). SMS
201–995 is proven to stimulate prostatic tumor growth and may
sensitize tumor cells to subsequent chemotherapy (67). Adenosine monophosphate may
participate in drug resistance of ovarian cancer through adenosine
monophosphate-activated protein kinase pathway (68). The ruthenium complexes are
effective tumor-inhibiting drugs in experimental therapy of
autochthonous colorectal carcinomas in rats, and they can be
promising candidate drugs in the second-line treatment of
colorectal cancers resistant to other cytostatic drugs (69). Thioredoxin has much higher levels
in all cisplatin-resistant human bladder and prostatic cancer cell
lines compared with their drug-sensitive parental counterpart, and
downregulation of its expression can increase cell sensitivity to
cisplatin and also to other superoxide-generating agents including
hydrogen peroxide, suggesting that hydrogen peroxide may also
relate to drug resistance (70);
2,2′-bipyridine is a well-characterized chelating agent known to
have anti-proliferative activity that links to drug resistance
(71).
Function prediction and analysis based
on KEGG pathways modulated by miRNAs
Total of 37 and 31 miRNAs were predicted to be the
transcriptional targets of CCL21 and SPARCL1 through miRWalk,
respectively. The pathway enrichment analysis of those miRNAs was
performed with DIANA-mirPath, and an overview of the parts of the
pathway modulated by miRNAs was integrated. Among all the pathways
modulated by miRNAs targeted CCL21 and SPARCL1, 11 of them are
involved in drug resistance in ovarian and many other cancers
(Table IV).
 | Table IVThe drug resistance-related pathways
modulated by miRNAs targeted CCL21 |
Table IV
The drug resistance-related pathways
modulated by miRNAs targeted CCL21
| Gene | miRNAa | KEGG
pathwaysb | Pathway ID | - ln (p-value)
(union) | Pathways
contributing to drug resistance in cancers |
|---|
| CCL21 |
hsa-miR-331-5p, | Wnt signaling
pathway | hsa04310 | 17.64 | Ovarian cancer
(77,78); colon cancer cells (91) |
| 338-3p, 608,
631, | MAPK signaling
pathway | hsa04010 | 9.16 | Ovarian cancer
(76); |
| 205, 330-5p,
574-5p, 876-3p, | Cell adhesion
molecules (CAMs) | hsa04514 | 6.78 | Ovarian cancer
(79) |
| 125b, 492,
637, | p53 signaling
pathway | hsa04115 | 3.6 | Ovarian cancer
(10); lung cancer (92) |
| 138, 498, 644, 939,
647, 604, | Cell cycle | hsa04110 | 1.71 | Ovarian cancer
(82); lung cancer (92); breast cancer cells (92,93) |
| 518e, 654-5p, | Cell
Communication | hsa01430 | 1.68 | Ovarian cancer
(80) |
| 484, 296-5p, | mTOR signaling
pathway | hsa04150 | 1.59 | Ovarian cancer
(11); lung cancer cells (94,95) |
| 767-3p, 138, | Apoptosis | hsa04210 | 1.51 | Ovarian cancer
(85,86) |
| 485-5p, 370,
541, | VEGF signaling
pathway | hsa04370 | 1.45 | Ovarian cancer
(87); other human cancer
(96,97) |
| 125a-5p, 487a, 7,
331-5p, | Regulation of
autophagy | hsa04140 | 0.66 | Ovarian cancer
(88); hematological cancers
(98,99) |
| (30, 1248, 1279,
1178, 1237, 1293)c | ABC transporters -
General | hsa02010 | 0 | Ovarian cancer
(89,90); other human cancers (100,101) |
| SPARCL1 |
Has-miR-450b-5p | Wnt signaling
pathway | hsa04310 | 24.87 | Ovarian cancer
(77,78); colon cancer cells (91) |
| 431, 586, 448, | MAPK signaling
pathway | hsa04010 | 15.93 | Ovarian cancer
(76) |
| 101, 519b-3p, | mTOR signaling
pathway | hsa04150 | 7.36 | Ovarian cancer
(11); lung cancer cells (94,95) |
| 569, 875-3p, | p53 signaling
pathway | hsa04115 | 3.29 | Ovarian cancer
(10); lung cancer (92) |
| 519a, 140-5p, 633,
369-3p, 144, | Cell cycle | hsa04110 | 2.41 | Ovarian cancer
(82); lung cancer (92); breast cancer cells (92,93) |
| 485-3p, 655,
105, | ABC transporters -
General | hsa02010 | 1.68 | Ovarian cancer
(89,90); other human cancers (100,101) |
| 373, 507, 153, 656,
338-5p, 561, | Apoptosis | hsa04210 | 1.34 | Ovarian cancer
(85,86) |
| 448, 569, | Cell
Communication | hsa01430 | 1.24 | Ovarian cancer
(80) |
| 519c-3p, | Cell adhesion
molecules (CAMs) | hsa04514 | 0.55 | Ovarian cancer
(79) |
| (25, 1179, 1287,
1290, 1283, 513b)c | Regulation of
autophagy | hsa04140 | 0.28 | Ovarian cancer
(88); hematological cancers
(98,99) |
Van Jaarsveld et al(72) systematically reviewed the miRNAs
related to drug resistance in ovarian cancer. Among the miRNAs,
some were the transcriptional targets of CCL21 and SAPRCL1
(Table IV). For instance,
hsa-miR-125b and hsa-miR-370 were the targets of CCL21.
Hsa-miR-125b is downregulated in paclitaxel resistant A2780 cell
lines, therefore suggesting a direct involvement in the development
of chemoresistance (73).
Hsa-miR-370 is upregulated in platinum resistant EOC (74), suggesting that hsa-miR-370 may
contribute to drug resistance through downregulation its target
genes. Similarly, hsa-miR-431, the transcriptional target of
SPARCL1, is also upregulated in topotecan resistant ovarian cancer
cells (75). The pathway
enrichment analysis of miRNAs related to drug resistance in ovarian
cancer has also been studied. For example, it has been reported
that 11 miRNAs are differentially expressed in cisplatin resistant
ovarian cancer cells, which potentially target many important
pathways comprising MAPK, Wnt signaling, mTOR signaling, apoptosis
and many other signaling pathways which are all related to drug
resistance in cancers (76).
All of these 11 drug resistance-related pathways
modulated by the miRNAs targeted CCL21 and SPARCL1 (Table IV) were proven to be involved in
drug resistance in ovarian cancer. The Wnt signaling pathway
participates in drug resistance through inducing apoptosis and
inhibiting tumor growth (77,78).
Cell adhesion molecule is overexpressed in ovarian cancer,
especially in recurrent/chemotherapy-resistant epithelial ovarian
cancer, suggesting that cell adhesion molecule and its pathway may
play a role in drug resistance (79). p53 signaling pathway is a
well-studied and contributes to the whole process of cancer
developments and it is involved in drug resistance in ovarian
cancer through regulating cell proliferation following DNA damage
(10). Cell communication is
important to tumor mechanisms and relevant to the acquisition of
drug resistance in ovarian cancer (80). With the better understanding of the
relationship between cell cycle and the impact of chemotherapeutic
agents on the cell cycle, it becomes apparent that this physiology
can create drug resistance, therefore reducing combination
chemotherapeutic efficacy (81,82).
Amplified PI3K and activated Akt have been observed in 12–68% of
tumors, and are closely associated with upregulation of mTOR
signaling (83), therefore,
activation of the PI3K/Akt pathway and its downstream mTOR
signaling appear to represent drug resistance and poor prognosis
(11,83); apoptosis plays an important role in
the maintenance of physiological homeostasis in response to
stimuli. When the apoptosis machinery fails, abnormal cells can
survive, resulting in unopposed tissue growth and eventually fatal
disease such as cancer (84).
Apoptosis has been demonstrated to be involved in drug resistance
in many solid tumors including ovarian cancer (85,86).
VEGF signaling pathway is a key pathway in normal ovarian
physiology and ovarian cancer, and closely related to drug
resistance (87). Autophagy is
involved in nucleus accumbens-1 mediated resistance to cisplatin,
which is known to have important roles in proliferation, growth of
tumor cells and chemotherapy resistance. Thus, the regulation of
autophagy is considered to be involved in drug resistance in
ovarian cancer (88). ABC
transporters participated in drug resistance through controlling
the drug transportation (89,90).
Taken together, we found that all the 11 pathways
modulated by the miRNAs targeted CCL21 and SPARCL1 contributing to
drug resistance in ovarian cancer, suggesting that CCL21 and
SPARCL1 may also be involved in drug resistance in ovarian
cancer.
Discussion
Drug resistance, comprising both intrinsic and
acquired resistance, is believed to cause treatment failure in over
90% of patients with metastatic cancer (102). Apparently, the survival of cancer
patients would be highly increased if drug resistance could be
overcome. There are many factors affecting drug sensitivity and
cancer cell resistance to chemotherapy can occur at many levels,
including drug transportation, drug inactivation, alterations in
drug target, processing of drug-induced damage and failure of
apoptosis (102). In ovarian
cancer, some mechanisms on drug resistance have been revealed. A
decrease in cell-associated drug, altered GSH-mediated metabolism
and enhanced DNA repair may play roles in cellular resistance to
cisplatin and alkylating agents (103). Further studies show that the
increased anti-apoptotic regulator activity, increased DNA repair
activity, defective DNA damage response, deregulation of growth
factor receptor and post-translational modification or altered
expression of β-tubulin and other microtubule regulatory proteins
may be involved in drug resistance in ovarian cancer (73). Among all these mechanisms and
factors which contribute to drug resistance, some are essentially
involved in aberrant expression of genes. Thus, mining and
exploring of potentially drug resistance-related genes would be a
feasible and reasonable way to solve the drug resistance in ovarian
cancer.
Gene function prediction based on bioinformatics
analysis is a potential, feasible and valuable way for gene
function mining, and many large-scale networks of molecular
interactions within the cell have made it possible to go beyond one
dimensional approaches to study protein function in the context of
a network (104). Pubgene is a
gene/protein database and web-based tool for literature mining. It
carries out automated extraction of experimental and theoretical
biomedical knowledge from publicly available gene and text
databases to create a gene-to-gene co-citation network for 13,712
named human genes by automated analysis of titles and abstracts in
over 10 million MEDLINE records (20). Therefore, gene and protein names
are cross-referenced to each other and to terms that are relevant
to understanding their biological function and importance in
disease. GeneMANIA is a web-based database and tool for prediction
of genes function on the basis of multiple networks derived from
different genomic or proteomic data/sources. It is fast enough to
predict gene function with great accuracy (21). BiologicalNetworks server allows
easy retrieval, construction and visualization of complex
biological networks, including genome-scale integrated networks of
protein-DNA, protein-protein and genetic interactions. Most
importantly, BiologicalNetworks satisfy the need for analysis of
expression profiles of genes or proteins simultaneously on to small
molecules (metabolic) and cellular networks (22). Thus, the predicted functions of
CCL21 and SPARCL1 based on these networks were reasonable and
reliable.
Based on the network analyses, we found that 8 of
the 10 genes which co-occurred with CCL21 and SPARCL1 in ovarian
cancer were drug resistance-related genes (Fig. 1). Among these genes, p53 (5,6),
BRCA1 (7,8), BRCA2 (8), and ERBB2 (9) are already proven to be involved in
drug resistance in ovarian cancer. CCL21 and SPARCL1 were
co-expressed, co-localized, physically interacted and shared
protein domains and pathways with other genes/proteins according to
GeneMANIA network (Fig. 2).
Annotated functions (Table III)
suggested that CCL21 might participate in drug resistance through
regulation of leukocytes, neutrophil chemotaxis, G-protein coupled
receptor activity and calcium ion, therefore, CCL21 might be a
potential drug resistance-related gene. Even though SPARCL1 had no
annotated functions, it was co-expressed with RAMP3 and VWF, and
shared protein domains with IGFBP7 (Fig. 2), which are all reported to be
associated with drug resistance (58,59,61).
SPARCL1 was co-expressed with CCL21, and interacted with each other
through other genes, indicating that SPARCL1 might have a close
relationship with CCL21 in functions. In addition, SPARCL1 exhibits
62% identity (63) and shares four
domains (FOLN, Kazal_1, Kazal_2 and SPARC_Ca_bdg) with SPARC
(Table II), which plays an
important part in drug resistance in ovarian cancer (64,65).
CCL21 had co-citations with 12 small molecules according to
BiologicalNetworks (Fig. 3). Among
them, omeprazole, SMS 201–995, ruthenium complex, hydrogen peroxide
and 2,2′-bipyridine which are demonstrated to be related to drug
resistance in cancers (66,67,69–71)
and adenosine monophosphate is associated with drug resistance in
ovarian cancer (68). Among all
the genes, proteins and small molecules which had interactions with
CCL21 and SPARCL1, most of them are participating in drug
resistance of cancers, and some of them contribute to drug
resistance in ovarian cancer. Therefore, we concluded that both
CCL21 and SPARCL1 might have close relationships with drug
resistance in ovarian cancer.
MicroRNAs (miRNAs) are a class of small (22 bp)
endogenous non-coding RNAs which regulate gene expression mainly by
its binding to the 3′-UTR of the target mRNA, and causing mRNA
cleavage, destabilization or translational repression (105,106). miRNA-mediated
post-transcriptional gene regulation is considered as a significant
regulator of many cellular processes, both physiological and
pathological (107,108). It has been proven that miRNAs
play important roles in drug resistance of many cancers including
ovarian cancer (73). Because
miRNAs perform their functions through the regulation on their
target genes, and it has been well established that miRNAs
represent a class of genes with a great potential for use in
diagnostics, prognosis and therapy (109), therefore, we can predict the gene
function through the functions of miRNAs targeting the gene.
MiRWalk is a comprehensive database on miRNAs, which
gathers predicted and validated miRNA binding sites on all mRNAs,
mitochondrial genes and 10 kb upstream flanking regions of all
known genes of human, mouse and rat. More importantly, the miRWalk
is a real-time database to some extent, in which the ‘Validated
Target module’ is updated every month and the ‘Predicted Target
module’ is updated every 6 months (23). DIANA-mirPath is a web-based
computational tool developed to identify molecular pathways
potentially modulated by the expression of miRNAs. The software
performs an enrichment analysis of multiple miRNA target genes
comparing each set of miRNA targets to all known KEGG pathways. The
output of the program shows an overview of the parts of the pathway
modulated by miRNAs, facilitating the interpretation and
presentation of the results of the analysis and genes (24).
Based on the analysis of miRWalk and DIANA-mirPath,
we found that among all the pathways enriched by multiple miRNAs
targeted CCL21 and SPARCL1, there were 11 pathways (Table IV) closely associated with drug
resistance in ovarian cancer, indicating that CCL21 and SPARCL1
might contribute to drug resistance through those miRNAs to
modulate drug resistance-related pathways.
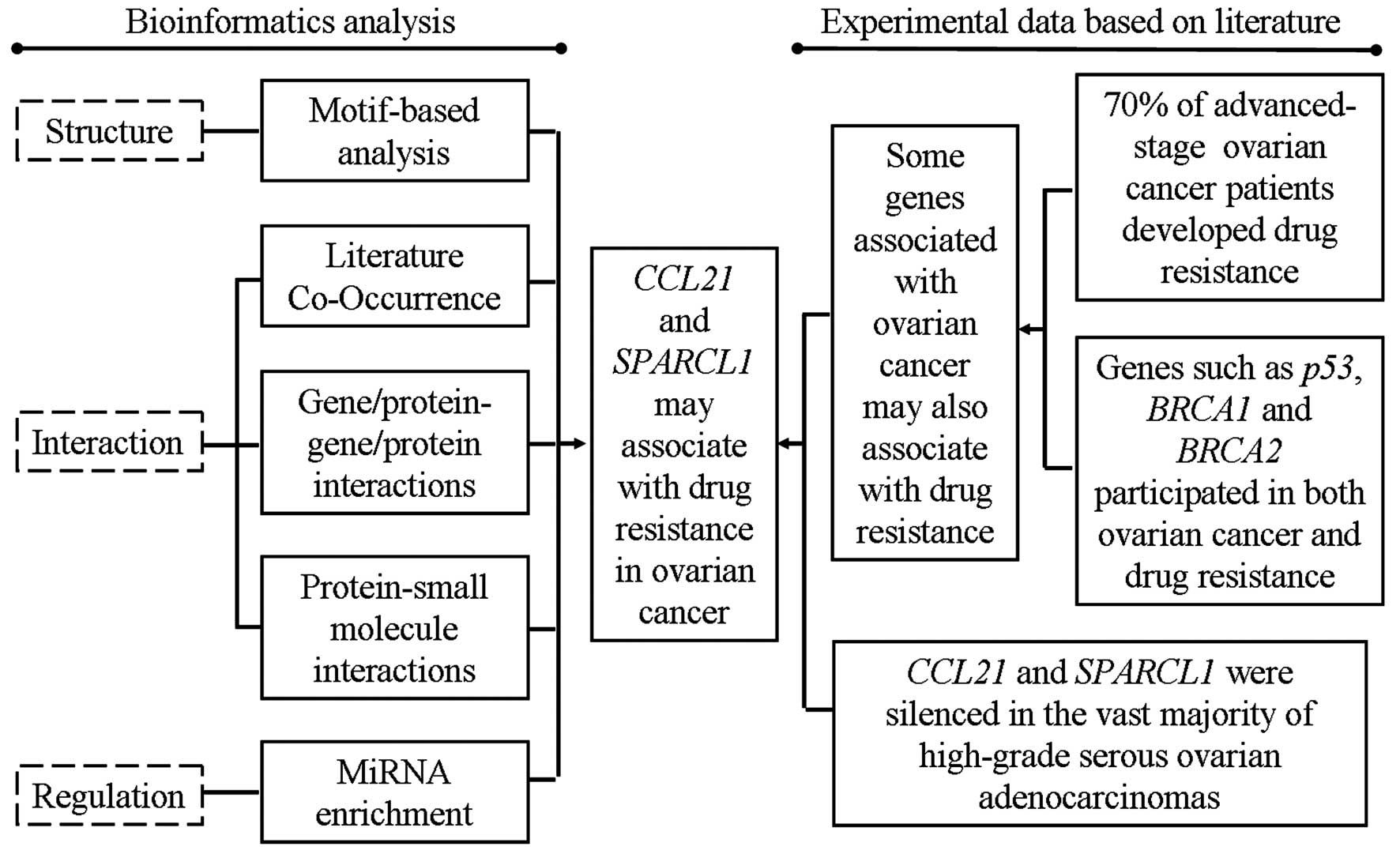
Collectively, based on the function prediction using
motif-based approaches, network interactions, pathway enrichment
analysis of miRNAs and function predictions on the basis of
experimental data from literature (Fig. 4), we concluded that CCL21 and
SPARCL1 might contribute to drug resistance in ovarian cancer. This
is the first report of the drug resistance-functions of CCL21 and
SPARCL1 in ovarian cancer, and thus this study might set the stage
for further experimental studies of CCL21 and SPARCL1 with their
drug resistance associations in ovarian cancer. This study provided
important information for further investigation of drug
resistance-related functions of CCL21 and SPARCL1 in ovarian
cancer.
Acknowledgements
We thank Keqiang Wu at National Taiwan
University for revising the manuscript.
References
|
1
|
Balch C, Huang TH, Brown R and Nephew KP:
The epigenetics of ovarian cancer drug resistance and
resensitization. Am J Obstet Gynecol. 191:1552–1572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
3
|
Cannistra SA: Cancer of the ovary. N Engl
J Med. 351:2519–2529. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bast RC Jr, Hennessy B and Mills GB: The
biology of ovarian cancer: new opportunities for translation. Nat
Rev Cancer. 9:415–428. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fraser M, Bai T and Tsang BK: Akt promotes
cisplatin resistance in human ovarian cancer cells through
inhibition of p53 phosphorylation and nuclear function. Int J
Cancer. 122:534–546. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hagopian GS, Mills GB, Khokhar AR, Bast RC
Jr and Siddik ZH: Expression of p53 in cisplatin-resistant ovarian
cancer cell lines: modulation with the novel platinum analogue (1R,
2R-diaminocyclohexane)(trans-diacetato)(dichloro)-platinum(IV).
Clin Cancer Res. 5:655–663. 1999.
|
|
7
|
Zhou C, Smith JL and Liu J: Role of BRCA1
in cellular resistance to paclitaxel and ionizing radiation in an
ovarian cancer cell line carrying a defective BRCA1. Oncogene.
22:2396–2404. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang D, Khan S, Sun Y, Hess K, Shmulevich
I, Sood AK and Zhang W: Association of BRCA1 and BRCA2 mutations
with survival, chemotherapy sensitivity, and gene mutator phenotype
in patients with ovarian cancer. JAMA. 306:1557–1565. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wu L, Wu A and Jiang K: Effect of
antisense c-erbB2 on biologic behaviour and chemotherapeutic drug
sensitivity in human ovarian cancer cells. Zhonghua Fu Chan Ke Za
Zhi. 31:169–172. 1996.(In Chinese).
|
|
10
|
Benoit DS, Henry SM, Shubin AD, Hoffman AS
and Stayton PS: pH-responsive polymeric sirna carriers sensitize
multidrug resistant ovarian cancer cells to doxorubicin via
knockdown of polo-like kinase 1. Mol Pharm. 7:442–455. 2010.
View Article : Google Scholar
|
|
11
|
Itamochi H: Targeted therapies in
epithelial ovarian cancer: molecular mechanisms of action. World J
Biol Chem. 1:209–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cancer Genome Atlas Research Network:
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar
|
|
13
|
Narod S, Moody J, Rosen B, Fan I, Risch A,
Sun P and McLaughlin J: Estimating survival rates after ovarian
cancer among women tested for BRCA1 and BRCA2 mutations. Clin
Genet. June 8–2012.(Epub ahead of print).
|
|
14
|
Szabova L, Yin C, Bupp S, et al:
Perturbation of Rb, p53, and Brca1 or Brca2 cooperate in inducing
metastatic serous epithelial ovarian cancer. Cancer Res. 72:1–13.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Thangaraju M, Kaufmann SH and Couch FJ:
BRCA1 facilitates stress-induced apoptosis in breast and ovarian
cancer cell lines. J Biol Chem. 275:33487–33496. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Connor JP, Felder M, Kapur A and Onujiogu
N: DcR3 binds to ovarian cancer via heparan sulfate proteoglycans
and modulates tumor cells response to platinum with corresponding
alteration in the expression of BRCA1. BMC Cancer. 12:1762012.
View Article : Google Scholar
|
|
17
|
Quinn JE, James CR, Stewart GE, et al:
BRCA1 mRNA expression levels predict for overall survival in
ovarian cancer after chemotherapy. Clin Cancer Res. 13:7413–7420.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yellaboina S, Tasneem A, Zaykin DV,
Raghavachari B and Jothi R: DOMINE: a comprehensive collection of
known and predicted domain-domain interactions. Nucleic Acids Res.
39:D730–D735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Raghavachari B, Tasneem A, Przytycka TM
and Jothi R: DOMINE: a database of protein domain interactions.
Nucleic Acids Res. 36:D656–D661. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jenssen TK, Laegreid A, Komorowski J and
Hovig E: A literature network of human genes for high-throughput
analysis of gene expression. Nat Genet. 28:21–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: a real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baitaluk M, Sedova M, Ray A and Gupta A:
BiologicalNetworks: visualization and analysis tool for systems
biology. Nucleic Acids Res. 34:W466–W471. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011.
|
|
24
|
Papadopoulos GL, Alexiou P, Maragkakis M,
Reczko M and Hatzigeorgiou AG: DIANA-mirPath: integrating human and
mouse microRNAs in pathways. Bioinformatics. 25:1991–1993. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Baggiolini M, Dewald B and Moser B: Human
chemokines: an update. Annu Rev Immunol. 15:675–705. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu Y, Liu L, Qiu X, et al: CCL21/CCR7
prevents apoptosis via the ERK pathway in human non-small cell lung
cancer cells. PLoS One. 7:e332622012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hwang TL, Lee LY, Wang CC, Liang Y, Huang
SF and Wu CM: CCL7 and CCL21 overexpression in gastric cancer is
associated with lymph node metastasis and poor prognosis. World J
Gastroenterol. 18:1249–1256. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Y, Liu L, Qiu X, et al: CCL21/CCR7
promotes G2/M phase progression via the ERK pathway in human
non-small cell lung cancer cells. PLoS One. 6:e211192011.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yousefieh N, Hahto SM, Stephens AL and
Ciavarra RP: Regulated expression of CCL21 in the prostate tumor
microenvironment inhibits tumor growth and metastasis in an
orthotopic model of prostate cancer. Cancer Microenviron. 2:59–67.
2009. View Article : Google Scholar
|
|
30
|
Zhang H, Widegren E, Wang DW and Sun XF:
SPARCL1: a potential molecule associated with tumor diagnosis,
progression and prognosis of colorectal cancer. Tumour Biol.
32:1225–1231. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hambrock HO, Nitsche DP, Hansen U,
Bruckner P, Paulsson M, Maurer P and Hartmann U: SC1/hevin. An
extracellular calcium-modulated protein that binds collagen I. J
Biol Chem. 278:11351–11358. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Girard JP and Springer TA: Modulation of
endothelial cell adhesion by hevin, an acidic protein associated
with high endothelial venules. J Biol Chem. 271:4511–4517. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li P, Qian J, Yu G, Chen Y, Liu K, Li J
and Wang J: Down-regulated SPARCL1 is associated with clinical
significance in human gastric cancer. J Surg Oncol. 105:31–37.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yu SJ, Yu JK, Ge WT, Hu HG, Yuan Y and
Zheng S: SPARCL1, Shp2, MSH2, E-cadherin, p53, ADCY-2 and MAPK are
prognosis-related in colorectal cancer. World J Gastroenterol.
17:2028–2036. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sullivan MM and Sage EH: Hevin/SC1, a
matricellular glyco-protein and potential tumor-suppressor of the
SPARC/BM-40/Osteonectin family. Int J Biochem Cell Biol.
36:991–996. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mintz MB, Sowers R, Brown KM, et al: An
expression signature classifies chemotherapy-resistant pediatric
osteosarcoma. Cancer Res. 65:1748–1754. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Song J, Wang X, Lei C, et al: Fusion of
chemotactic peptide to a single-chain bi-specific antibody (scBsAb)
potentiates its cytotoxicity to target tumour cells. Biotechnol
Appl Biochem. 45:147–154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Biade S, Marinucci M, Schick J, et al:
Gene expression profiling of human ovarian tumours. Br J Cancer.
95:1092–1100. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu X, Zhai C, Gopalakrishnan V and
Buchanan BG: Automatic annotation of protein motif function with
Gene Ontology terms. BMC Bioinformatics. 5:1222004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang W, Chen LP, Huang R and Huang RP:
Inhibition of IL-6 and IL-8 enhances chemosensitization in
multidrug resistant human breast cancer cells. AACR Meeting
Abstracts. 2005:1199-c2005.
|
|
41
|
Duan Z, Feller AJ, Penson RT, Chabner BA
and Seiden MV: Discovery of differentially expressed genes
associated with paclitaxel resistance using cDNA array technology:
analysis of interleukin (IL) 6, IL-8, and monocyte chemotactic
protein 1 in the paclitaxel-resistant phenotype. Clin Cancer Res.
5:3445–3453. 1999.
|
|
42
|
Soon WW, Miller LD, Black MA, et al:
Combined genomic and phenotype screening reveals secretory factor
SPINK1 as an invasion and survival factor associated with patient
prognosis in breast cancer. EMBO Mol Med. 3:451–464. 2011.
View Article : Google Scholar
|
|
43
|
Chen T, Lee TR, Liang WG, Chang WS and Lyu
PC: Identification of trypsin-inhibitory site and structure
determination of human SPINK2 serine proteinase inhibitor.
Proteins. 77:209–219. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kutuzov MA, Bennett N and Andreeva AV:
Protein phosphatase with EF-hand domains 2 (PPEF2) is a potent
negative regulator of apoptosis signal regulating kinase-1 (ASK1).
Int J Biochem Cell Biol. 42:1816–1822. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gibadulinova A, Tothova V, Pastorek J and
Pastorekova S: Transcriptional regulation and functional
implication of S100P in cancer. Amino Acids. 41:885–892. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhu W, Xu H, Zhu D, et al: miR-200bc/429
cluster modulates multidrug resistance of human cancer cell lines
by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 69:723–731.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hong JH, Lee E, Hong J, Shin YJ and Ahn H:
Antisense Bcl2 oligonucleotide in cisplatin-resistant bladder
cancer cell lines. BJU Int. 90:113–117. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Dhar DK, Nagasue N, Yoshimura H, et al:
Overexpression of P-glycoprotein in untreated AFP-producing gastric
carcinoma. J Surg Oncol. 60:50–54. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xu Z, Wang M, Wang L, Wang Y, Zhao X, Rao
Q and Wang J: Aberrant expression of TSC2 gene in the newly
diagnosed acute leukemia. Leuk Res. 33:891–897. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu Z and Chen S: ER regulates an
evolutionarily conserved apoptosis pathway. Biochem Biophys Res
Commun. 400:34–38. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Perou CM, Sorlie T, Eisen MB, et al:
Molecular portraits of human breast tumours. Nature. 406:747–752.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dabholkar M, Bradshaw L, Parker RJ, Gill
I, Bostick-Bruton F, Muggia FM and Reed E: Cisplatin-DNA damage and
repair in peripheral blood leukocytes in vivo and in vitro. Environ
Health Perspect. 98:53–59. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jo H, Loison F, Hattori H, Silberstein LE,
Yu H and Luo HR: Natural product Celastrol destabilizes tubulin
heterodimer and facilitates mitotic cell death triggered by
microtubule-targeting anti-cancer drugs. PLoS One. 5:e103182010.
View Article : Google Scholar
|
|
54
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Herr DR: Potential use of G
protein-coupled receptor-blocking monoclonal antibodies as
therapeutic agents for cancers. Int Rev Cell Mol Biol. 297:45–81.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ma Q, Zhang ZS, Zhang YL and Lai ZS:
Relationship between multidrug resistance in human colon carcinoma
LoVo/Adr cell line and intracellular calcium ion concentration. Ai
Zheng. 21:846–849. 2002.(In Chinese).
|
|
57
|
Liang X and Huang Y: Intracellular free
calcium concentration and cisplatin resistance in human lung
adenocarcinoma A549 cells. Biosci Rep. 20:129–138. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Nan A, Ghandehari H, Hebert C, Siavash H,
Nikitakis N, Reynolds M and Sauk JJ: Water-soluble polymers for
targeted drug delivery to human squamous carcinoma of head and
neck. J Drug Target. 13:189–197. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Westhoff MA and Fulda S: Adhesion-mediated
apoptosis resistance in cancer. Drug Resist Updat. 12:127–136.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Benatar T, Amemiya Y, Yang W and Seth A:
Insulin-like-growth factor-binding-protein 7: an antagonist to
breast cancer. Breast Cancer - Focusing Tumor Microenvironment,
Stem cells and Metastasis. Gunduz M: InTech; Rijeka: pp. 39–68.
2011
|
|
61
|
Heesch S, Schlee C, Neumann M, et al:
BAALC-associated gene expression profiles define IGFBP7 as a novel
molecular marker in acute leukemia. Leukemia. 24:1429–1436. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lane TF, Iruela-Arispe ML, Johnson RS and
Sage EH: SPARC is a source of copper-binding peptides that
stimulate angiogenesis. J Cell Biol. 125:929–943. 1994.PubMed/NCBI
|
|
63
|
Girard JP and Springer TA: Cloning from
purified high endothelial venule cells of hevin, a close relative
of the antiadhesive extracellular matrix protein SPARC. Immunity.
2:113–123. 1995. View Article : Google Scholar
|
|
64
|
Socha MJ, Said N, Dai Y, et al: Aberrant
promoter methylation of SPARC in ovarian cancer. Neoplasia.
11:126–135. 2009.PubMed/NCBI
|
|
65
|
Tai IT, Dai M, Owen DA and Chen LB:
Genome-wide expression analysis of therapy-resistant tumors reveals
SPARC as a novel target for cancer therapy. J Clin Invest.
115:1492–1502. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
De Milito A and Fais S: Proton pump
inhibitors may reduce tumour resistance. Expert Opin Pharmacother.
6:1049–1054. 2005.PubMed/NCBI
|
|
67
|
Logothetis CJ, Hossan EA and Smith TL: SMS
201–995 in the treatment of refractory prostatic carcinoma.
Anticancer Res. 14:2731–2734. 1994.
|
|
68
|
Matrone A, Grossi V, Chiacchiera F, et al:
p38alpha is required for ovarian cancer cell metabolism and
survival. Int J Gynecol Cancer. 20:203–211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kapitza S, Pongratz M, Jakupec MA, et al:
Heterocyclic complexes of ruthenium (III) induce apoptosis in
colorectal carcinoma cells. J Cancer Res Clin Oncol. 131:101–110.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Yokomizo A, Ono M, Nanri H, et al:
Cellular levels of thioredoxin associated with drug sensitivity to
cisplatin, mitomycin C, doxorubicin, and etoposide. Cancer Res.
55:4293–4296. 1995.PubMed/NCBI
|
|
71
|
Turk D, Hall MD, Chu BF, Ludwig JA, Fales
HM, Gottesman MM and Szakacs G: Identification of compounds
selectively killing multidrug-resistant cancer cells. Cancer Res.
69:8293–8301. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Van Jaarsveld MT, Helleman J, Berns EM and
Wiemer EA: MicroRNAs in ovarian cancer biology and therapy
resistance. Int J Biochem Cell Biol. 42:1282–1290. 2010.PubMed/NCBI
|
|
73
|
Sorrentino A, Liu CG, Addario A, Peschle
C, Scambia G and Ferlini C: Role of microRNAs in drug-resistant
ovarian cancer cells. Gynecol Oncol. 111:478–486. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Yang N, Kaur S, Volinia S, et al: MicroRNA
microarray identifies Let-7i as a novel biomarker and therapeutic
target in human epithelial ovarian cancer. Cancer Res.
68:10307–10314. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Boren T, Xiong Y, Hakam A, et al:
MicroRNAs and their target messenger RNAs associated with ovarian
cancer response to chemotherapy. Gynecol Oncol. 113:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kumar S, Kumar A, Shah PP, Rai SN,
Panguluri SK and Kakar SS: MicroRNA signature of cis-platin
resistant vs. cisplatin sensitive ovarian cancer cell lines. J
Ovarian Res. 4:172011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hilliard TS, Gaisina IN, Muehlbauer AG,
Gaisin AM, Gallier F and Burdette JE: Glycogen synthase kinase
3beta inhibitors induce apoptosis in ovarian cancer cells and
inhibit in-vivo tumor growth. Anticancer Drugs. 22:978–985.
2011.PubMed/NCBI
|
|
78
|
Su HY, Lai HC, Lin YW, et al: Epigenetic
silencing of SFRP5 is related to malignant phenotype and
chemoresistance of ovarian cancer through Wnt signaling pathway.
Int J Cancer. 127:555–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bellone S, Siegel ER, Cocco E, et al:
Overexpression of epithelial cell adhesion molecule in primary,
metastatic, and recurrent/chemotherapy-resistant epithelial ovarian
cancer: implications for epithelial cell adhesion molecule-specific
immunotherapy. Int J Gynecol Cancer. 19:860–866. 2009. View Article : Google Scholar
|
|
80
|
Chen JY, Shen C, Yan Z, Brown DP and Wang
M: A systems biology case study of ovarian cancer drug resistance.
Comput Syst Bioinformatics Conf. 389–398. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Shah MA and Schwartz GK: Cell
cycle-mediated drug resistance: an emerging concept in cancer
therapy. Clin Cancer Res. 7:2168–2181. 2001.PubMed/NCBI
|
|
82
|
Miller DH, Fischer AK, Chu KF, Burr R,
Hillenmeyer S, Brard L and Brodsky AS: T0901317 inhibits
cisplatin-induced apoptosis in ovarian cancer cells [corrected].
Int J Gynecol Cancer. 21:1350–1356. 2011.PubMed/NCBI
|
|
83
|
Trinh XB, van Dam PA, Dirix LY, Vermeulen
PB and Tjalma WA: The rationale for mTOR inhibition in epithelial
ovarian cancer. Expert Opin Investig Drugs. 18:1885–1891. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Li J, Feng Q, Kim JM, et al: Human ovarian
cancer and cisplatin resistance: possible role of inhibitor of
apoptosis proteins. Endocrinology. 142:370–380. 2001.PubMed/NCBI
|
|
85
|
Luo T, Yu J, Nguyen J, et al: Electron
transfer-based combination therapy of cisplatin with
tetramethyl-p-phenylenediamine for ovarian, cervical, and lung
cancers. Proc Natl Acad Sci USA. 109:10175–10180. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Nessa MU, Beale P, Chan C, Yu JQ and Huq
F: Combinations of resveratrol, cisplatin and oxaliplatin applied
to human ovarian cancer cells. Anticancer Res. 32:53–59.
2012.PubMed/NCBI
|
|
87
|
Kumaran GC, Jayson GC and Clamp AR:
Antiangiogenic drugs in ovarian cancer. Br J Cancer. 100:1–7. 2009.
View Article : Google Scholar
|
|
88
|
Zhang Y, Cheng Y, Ren X, et al: NAC1
modulates sensitivity of ovarian cancer cells to cisplatin by
altering the HMGB1-mediated autophagic response. Oncogene.
31:1055–1064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Buys TP, Chari R, Lee EH, et al: Genetic
changes in the evolution of multidrug resistance for cultured human
ovarian cancer cells. Genes Chromosomes Cancer. 46:1069–1079. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Auner V, Sehouli J, Oskay-Oezcelik G,
Horvat R, Speiser P and Zeillinger R: ABC transporter gene
expression in benign and malignant ovarian tissue. Gynecol Oncol.
117:198–201. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Chikazawa N, Tanaka H, Tasaka T, Nakamura
M, Tanaka M, Onishi H and Katano M: Inhibition of Wnt signaling
pathway decreases chemotherapy-resistant side-population colon
cancer cells. Anticancer Res. 30:2041–2048. 2010.PubMed/NCBI
|
|
92
|
Wang S, Li W, Xue Z, et al: Molecular
imaging of p53 signal pathway in lung cancer cell cycle arrest
induced by cisplatin. Mol Carcinog. Jun 5–2012.(Epub ahead of
print). View Article : Google Scholar
|
|
93
|
Leon-Galicia I, Diaz-Chavez J,
Garcia-Villa E, et al: Resveratrol induces downregulation of DNA
repair genes in MCF-7 human breast cancer cells. Eur J Cancer Prev.
22:11–20. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Han W, Pan H, Chen Y, et al: EGFR tyrosine
kinase inhibitors activate autophagy as a cytoprotective response
in human lung cancer cells. PLoS One. 6:e186912011. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu LZ, Zhou XD, Qian G, Shi X, Fang J and
Jiang BH: AKT1 amplification regulates cisplatin resistance in
human lung cancer cells through the mammalian target of
rapamycin/p70S6K1 pathway. Cancer Res. 67:6325–6332. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Chung AS, Kowanetz M, Wu X, et al:
Differential drug class-specific metastatic effects following
treatment with a panel of angiogenesis inhibitors. J Pathol.
227:404–416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Waldner MJ and Neurath MF: Targeting the
VEGF signaling pathway in cancer therapy. Expert Opin Ther Targets.
16:5–13. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Ishdorj G, Li L and Gibson SB: Regulation
of autophagy in hematological malignancies: role of reactive oxygen
species. Leuk Lymphoma. 53:26–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Liu L, Yang M, Kang R, et al:
DAMP-mediated autophagy contributes to drug resistance. Autophagy.
7:112–114. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fisher C, Coleman T and Plant N:
Probabilistic orthology analysis of the ATP-binding cassette
transporters: implications for the development of multiple drug
resistance phenotype. Drug Metab Dispos. 40:1397–1402. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Shukla S, Chen ZS and Ambudkar SV:
Tyrosine kinase inhibitors as modulators of ABC
transporter-mediated drug resistance. Drug Resist Updat. 15:70–80.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Longley DB and Johnston PG: Molecular
mechanisms of drug resistance. J Pathol. 205:275–292. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Johnson SW, Ozols RF and Hamilton TC:
Mechanisms of drug resistance in ovarian cancer. Cancer.
71:644–649. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Behm-Ansmant I, Rehwinkel J and Izaurralde
E: MicroRNAs silence gene expression by repressing protein
expression and/or by promoting mRNA decay. Cold Spring Harb Symp
Quant Biol. 71:523–530. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Tili E, Michaille JJ, Gandhi V, Plunkett
W, Sampath D and Calin GA: miRNAs and their potential for use
against cancer and other diseases. Future Oncol. 3:521–537. 2007.
View Article : Google Scholar : PubMed/NCBI
|