Introduction
Hepatocellular carcinoma (HCC) is one of the top
five causes of cancer-related deaths worldwide (1). Hepatitis B and C viruses are major
risk factors for HCC and many studies of HCC associated with
viruses have been undertaken. Recent developments in imaging have
enabled the detection of early-stage HCC and multidisciplinary
treatment of HCC has greatly improved the survival rate; however,
recurrence of HCC remains prominent. One reason for this high rate
of recurrence is that cancer is a genetic disease of somatic cells
arising from an accumulation of genetic mutations. Therefore, in
order to improve the prognosis, it is important to find the genetic
markers occurring in recurrent or metastatic HCC.
The development of high-throughput technologies such
as gene expression microarrays and single nucleotide polymorphism
(SNP) arrays that can simultaneously screen thousands of genes has
enabled wide and comprehensive identification of alterations in
gene expression caused by oncogenesis (2–4).
These technologies have revealed an enhanced characterization of
individual tumors concerning metastatic potential, compared with
that provided by traditional clinicopathological methods.
We have previously reported a novel method named
double-combination array analysis that combines SNP arrays with
gene expression arrays (5–10). In addition, we have hypothesized
that the decrease in gene expression is due to hypermethylation of
the CpG islands. Aberrant DNA methylation of the promoter and other
genomic regions can lead to changes in gene expression. However,
our double array analysis does not focus on the presence of
hypermethylation. Therefore, we additionally performed methylation
array analysis by using the IlluminaInfiniumHumanMethylation 27
BeadChip platform (Illumina, San Diego, CA, USA) to comprehensively
evaluate the methylation status. We used data from all three
analyses to detect the aberrant gene expression in tumor tissue. We
called this approach triple-combination array analysis.
In the assessment of the data from the
triple-combination array analysis, estrogen receptor 1 (ESR1) gene
was identified as one of promising candidates for a novel tumor
suppressor gene. Estrogens play important roles, including
oncogenic role in various organs of which breast cancer is a
well-known example. Moreover, estrogens have antifibrotic effects
and are protective factors for the progression of fibrosis in
patients with chronic hepatitis. Thus, we selected ESR1 as a
candidate tumor suppressor gene in the present study.
Materials and methods
Sample collection and DNA
preparation
Nine HCC cell lines (HuH1, HuH2, HuH7, HepG2, Hep3B,
HLE, HLF, SK-Hep1 and PLC/PRF/5) were obtained from the American
Type Culture Collection (Manassas, VA, USA). The cell lines were
cultured in RPMI-1640, supplemented with 10% fetal bovine serum and
incubated in 5% CO2 at 37°C.
A 68-year-old woman with chronic hepatitis C was
diagnosed as having HCC in the right lobe and underwent liver
resection, and the tumor was pathologically diagnosed as HCC. The
total RNA and DNA were extracted from her tumor and non-tumor
tissues. Total RNA was sent to the manufacturer of Affymetrix to
prepare it for expression array analysis, and genomic DNA was used
for SNP array analysis, and bisulfite-converted DNA was used for
the IlluminaInfiniumHumanMethylation 27 BeadChip methylation array
analysis.
HCC tissues and corresponding normal tissues were
obtained from 48 patients who had undergone liver resection at
Nagoya University Hospital, Japan, between 1994 and 2001. Their
ages ranged from 39 to 77 years (62.4±7.9 years: means ± standard
deviation) and the male-to-female ratio was 43:5. Thirty-eight
patients had hepatitis C and seven had hepatitis B. The median
length of follow-up was 64.5 months (range 17.9–105.9 months). All
tissues collected were diagnosed pathologically as HCC. Written
informed consent, as required by the institutional review board,
was obtained from all patients. The tissue samples were immediately
frozen in liquid nitrogen and stored at −80°C until analysis.
Genomic DNA was obtained from the tissue samples by digestion with
proteinase K, followed by phenol/chloroform extraction.
RNA isolation, microarray and gene chip
Affymetrix procedures
The expression array and SNP array analysis were
performed, as previously described (5–10),
using total RNA and DNA extracted from the 68-year-old woman’s
tissue samples.
Methylation array platform
Methylation array analysis was performed as
described previously (12,13).
RT-PCR
The expression of ESR1 mRNA was analyzed by RT-PCR
and real-time RT-PCR. After total RNA (10 μg) was isolated
from nine HCC cell lines and primary HCC tissues and normal tissues
were used to generate cDNA, they were amplified by PCR primers for
ESR1 sense (S) (5′-CCGGCTCCGTAAATGCTACG-3′ in exon 10) and
antisense (AS) (5′-TCC AGCAGACCCCACTTCAC-3′ in exon 11), which
amplified a 133-bp product. RT-PCR amplification consisted of 36
cycles of 94°C for 12 sec, 60°C for 8 sec and 72°C for 8 sec, after
the initial denaturation step (94°C for 5 min). GAPDH (TaqMan,
GAPDH Control Reagents; Applied Biosystems) was used as a control
reference. Each PCR product was loaded directly onto 3% agarose
gels, stained with ethidium bromide and visualized under
ultraviolet (UV) illumination.
Quantitative PCR
PCR reactions were performed by the SYBR Green PCR
Core Reagents kit (Applied Biosystems) under the following
conditions: 1 cycle at 95°C for 10 min, then 40 cycles at 95°C for
15 sec and at 60°C for 30 sec. Real-time detection of the SYBR
Green emission intensity was conducted with an ABI PRISM 7000
Sequence Detector (Applied Biosystems). The primers used for this
PCR were the same primer pairs as used for the RT-PCR described
above. For standardization, expression of GAPDH in each sample was
quantified. Quantitative RT-PCR was performed at least three times,
including no-template samples serving as a negative control. The
expression of ESR1 was normalized by dividing the amount of ESR1
expression by GAPDH expression for each sample.
Methylation-specific PCR (MSP) and
unmethylation-specific PCR (UNMSP)
DNA from HCC cell lines, the primary tumor and
corresponding normal specimens were subjected to bisulfite
treatment. Briefly, 2 μg DNA was denatured by NaOH and
modified by sodium bisulfite. Then, DNA samples were purified with
the Wizard purification resin (Promega, Madison, WI, USA), treated
again with NaOH, precipitated with ethanol and resuspended in
water. The primer pairs for detecting methylation targeted the ESR1
promoter region near exon 3 were as follows: S
(5′-TTCGTCGGGTCGTTCGGTTT-3′) and AS (5′-ATATCCCGCCGACACGCGAA-3′),
which amplified an 81-bp product. Primers for unmethylated
detection targeted the same promoter region: S (5′-AGTTGGTGGAGGGTGT
TTGT-3′) and AS (5′-CACATATCCCACCAACACAC-3′) and amplified a 122-bp
product. Each PCR product was loaded directly onto 3% agarose gels,
stained with ethidium bromide and visualized under UV
illumination.
5-Aza-2’-deoxycytidine (5-aza-dC)
treatment
To confirm that the promoter hypermethylation was
responsible for the silencing of the gene expression, nine HCC cell
lines were treated with a DNA methylation inhibitor, 5-aza-dC
(Sigma-Aldrich, St. Louis, MO, USA). Cells were cultured for 6 days
with the medium replaced on days 1, 3 and 5. After incubation,
cells were collected, RNA was extracted and RT-PCR was performed as
described above.
Sequence analysis
Genomic bisulfite-treated DNA of HCC cell lines was
sequenced and PCRs were performed on each sample. The primer pair
for the sequence was in the ESR1 promoter region for forward primer
and in exon 3 for reverse primer: S (5′-TTTGGAGTGATGTTTAAGTT-3′)
and AS (5′-CCACCTAAAAAAAAAACACA-3′), which amplified a 288-bp
product. The PCR amplification consisted of 25 cycles of 96°C for
10 sec, 50°C for 5 sec and 60°C for 4 min, following the initial
denaturation step (96°C for 1 min). PCR products were purified
directly using the QIA Quick Gel extraction kit (Qiagen, Hilden,
Germany). The purified DNA fragments were subcloned into a TA
cloning vector (Invitrogen, Carlsbad, CA, USA). Six cloning samples
were picked out from two HCC cell lines (HuH7 and SK-Hep1). Each
cloning DNA was mixed with the specific primer (M13) and Cycle
Sequence Mix (ABI PRISM Terminator v1. 1 Cycle Sequencing kit;
Applied Biosystems, Foster City, CA, USA). We performed sequence
analysis using an ABI PRISM 310 genetic analyzer (Applied
Biosystems) and sequence electropherograms were generated by ABI
Sequence Analysis software 5.1.
Statistical analysis
Continuous variables are expressed as medians
(range) and comparisons were made using the Mann-Whitney U test.
Categorical variables were compared using χ2 or Fisher’s
exact tests, where appropriate. Overall and disease-free survival
rates were analyzed by Kaplan-Meier and log-rank tests. All
statistical analyses were performed using Jump 9 (SAS Institute
Inc. Cary, NC, USA). The level of statistical significance was set
at P<0.05.
Results
Expression, SNP and methylation
arrays
To reveal potential tumor suppressor genes involved
in HCC, expression array chips were used to identify genes for
which expression was lower in HCC compared with normal tissue.
Aberrant gene expression between HCC and normal tissue was ranked
on the basis of change in intensity. We consequently identified
that the ESR1 gene had greatly reduced expression in HCC tissues at
the high level of −2.5 (log2 ratio) (Table I). RT-PCR that used cDNA obtained
from the 68-year-old woman confirmed ESR1 gene was downregulated
(Fig. 1A). Focusing on the ESR1
gene, by means of the SNP-Chip array, we detected deletions in 3q,
8p, 11q, 12q, 16p, 17p and 19p, as well as in the X chromosomes and
chromosomal gains in 1q, 3q, 11q, 12p and 12q. Interestingly, there
were no abnormalities in the copy number of chromosome 6, where
ESR1 is located (Fig. 2). SNP
array data of the ESR1 locus were analyzed and displayed a high
number of SNPs. Twenty-five SNPs showed a heterozygous AB allele in
both normal and tumor tissues, indicating that both alleles were
retained at the locus (Table II).
These results suggest that ESR1 expression was reduced without loss
of heterozygosity or deletion. In the methylation array analysis,
the continuous β values were 0.775 for HCC tissue versus 0.093 for
normal tissue, indicating high methylation in the former (Table III). Based on these findings, many
CpG islands were detected when the sequence of the promoter region
of ESR1 was examined, leading to the hypothesis that
hypermethylation of the CpG islands was the mechanism responsible
for decreasing ESR1 expression in tumor tissue. We subsequently
confirmed hypermethylation by MSP in tumor tissue obtained from the
68-year-old woman (Fig. 1B).
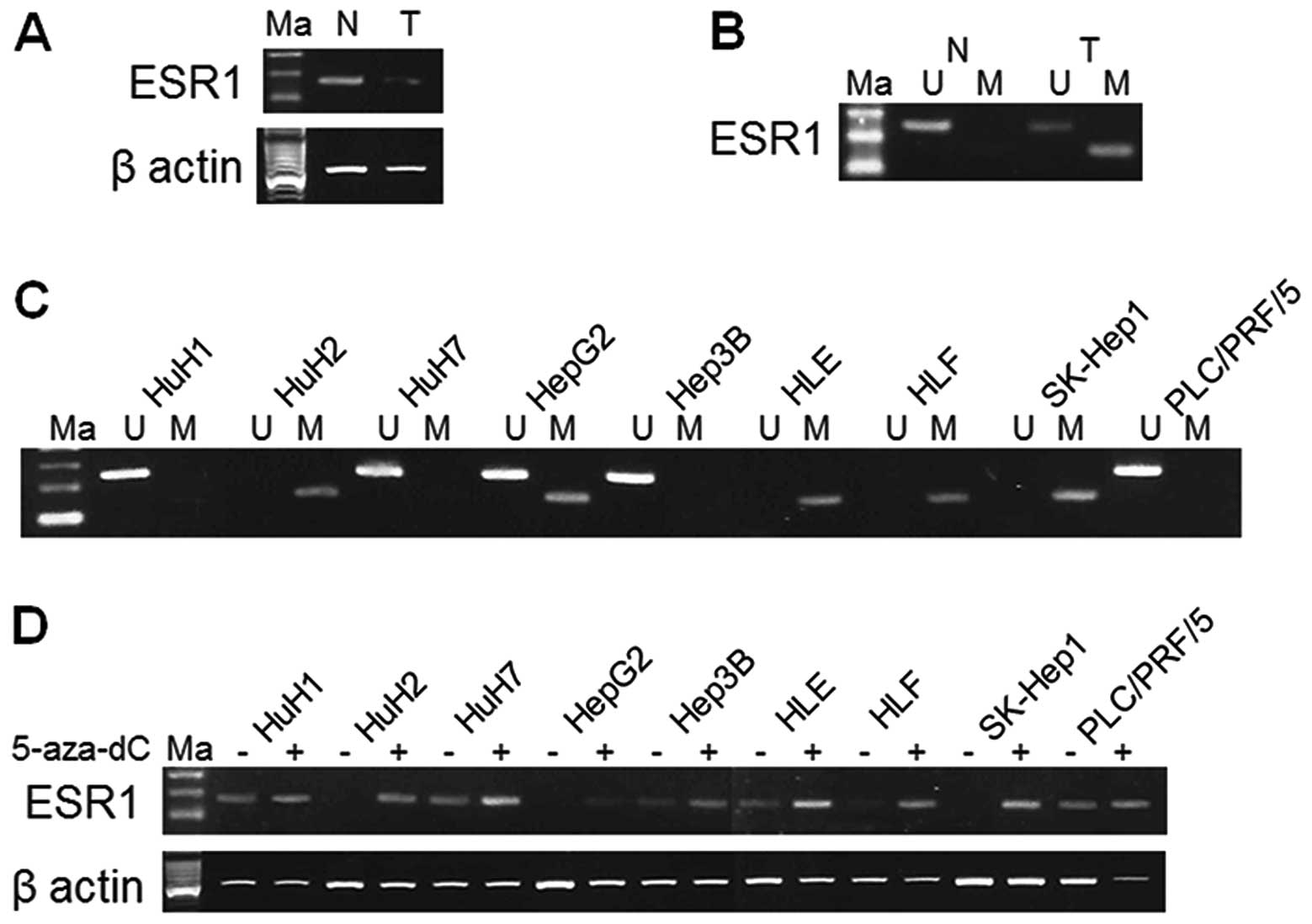 | Figure 1.Results of RT-PCR, MSP and UNMSP in
ESR1 gene, which produced a product of 133, 81 and 122 bp,
respectively. (A) RT-PCR used cDNA obtained from the 68-year-old
woman confirmed that ESR1 gene was downregulated. (B)
Hypermethylation was confirmed by MSP in tumor tissue obtained from
the 68-year-old woman donor. (C) Methylation status of ESR1 gene in
HCC cell lines. HuH2, HepG2, HLE, HLF and SK-Hep1 cells showed
hypermethylation. Unmethylation was shown in HuH1, HuH7, HepG2,
Hep3B and PLC/PRF/5 cells. (D) Expression of ESR1 in HuH2, HepG2,
HLF and SK-Hep1 cells displayed reactivation after 5-aza-dC
treatment. ESR1, estrogen receptor 1; Ma, marker; N, normal tissue;
T, tumor tissue; M, MSP; U, UNMSP. |
 | Table I.Expression array analysis of ESR1
gene. |
Table I.
Expression array analysis of ESR1
gene.
| Gene symbol | Log2
ratio | Normal signal | Detection | Tumor signal | Detection | Probe ID | Chromosomal
location |
|---|
| ESR1 | −2.5 | 295.8 | P | 55 | P | HU133p2_14673 | 6q25.1 |
 | Table II.SNP signal at ESR1 gene locus. |
Table II.
SNP signal at ESR1 gene locus.
| Probe_Set_ID | Chromosome | Physical
position | Normal cell | Confidence | Tumor cell | Confidence |
|---|
| SNP_A-4212066 | 6 | 152066213 | AB | 0.09375 | AB | 0.09375 |
| SNP_A-4212068 | 6 | 152084195 | AB | 0.007813 | AB | 0.015625 |
| SNP_A-2138703 | 6 | 152109562 | AB | 0.0625 | AB | 0.1875 |
| SNP_A-2241225 | 6 | 152124339 | AB | 0.039063 | AB | 0.023438 |
| SNP_A-4302034 | 6 | 152126286 | AB | 0.015625 | AB | 0.015625 |
| SNP_A-2043771 | 6 | 152132228 | AB | 0.007813 | AB | 0.1875 |
| SNP_A-2185504 | 6 | 152221934 | AB | 0.0625 | AB | 0.03125 |
| SNP_A-1856863 | 6 | 152241822 | AB | 0.007813 | AB | 0.039063 |
| SNP_A-2012176 | 6 | 152307215 | AB | 0.1875 | AB | 0.125 |
| SNP_A-2131360 | 6 | 152349399 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-2032260 | 6 | 152350666 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-2182907 | 6 | 152370309 | AB | 0.09375 | AB | 0.015625 |
| SNP_A-2126916 | 6 | 152389551 | AB | 0.039063 | AB | 0.09375 |
| SNP_A-2055019 | 6 | 152413084 | AB | 0.1875 | AB | 0.078125 |
| SNP_A-2021652 | 6 | 152414235 | AB | 0.0625 | AB | 0.039063 |
| SNP_A-2166370 | 6 | 152418873 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-4233902 | 6 | 152424004 | AB | 0.0625 | AB | 0.125 |
| SNP_A-4202504 | 6 | 152424018 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-1951801 | 6 | 152426493 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-2254273 | 6 | 152426929 | AB | 0.007813 | AB | 0.007813 |
| SNP_A-2230180 | 6 | 152428321 | AB | 0.132813 | AB | 0.007813 |
| SNP_A-1988124 | 6 | 152431055 | AB | 0.015625 | AB | 0.015625 |
| SNP_A-4232738 | 6 | 152434853 | AB | 0.039063 | AB | 0.09375 |
| SNP_A-4219387 | 6 | 152435454 | AB | 0.007813 | AB | 0.016602 |
| SNP_A-2043027 | 6 | 152449819 | AB | 0.007813 | AB | 0.1875 |
 | Table III.Methylation array analysis of ESR1
gene. |
Table III.
Methylation array analysis of ESR1
gene.
| | | | | Status
| | |
|---|
| Probe ID | Gene symbol | Sample | Methylation
value | Total | Unmethylated | Methylated | Confidence | Chromosomal
location |
|---|
| cg00655307 | ESR1 | Normal | 0.093132 | 3948 | 3571 | 377 | 3.68E-38 | 6q25.1 |
| | Tumor | 0.774897 | 3525 | 716 | 2809 | 3.68E-38 | |
MSP and UNMSP of nine cell lines
We designed primers for MSP and UNMSP and checked
the methylation status of the HCC samples used for the arrays and
nine HCC cell lines. We obtained bands of appropriate size in the
lanes of tumor tissue for HuH2, HepG2, HLE, HLF and SK-Hep1 cells
by electrophoresis of MSP. In UNMSP, however, there were bands in
lanes of HuH1, HuH7, HepG2, Hep3B and PLC/PRF/5 cells (Fig. 1C). We consider that complete
methylation existed in HuH2, HLE, HLF and SK-Hep1 cells, complete
unmethylation in HuH1, HuH7, Hep3B and PLC/PRF/5 cells and partial
methylation in HepG2 cells.
5-Aza-dC treatment of nine cell
lines
The ESR1 expression of HuH2, HepG2, HLE, HLF and
SK-Hep1 cells were reduced by promoter hypermethylation and were
reactivated after 5-aza-dC treatment. HuH2, HLF and SK-Hep1 cells
that underwent complete promoter hypermethylation showed increase
in expression after 5-aza-dC treatment (Fig. 1D).
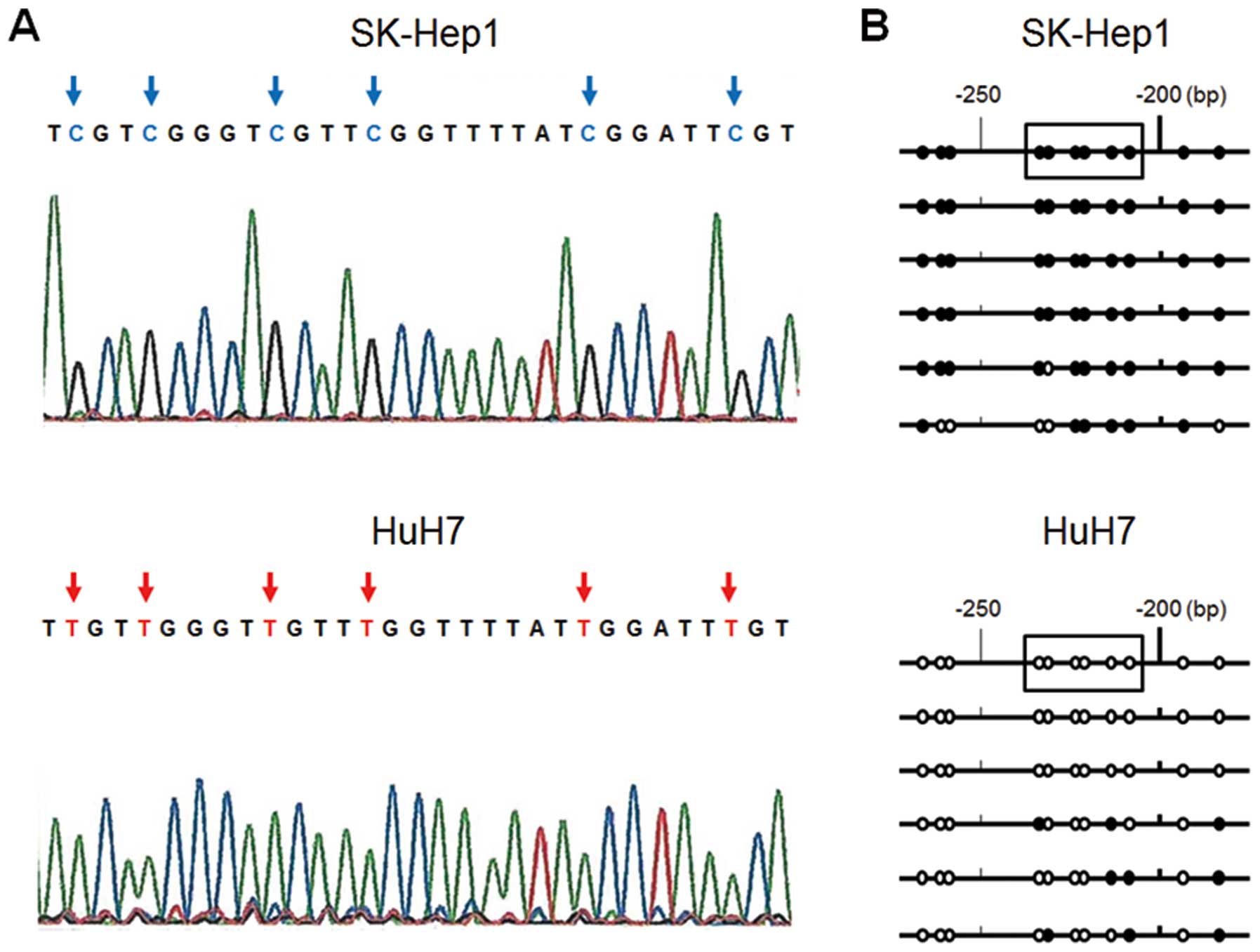
Sequence analysis
To confirm that the amplification of MSP was
correctly performed, sequence analysis of the ESR1 promoter region
was performed in six colonies by TA cloning in HuH7 and Sk-Hep1
cells. In most cases, we found all CpG islands in the fragment of
SK-Hep1 cells to be CG, whereas all those of HuH7 cells were TG
(Fig. 3A). Some of the six clones
showed a different methylation status in both cell lines (Fig. 3B) and this result suggested that
they were partially methylated and reflected the MSP and UMSP
results.
MSP and UNMSP of samples from 48 HCC
patients
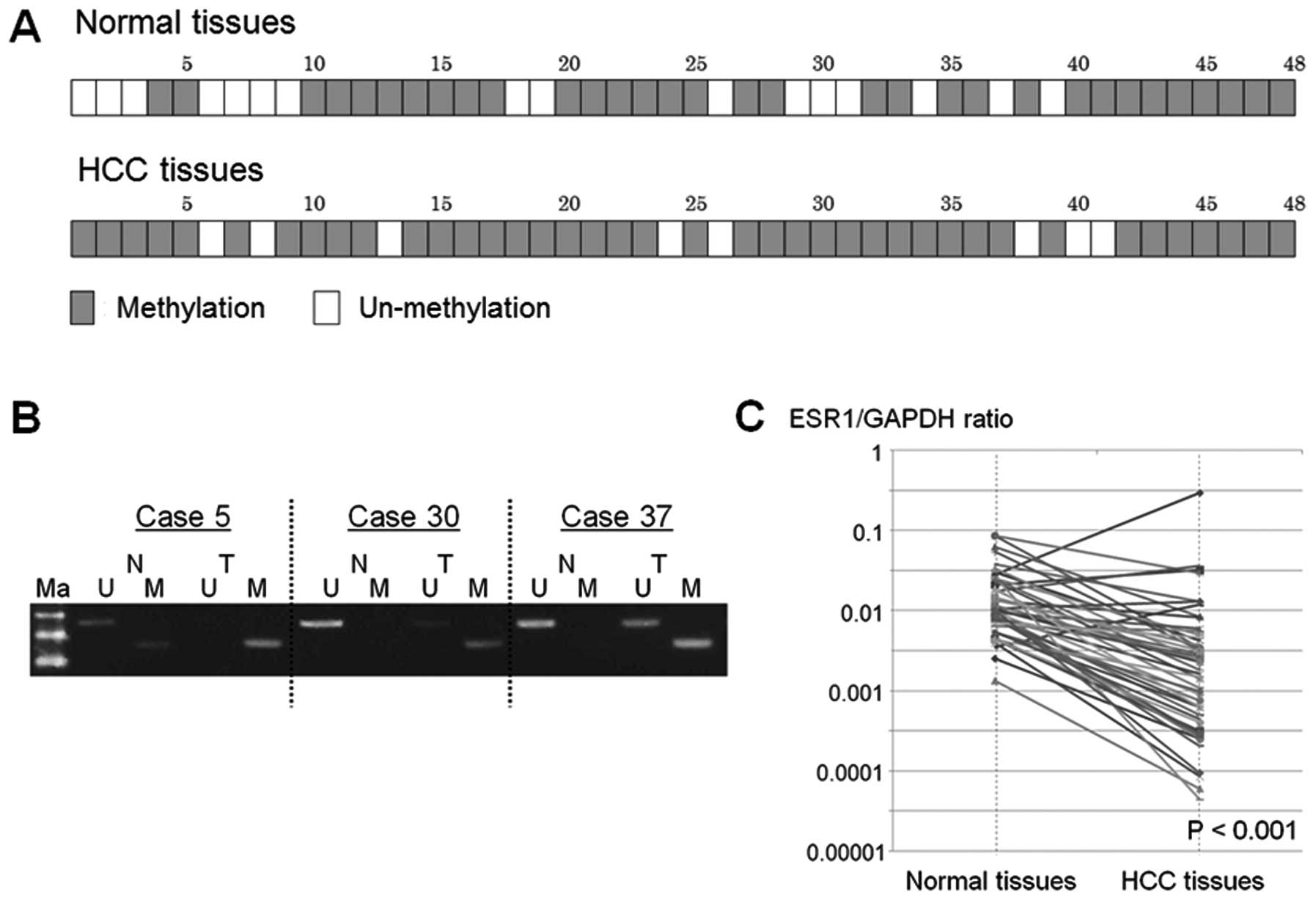
A total of 40 (83.3%) of the 48 HCC tissues
displayed promoter hypermethylation, but 32 (66.7%) of 48 normal
tissues showed promoter hypermethylation of the ESR1 gene (Fig. 4A and B).
Quantitative PCR of samples from 48 HCC
patients
Total ESR1 expression in HCC tissue was
significantly lower than in normal tissue, relative to GAPDH
(median ESR1/GAPDH ratio, 0.0175 vs. 0.0107; P<0.001) (Fig. 4C). In 24 (50%) of 48 HCC samples,
the expression level of ESR1 gene decreased by >90% in HCC
compared to normal tissue.
Correlation between the
clinicopathological factors in 48 HCC patients and results of our
findings
No correlation was found between promoter
hypermethylation of ESR1 and patient clinicopathological factors in
either normal or HCC tissue (data not shown). Disease-free and
overall survival of patients with promoter hypermethylation of ESR1
in HCC tissue were not significantly shorter than in those without
promoter hypermethylation of ESR1 (P=0.2378 and P=0.5402,
respectively). On the other hand, In 24 (50%) HCC samples, the
expression level of ESR1 gene was decreased by >90%. The
decreased expression was significantly related to high liver damage
score (P= 0.0388), pathological invasion of the intrahepatic portal
vein (P= 0.0236), the size of tumor (>3 cm in diameter)
(P=0.0417) and hepatitis B virus (HBV) infection (P= 0.011). In
contrast, disease-free survival and overall survival of patients
with reduced ESR1 expression in HCC tissue were not significantly
shorter than in those without reduced ESR1 expression in HCC tissue
(P=0.4854 and P=0.4612, respectively).
Discussion
Epidemiological reports indicate that the incidence
of HCC is higher in male than in female patients (14). It has been observed that chronic
liver disease progresses more rapidly to cirrhosis and HCC in male
than female patients (15).
Therefore, estrogens are considered to play an important role in
liver diseases. Some studies report that estrogens have
antifibrotic effects and are protective factors for the progression
of fibrosis in patients with chronic hepatitis (16). Naugler et al have indicated
in diethylnitrosamine-treated male mice that estrogen reduces
circulating concentrations of interleukin (IL)-6, a proinflammatory
molecule released from Kupffer cells upon exposure to necrotic
hepatocytes. They have proposed that estrogen-mediated inhibition
of IL-6 production reduces liver cancer risk (17). Estrogen has also been reported to
suppress generation of oxidative-stress-induced reactive oxygen
species (ROS), lipid peroxidation, activation of activator protein
(AP)-1 and nuclear factor (NF)-κB, which are transcription factors
(18).
We chose ESR1 gene as a tumor suppressor gene from
the candidates, because of the above-mentioned functions of
estrogens. ESR1 encodes estrogen receptor (ER)α, a ligand-activated
transcription factor composed of several domains important for
hormone binding, DNA binding and activation of transcription.
Furthermore, the relation between ESR1 and malignant disease has
been discussed in a variety of tissues including breast, colon,
blood, bladder and liver (19–27).
ERα plays key roles in cell development and
differentiation. Many studies have investigated these roles in
relation to breast cancer. In the presence of ligand, ERα may
inhibit invasion through mechanisms involving transcriptional
activation of estrogen response element regulating target genes,
such as E-cadherin, which increases cell-cell adhesion (28). In the absence of ligand, ERα also
inhibits invasion through a distinct mechanism involving
protein-protein interaction with the region of the first zinc
finger of ERα (28). It is
considered that these roles of ERα are also applicable to HCC and
result in a significant association between the decrease in ERα and
pathological invasion of HCC.
In a recent Japanese nationwide investigation, the
rate of HCV infection among the HCC patients was 72.3% and that of
HBV infection was 16.8% (29). In
the present study, the infection rates were compatible at 79.2 and
14.6%, respectively. The expression of ESR1 gene was decreased in
all of the seven patients infected with HBV, implicating that it
was inversely correlated with the HBV infection. Some reports
actually discussed the relationship between ERs and HBV infection.
The level of ERs in the cytosol of peripheral blood mononuclear
cells is significantly lower in asymptomatic HBV carriers and
patients with chronic hepatitis than in healthy controls (30,31).
In contrast, estrogen can repress transcription of HBV genes by
upregulating ERα, which interacts with and alters binding of
hepatocyte nuclear factor-4α to HBV enhancer I (32). There is a possibility that the
decrease in ERα (ESR1 expression) in liver tissue enhances the
adverse influence of HBV infection and this may be the reason that
the ESR1 expression was found in the present study to be lower in
HCC than in normal tissue.
In this study, there were some cases that had
hypermethylation of the promoter region in both non-cancer and HCC
tissues. However, in the electrophoresis of MSP, the density of the
band in tumor tissue tended to be stronger than the band in
non-cancer tissue. Therefore, it can be speculated that some
promoter hypermethylated regions already existed in precancer liver
tissue in the form of chronic hepatitis or cirrhosis and the
hypermethylation status becomes further enhanced as these tissues
develop into cancer.
The size of tumor with reduced ESR1 expression in
HCC tissue was significantly larger than in those without reduced
ESR1 expression in HCC tissue. Regarding the pathological
diagnosis, however, the HCC with the reduced ESR1 expression tended
to be well differentiated phenotype (P= 0.1576) which is associated
with more indolent biology. This could be the reason that the
disease-free survival and overall survival of patients with the
reduced ESR1 expression were not significantly shorter than in
those without the reduced ESR1 expression.
In conclusion, our results indicate that ESR1 acts
as a tumor suppressor gene and that one of the mechanisms of ESR1
silencing is related to promoter hypermethylation in human HCC. The
novel method of triple-combination array analysis that was applied
in this study is useful for detecting new suppressor oncogenes and
their mechanisms and further investigations using this method is
warranted.
Acknowledgements
This study was supported by Japan
Society for the Promotion of Science (JSPS) KAKENHI Grant-in-Aid
for Scientific Research (C) no. 22591427.
References
|
1.
|
Parkin DM, Bray F, Ferlay J, et al: Global
cancer statistics, 2002. CA Cancer J Clin. 55:74–108. 2005.
View Article : Google Scholar
|
|
2.
|
Schena M, Shalon D, Davis RW, et al:
Quantitative monitoring of gene expression patterns with a
complementary DNA microarray. Science. 270:467–470. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Lau WY, Lai PB, Leung MF, et al:
Differential gene expression of hepatocellular carcinoma using cDNA
microarray analysis. Oncol Res. 12:59–69. 2002.PubMed/NCBI
|
|
4.
|
Wang DG, Fan JB, Siao CJ, et al:
Large-scale identification, mapping and genotyping of
single-nucleotide polymorphisms in the human genome. Science.
280:1077–1082. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kanda M, Nomoto S, Okamura Y, et al:
Detection of metallothionein 1G as a methylated tumor suppressor
gene in human hepatocellular carcinoma using a novel method of
double combination array analysis. Int J Oncol. 35:477–483.
2009.
|
|
6.
|
Okamura Y, Nomoto S, Kanda M, et al:
Leukemia inhibitory factor receptor (LIFR) is detected as a novel
suppressor gene of hepatocellular carcinoma using
double-combination array. Cancer Lett. 289:170–177. 2010.
View Article : Google Scholar
|
|
7.
|
Nomoto S, Kanda M, Okamura Y, et al:
Epidermal growth factor-containing fibulin-like extracellular
matrix protein 1, EFEMP1, a novel tumor-suppressor gene detected in
hepatocellular carcinoma using double combination array analysis.
Ann Surg Oncol. 17:923–932. 2010. View Article : Google Scholar
|
|
8.
|
Okamura Y, Nomoto S, Kanda M, et al:
Reduced expression of reelin (RELN) gene is associated with high
recurrence rate of hepatocellular carcinoma. Ann Surg Oncol.
18:572–579. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kanda M, Nomoto S, Okamura Y, et al:
Promoter hypermethylation of fibulin 1 gene is associated with
tumor progression in hepatocellular carcinoma. Mol Carcinog.
50:571–579. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Hayashi M, Nomoto S, Kanda M, et al:
Identification of the A kinase anchor protein 12 (AKAP12) gene as a
candidate tumor suppressor of hepatocellular carcinoma. J Surg
Oncol. 105:381–386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Taira S and Yokoyama K: Self-assembly
DNA-conjugated polymer for detection of single nucleotide
polymorphism. Biotechnol Bioeng. 88:35–41. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Bibikova M and Fan JB: GoldenGate assay
for DNA methylation profiling. Methods Mol Biol. 507:149–163. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Okamura Y, Nomoto S, Hayashi M, et al:
Identification of the bleomycin hydrolase gene as a methylated
tumor suppressor gene in hepatocellular carcinoma using a novel
triple-combination array method. Cancer Lett. 312:150–157. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Shimizu I: Impact of oestrogens on the
progression of liver disease. Liver Int. 23:63–69. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
DiMartino V, Lebray P, Myers RP, et al:
Progression of liver fibrosis in women infected with hepatitis C:
long-term benefit of estrogen exposure. Hepatology. 40:1426–1433.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Naugler WE, Sakurai T, Kim S, et al:
Gender dis:parity in liver cancer due to sex differences in
MyD88-dependent IL-6 production. Science. 317:121–124. 2007.
View Article : Google Scholar
|
|
18.
|
Omoya T, Shimizu I, Zhou Y, et al: Effects
of idoxifene and estradiol on NF-kappaB activation in cultured rat
hepatocytes undergoing oxidative stress. Liver. 21:183–191. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Teng J, Wang ZY, Jarrard DF, et al: Roles
of estrogen receptor α and β in modulating urothelial cell
proliferation. Endocr Relat Cancer. 15:351–364. 2008.
|
|
20.
|
Danel L, Cordier G, Revillard JP, et al:
Presence of estrogen binding sites and growth-stimulating effect of
estradiol in the human myelogenous cell line HL60. Cancer Res.
42:4701–4705. 1982.PubMed/NCBI
|
|
21.
|
Hatfill SJ, Brusnicky J and Fester E:
Immunocytochemical identification of nuclear estrogen-receptors in
human acute myeloid leukemia. Leuk Res. 15:315–320. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Holst F, Stahl PR, Ruiz C, et al: Estrogen
receptor alpha (ESR1) gene amplification is frequent in breast
cancer. Nat Genet. 39:655–660. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Pancholi S, Lykkesfeldt AE, Hilmi C, et
al: ERBB2 influences the subcellular localization of the estrogen
receptor in tamoxifen-resistant MCF-7 cells leading to the
activation of AKT and RPS6KA2. Endocr Relat Cancer. 15:985–1002.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Boix L, Bruix J, Castells A, et al: Sex
hormone receptors in hepatocellular carcinoma. Is there a rationale
for hormonal treatment? J Hepatol. 17:187–191. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
van’t Veer LJ, Dai H, van de Vijver MJ, et
al: Gene expression profiling predicts clinical outcome of breast
cancer. Nature. 415:530–536. 2002.PubMed/NCBI
|
|
26.
|
van’t Veer LJ, Dai H, van de Vijver MJ, et
al: Expression profiling predicts outcome in breast cancer. Breast
Cancer Res. 5:57–58. 2003.PubMed/NCBI
|
|
27.
|
Archer KJ, Mas VR, Maluf DG, et al:
High-throughput assessment of CpG site methylation for
distinguishing between HCV-cirrhosis and HCV-associated
hepatocellular carcinoma. Mol Genet Genomics. 283:341–349. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Maynadier M, Nirde P, Ramirez JM, et al:
Role of estrogens and their receptors in adhesion and invasiveness
of breast cancer cells. AdvExp Med Biol. 617:485–491. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Ikai I, Itai Y, Okita K, et al: Report of
the 15th follow-up survey of primary liver cancer. Hepatol Res.
28:21–29. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Mizoguchi Y, Takeda H, Kobayashi K, et al:
Impairment in the response of peripheral blood mononuclear cells
from asymptomatic hepatitis B virus carriers to estradiol. Jpn J
Med. 27:183–186. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Mizoguchi Y, Takeda H, Sakagami Y, et al:
Estradiol receptors in the cytosol of peripheral blood mononuclear
cells in hepatitis B virus carriers treated with interferon-alpha.
Gastroenterol Jpn. 24:373–379. 1989.PubMed/NCBI
|
|
32.
|
Wang SH, Yeh SH and Lin WH: Estrogen
receptor α represses transcription of HBV genes via interaction
with hepatocyte nuclear factor 4α. Gastroenterology. 142:989–998.
2012.
|