Introduction
Testicular cancer is the most frequently occurring
malignancy in young men aged 20–39 years (1). In 2008 it was estimated that over
8,000 cases of testicular germ cell tumors (TGCT) were diagnosed in
the United States and Europe, a comparably rare tumor. Furthermore,
the overall incidence has increased worldwide since the turn of the
century and was associated to genetic predispositions and exposure
to environmental contaminants (2,3). The
biology of testicular germ cell tumors is diverse, arising from a
precursor lesion called intratubular germ cell neoplasia that can
be found growing in situ within seminiferous tubules and
which expresses transcription factors common to embryonic stem (ES)
cells, suggesting that the cell of origin is a pluripotent
gonocyte. Despite a common cell of origin, testicular cancers are
histologically and clinically separated into seminoma and
non-seminoma, comprising embryonal carcinoma, yolk sac tumor,
choriocarcinoma and teratoma. The core stemness transcription
factors POU5F1 and NANOG which are expressed in both, seminoma and
non-seminoma tumor cells are thought to be pivotal for the
identification of TGCT. Apart from these common markers, SOX2 has
been suggested to distinguish between the two histological
subtypes, expressed only in non-seminomas (4). The mammalian transcription factor
POU5F1 is expressed by early embryo cells and germ cells and is
essential for maintaining pluripotency (5). While lack of POU5F1 leads to
apoptosis, inappropriate high expression can promote tumorigenesis
(6,7). Similarly, NANOG, another
transcription-factor has been described to be essential for
self-renewal. Whereas NANOG disruption in ES cells results in
differentiation to endoderm lineages, knockdown leads to inhibition
of tumor development (8,9). A transcriptional regulatory circuitry
involving the transcription factors POU5F1, SOX2, NANOG and others
has been identified. Expressed specifically in pluripotent cells,
they may be essential for ES cells self-renewal and
differentiation. They are switched on/off by input environmental
signals and they are also regulated by themselves. When these genes
are expressed, the self-renewal genes are activated and the
differentiated genes are repressed so ES cells can maintain their
pluripotency (8). Experimental
studies revealed repressive epigenetic modification in the promoter
region of NANOG by histone deacetylase inhibitors (HDACi) resulting
in inhibition of the transcription factors NANOG, POU5F1 and SOX2.
The consequence of the knockdown of this ES-like gene signature was
cell cycle arrest and differentiation in all three germ layers
(10).
Phytoestrogens are of special interest in current
research for different reasons. On the one hand the epidemiological
incidence of malignancies is thought to be connected to the
abundance of (phyto-) estrogens (11). On the other hand, the popularity in
the population makes them attractive as potential drugs or
supportive medicine. Studies found that e.g. postmenopausal women
are more willing to take phytoestrogens instead of conventional
hormone-replacement therapy describing them as ‘unnatural’
(12). The rhizome of the leopard
lily Belamcanda chinensis is well known in traditional
Chinese medicine where it is utilized to treat various symptoms and
disease. Different compounds of the extract have been identified so
far, including several phytoestrogens, one of the major components
being tectorigenin (13).
Anti-cancerogenic effects of phytoestrogens, especially of
Belamcanda chinensis extract (BCE) and tectorigenin have
been shown in diverse types of cancer and cell lines. Lee et
al described a tumor inhibitory effect of tectorigenin in human
promyelocytic leukemia HL-60 cells (14). Later, Thelen et al reported
substantial data on the impact of tectorigenin and BCE on prostate
cancer (cell lines) focusing hormone pathways with notable results
(15,16).
The aim of this study was to elucidate the antitumor
activity of BCE and tectorigenin on TGCT cell lines represented by
TCam-2 (seminoma) cells and NTera-2 (non-seminoma). Furthermore, we
attempted to elucidate the mechanism of action of this herbal
drug.
Materials and methods
Cell culture and reagents
Human TGCT cell lines TCam-2 (seminoma) and NTera-2
(non-seminoma) were grown in RPMI-1640 (PAA Laboratories, Pasching,
Austria), supplemented with 10% fetal bovine serum (PAA
Laboratories), 1% penicillin/streptomycin (Invitrogen, Karlsruhe,
Germany), 1% glutamine (PAA Laboratories) and 2.5% HEPES-buffer
(PAA Laboratories). They were cultured in an incubator at 37°C and
5% CO2. After treatment for 24, 48 or 72 h cells were
harvested by scraping and washed three times with PBS. Cell
isolation for RNA and protein extraction was performed by
centrifugation at 1,200 × g for 4 min.
The cells were treated with various concentrations
of BCE (Christoffel Scientific Consulting, Buchberg-Sengenthal,
Germany) or tectorigenin (Girindus, Bergisch Gladbach, Germany)
solubilized in DMSO (Sigma-Aldrich Chemie, Steinheim, Germany),
which was adjusted to 0.1% in all experiments inclusive the
controls. Concentrations were selected according to Thelen et
al (15,16). Valproic acid (Sigma-Aldrich Chemie)
was prepared in sterile water and utilized at 5 mM. Trichostatin A
(Sigma-Aldrich Chemie) 500 nM was solubilized in DMSO.
Concentrations of these two HDACi were selected according to
Venkataramani et al (17).
Cell proliferation
Proliferation and viability of cultured cells after
treatment were colorimetrically measured with an MTT assay using
Cell Proliferation kit I (Roche Diagnostics, Mannheim, Germany).
Therefore 4,000 cells were cultured in 100 μl phenol
red-free RPMI on 96-well plates and stimulated with different
concentrations of BCE and tectorigenin. Further steps were carried
out according to the manufacturer’s protocol.
RNA extraction, quantification and
reverse transcription-PCR
Total RNA was extracted using QIAshredder and RNeasy
mini kit (Qiagen, Hilden, Germany), conducted according to the
producer’s instructions. Quantity and quality were examined by a
Bioanalyser 2100 utilizing the RNA 600 Nano LabChip-Kit (Agilent
Technologies, Waldbronn, Germany). Reverse transcription-PCR of 500
ng total cellular RNA with Random hexamer primers was done using
Omniscript RT kit (Qiagen).
Quantitative real-time PCR (qRT-PCR)
To analyze RNA expression, PCRs were run by using
gene-specific primers. To investigate the stem cell factors, PCR
amplification was performed using specific primer sets (Eurofins
MWG Operon, Ebersberg, Germany) for NANOG (upstream primer,
5′-TTCCTTCCTCCATGGATCTG; downstream primer,
5′-ATCTGCTGGAGGCTGAGGTA), POU5F1 (upstream primer,
5′-AGAAGGATGTGGTCCGAGTG; downstream primer,
5′-GTGAAGTGAGGGCTCCCATA) and SOX2 (upstream primer,
5′-CAAGATGCACAACTCGGAGA; downstream primer,
5′-CTCCGGGAAGCGTGTACTTA). A specific primer set for ARP (upstream
primer, 5′-CGACCTGGAAGTCCAACTAC; downstream primer,
5′-ATCTGCTGCATCTGCTTG) was used as a control. Each sample was
composed of 10 μl qPCR MasterMix Plus for SYBR-Green I
w/fluorescein (Eurogentec, Cologne, Germany), 0.15 μl
downstream and 0.15 μl upstream primer and 4.7 μl
RNase-free water. Individual PCR programs were designed and
amplification and fluorescence measurements were made with iCycler
iQ Real-Time PCR Detection System (Bio-Rad, Munich, Germany). The
achieved data were analyzed using appropriate software
(Bio-Rad).
Microarray analysis
RNA was extracted using the TRIzol method.
Microarrays were done using the Low RNA Input linear Amplification
Kit Plus, One Color protocol (Agilent Technologies, Waldbronn,
Germany). RNA was labeled (mono-color experiment) and hybridized to
the C. elegans 4×44K design array from Agilent Technologies
(Waldbronn, Germany). Quantity and Cy-dye incorporation rates of
the generated target material were measured using a NanoDrop
ND-100. Washing and staining of the arrays were done according to
the manufacturer’s recommendation. Cy3 intensities were detected by
one-color scanning using an Agilent DNA microarray scanner (G2505B)
at 5 micron resolution. Scanned image files were visually inspected
for artifacts and then analyzed. Intensity data were extracted
using Agilent’s Feature Extraction (FE) software, version 9.5 and
analyzed using the Limma package of Bioconductor (18,19).
To find over-represented functions we used DAVID (http://david.abcc.ncifcrf.gov/13.04.2010).
Connectivity map (cmap)
The UniGene IDs were translated into Affymetrix IDs.
Querying the connectivity map was performed by using version build
02 (http://www.broadinstitute.org/cmap/13.08.2011).
Nuclear protein extraction and
quantification
Nuclear lysates were gained by using NucBuster
Protein Extraction Kit (EMD Biosciences, Madison, WI, USA). The
quantification was performed after the well-established method
according to Bradford, utilizing ready-to-use Roti-Quant Kit (Roth,
Karlsruhe, Germany).
Western blot analysis
After preparing appropriate protein concentration of
25 μg, SDS-PAGE was performed using 4–12% Vario-Gels
(Anamed-Elektrophorese GmbH, Gross-Bieberau, Germany). Separation
of proteins by electrophoresis was followed by the transfer to
nitrocellulose membranes (GE Healthcare Europe GmbH, Munich,
Germany) and afterwards blocked for 1 h with 10% non-fat milk in
TBS-T. Primary antibodies were diluted in TBS-T/5% BSA and
incubated with membranes overnight at 4°C. The following primary
antibodies were used: NANOG, POU5F1, histone H3 (Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA), SOX2 (Abcam plc,
Cambridge, UK), acetyl-histone H4 (Upstate Millipore, Billerica,
MA, USA) and β-actin (Sigma-Aldrich Corporation, St. Louis, MO,
USA). The membranes were washed 3X with TBS-T before the secondary
antibody (Dako Denmark A/S, Glostrup, Denmark) was added. Proteins
were visualized on X-ray film (Hyperfilm EC, Amersham Biosciences,
Freiburg, Germany), scanned and analyzed using ImageJ software
(version 1.41o, National Institute of Health).
Results
BCE and tectorigenin inhibit
proliferation of TGCT cell lines
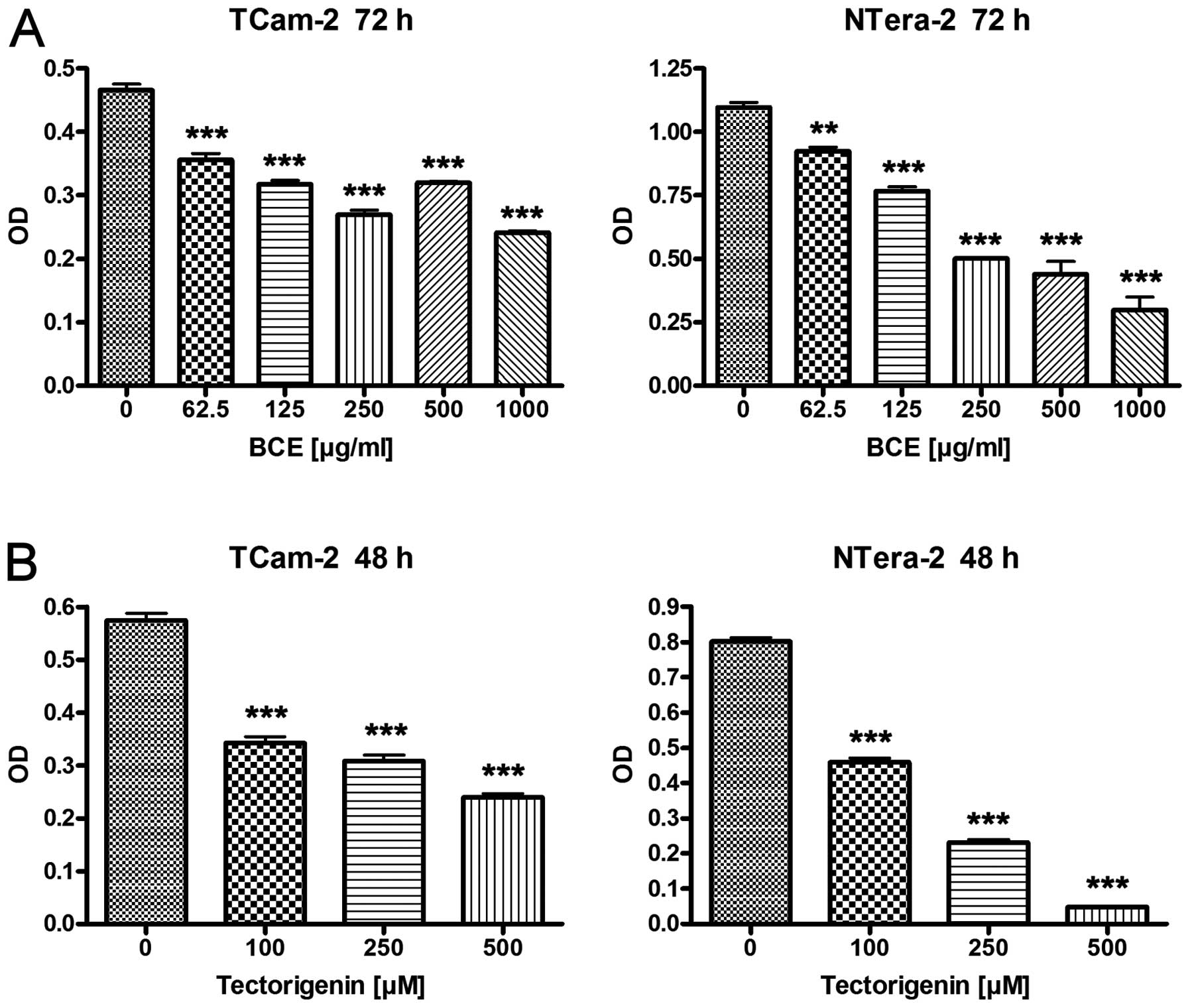
To evaluate the effects of BCE and tectorigenin on
proliferation the TGCT cell lines TCam-2 and NTera-2 were
stimulated with 62.5, 125, 250, 500 and 1,000 μg/ml of BCE
for 24 and 72 h, or 100, 250 and 500 μM of tectorigenin for
24 and 48 h, respectively. After 24 h stimulation with different
concentrations of BCE and tectorigenin cell lines TCam-2 and
NTera-2 showed no or minimal differences in the proliferation rate
compared to the controls, respectively (data not shown). In
contrast TCam-2 and NTera-2 showed a significant reduction of
proliferation in a dosage-dependent manner for tectorigenin after
48 h and BCE after 72 h, respectively (Fig. 1). Based on the morphological
observation that changes typical for apoptosis or cytotoxicity
could not be detected in the TGCT cell lines the results are
suggestive for changes in gene expression.
Differential expression of stem cell
factors in TGCT cell lines after phytoestrogen treatment
Various stem cell genes are important for
self-renewal and are also associated with poorly differentiated
tumors (20). Three important stem
cell genes the NANOG, POU5F1 and SOX2 are normally enriched in
embryonic stem cells. Based on these findings we investigated the
expression of these stem cell factors in TCGT cell lines after
stimulation with BCE.
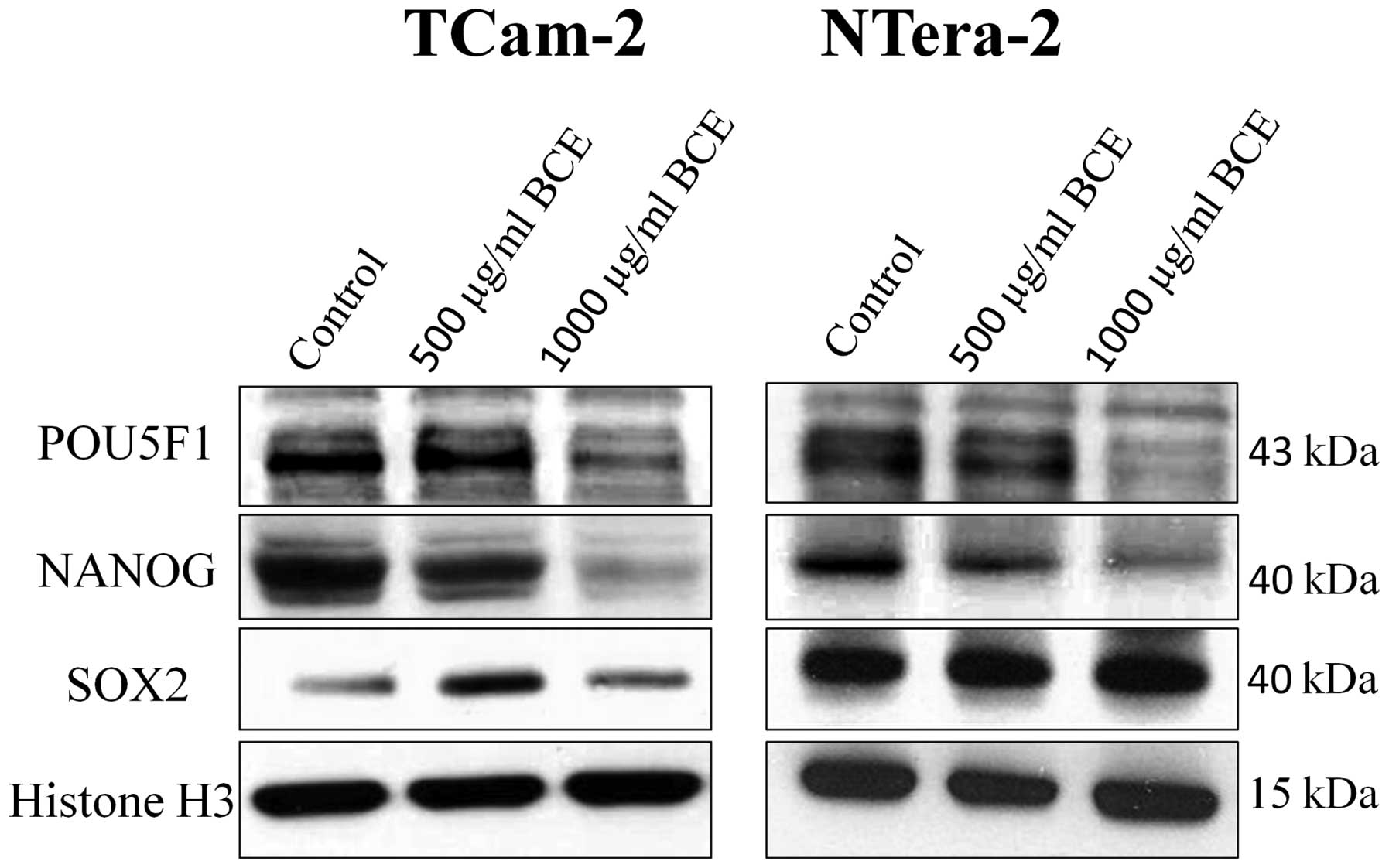
After stimulation with various concentrations of BCE
(62.5, 125, 250, 500 or 1,000 μg/ml) for 24 and 72 h, tumor
cells were analyzed by qRT-PCR or western blot analyses for the
expression of the stem cell genes NANOG, POU5F1 and SOX2,
respectively.
As shown in Fig. 2,
BCE stimulation caused a significant decrease of NANOG and POU5F1
mRNA expression in a dosage-dependent manner in the TGCT cell lines
TCam-2 and NTera-2, respectively. Depending on BCE concentration
NANOG mRNA expression is reduced up to 70% in TCam-2 and 64% in
NTera-2, respectively. In contrast, mRNA expression of SOX2
remained unchanged in both tumor cell lines. Consequently, we asked
whether BCE treatment of TGCT cell lines for 24 h alters the
protein expression of the stem cell genes. Western blot analyses
revealed that according to mRNA expression NANOG and POU5F1
proteins were significantly inhibited in a dosage-dependent manner.
In concordance with the mRNA expression, protein expression of SOX2
showed no different expression as compared to the control (Fig. 3).
Phytoestrogen-induced gene expression
profiling in TGCT cell lines
Gene expression profiling using microarray analysis
was performed after treatment of both TGCT cell lines TCam-2 and
NTera-2 with 1,000 μg/ml BCE for 72 h. Different numbers of
genes were up- or downregulated in TCam-2 and NTera-2 cells,
respectively. Further analyses were focused on differential
expression of genes important for differentiation as well as
carcinogenesis and proliferation. The results showed that genes
important for differentiation (e.g. β-catenin, AP-2γ) are induced
whereas genes being involved in carcinogenesis and proliferation
(e.g. phospholipase A2, GDF-3) are inhibited (Table I and II).
 | Table I.Phytoestrogen-induced gene expression
profiling in TCam-2 cells. |
Table I.
Phytoestrogen-induced gene expression
profiling in TCam-2 cells.
| Symbol | Description | Stimulated vs.
control |
|---|
| TDGF1 |
Teratocarcinoma-derived growth factor
1 | −3.6 |
| GDF3 | Growth
differentiation factor 3 | −3.3 |
| AICDA | Activation-induced
cytidine deaminase | −3.1 |
| MAL2 | Mal, T-cell
differentiation protein 2 | −2.9 |
| NANOG | Nanog homeobox | −2.8 |
| CALCA | Calcitonin-related
polypeptide alpha | −2.8 |
| SFRP2 | Secreted
frizzled-related protein 2 | −2.7 |
| GDF15 | Growth
differentiation factor 15 | −2.7 |
| SLC7A5 | Solute carrier
family 7, member 5 | −2.4 |
| AKT1 | V-akt murine
thymoma viral oncogene homolog 1 | −2.4 |
| TNP1 | Transition protein
1 | −2.3 |
| DIAPH2 | Diaphanous homolog
2 | −2.3 |
| ANGPTL4 | Angiopoietin-like
4 | −2.2 |
| PLP1 | Proteolipid protein
1 | −2.2 |
| EGFL6 | EGF-like-domain,
multiple 6 | −2.1 |
| IRX3 | Iroquois homeobox
3 | 2.0 |
| SOX3 | SRY(sex determining
region Y)-box 3 | 2.0 |
| EGR2 | Early growth
response 2 | 2.0 |
| TFAP2C | Transcription
factor AP-2γ | 2.1 |
| MYH9 | Myosin, heavy chain
9, non-muscle | 2.1 |
| MAFB | V-maf
musculoaponeurotic fibrosarcoma oncogene homolog B | 2.2 |
| DHRS2 |
Dehydrogenase/reductase (SDR family)
member 2 | 2.2 |
| NEUROG3 | Neurogenin 3 | 2.2 |
| HAND1 | Heart and neural
crest derivatives expressed 1 | 2.2 |
| GADD45B | Growth arrest and
DNA-damage-inducible, beta | 2.2 |
| FOXC1 | Forkhead box
C1 | 2.2 |
| JAG1 | Jagged 1 | 2.3 |
| ID3 | Inhibitor of DNA
binding 3, dominant negative helix-loop-helix protein | 2.3 |
| AXIN2 | Axin 2 | 2.3 |
| SEMA4D | Sema domain
(semaphorin) 4D | 2.4 |
| NEUROG2 | Neurogenin 2 | 2.5 |
| ZIC2 | Zic family member
2 | 2.6 |
| HES1 | Hairy and enhancer
of split 1 | 2.6 |
| NEUROG1 | Neurogenin 1 | 2.7 |
| TOB1 | Transducer of
ERBB2, 1 | 3.0 |
| CTNNB1 | Catenin
(cadherin-associated protein), beta 1 | 3.2 |
 | Table II.Phytoestrogen-induced gene expression
profiling in NTera-2 cells. |
Table II.
Phytoestrogen-induced gene expression
profiling in NTera-2 cells.
| Symbol | Description | Stimulated vs.
control |
|---|
| PLA2GA | Phospholipase A2,
group IIA | −4.5 |
| DAZL | Deleted in
azoospermia-like | −4.2 |
| GDF3 | Growth
differentiation factor 3 | −4.1 |
| GPNMB | Glycoprotein
(transmembrane) nmb | −3.5 |
| DAZ2 | Deleted in
azoospermia 2 | −3.1 |
| CALCA | Calcitonin-related
polypeptide alpha | −3.1 |
| GDF15 | Growth
differentiation factor 15 | −2.9 |
| TDGF1 |
Teratocarcinoma-derived growth factor
1 | −2.7 |
| GLI1 | GLI family zinc
finger 1 | −2.6 |
| DMRTB1 | DMRT-like family B
with proline-rich C-terminal, 1 | −2.5 |
| ASCL2 | Achaete-scute
complex homolog 2 | −2.4 |
| AICDA | Activation-induced
cytidine deaminase | −2.3 |
| LPL | Lipoprotein
lipase | −2.2 |
| THRB | Thyroid hormone
receptor, beta | −2.1 |
| EGFL6 | EGF-like-domain,
multiple 6 | −2.1 |
| HAND1 | Heart and neural
crest derivatives expressed 1 | 2.6 |
| HMX2 | H6 family homeobox
2 | 2.7 |
An attempt to identify the mode of action
of BCE on TGCT cell lines utilizing cmap
Connectivity map (cmap) is a reference collection of
gene-expression profiles from cultured human cells treated with
bioactive small molecules. The resource tries to provide a
systematic approach to discover functional connections for example
among diseases, drug action or any small molecules sharing a
mechanism of action (21).
Gene profiles of BCE-stimulated TCam-2 and NTera-2
cells were gained from microarray analysis (see above). Querying
the cmap revealed different substances causing similar and contrary
gene profiles in seminoma and non-semi-noma cell lines. High
connections were achieved with HDACi for both TGCT types
(NTera-2>TCam-2) but also with other different kind of drugs
(Table III and IV). Interestingly, estrogens and
antagonists, as well as genistein (as a phytoestrogen being
represented in the cmap) showed very low connection to BCE-induced
profiles, indicating different mechanism of action (data not
shown).
 | Table III.Results of the cmap query for
connections of BCE-stimulated TCam-2 cells with gene signatures
induced by other substances viewing the ‘permuted results’
table. |
Table III.
Results of the cmap query for
connections of BCE-stimulated TCam-2 cells with gene signatures
induced by other substances viewing the ‘permuted results’
table.
| Rank | Cmap name | n | Mean | P-value |
|---|
| 1 | Meticrane | 5 | −0.719 | 0.00004 |
| 2 | Lisruride | 5 | 0.624 | 0.00006 |
| 3 | Medrysone | 6 | −0.725 | 0.00006 |
| 4 | Doxorubicin | 3 | −0.819 | 0.00026 |
| 5 | Vigabatrin | 3 | 0.689 | 0.00036 |
| 6 | H-7 | 4 | −0.670 | 0.00060 |
| 7 | 0173570-0000 | 6 | −0.585 | 0.00068 |
| 8 | CP-690334-01 | 8 | 0.437 | 0.00076 |
 | Table IV.Results of the cmap query for
connections of BCE-stimulated NTera-2 cells with gene signatures
induced by other substances viewing the ‘permuted results’
table. |
Table IV.
Results of the cmap query for
connections of BCE-stimulated NTera-2 cells with gene signatures
induced by other substances viewing the ‘permuted results’
table.
| Rank | Cmap name | n | Mean | P-value |
|---|
| 1 | Vorinostat | 12 | 0.592 | 0.00000 |
| 2 | CP-690334-01 | 8 | 0.545 | 0.00000 |
| 3 | Trichostatin A | 182 | 0.394 | 0.00000 |
| 4 | Securinine | 4 | −0.588 | 0.00016 |
| 5 | Monensin | 6 | 0.557 | 0.00032 |
| 6 | Perphenazine | 5 | 0.444 | 0.00050 |
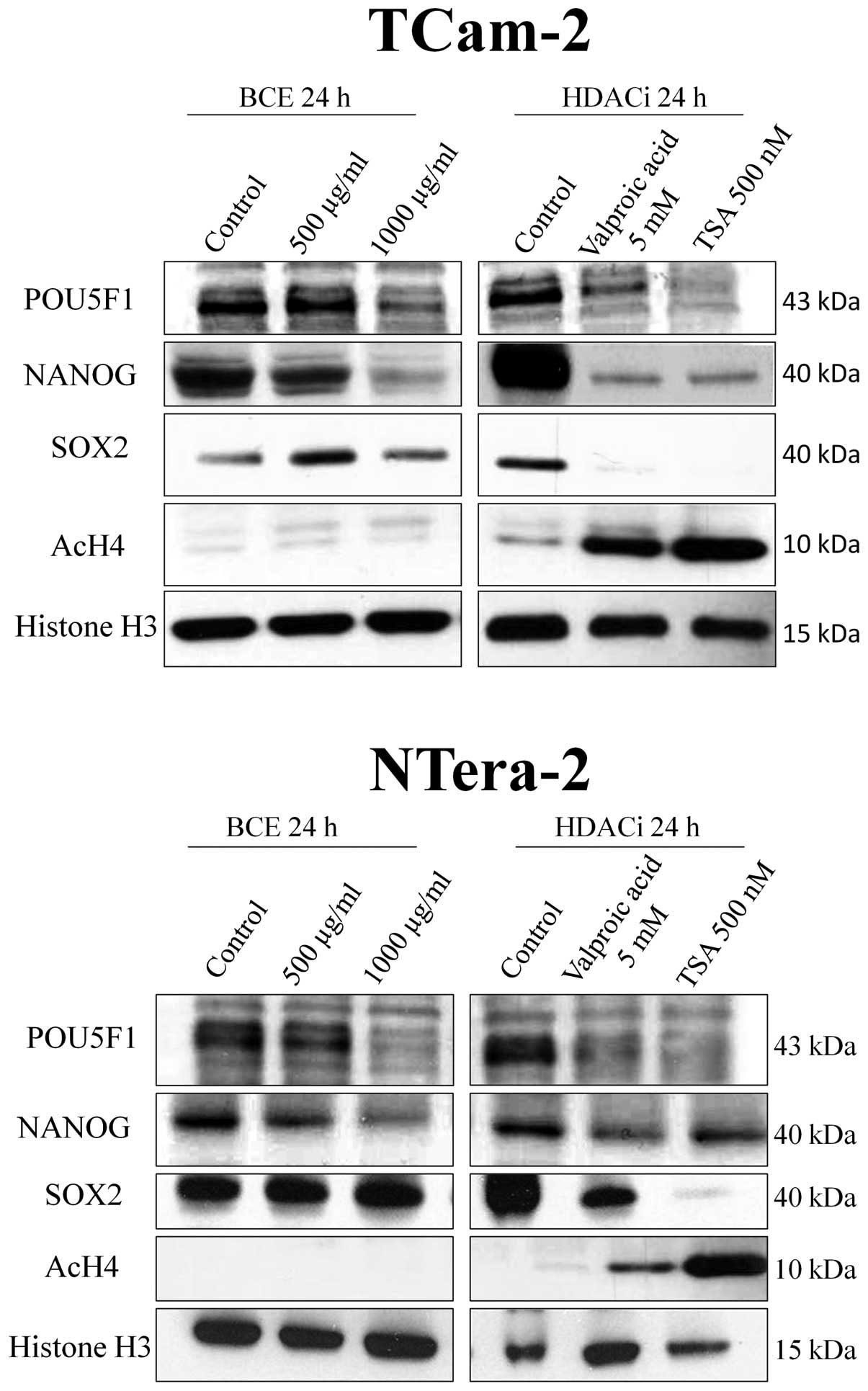
Comparing effects of HDAC inhibitors with
BCE
Based on our results of cmap and the published data
of the depletion of the embryonic stem cell signature by histone
deacetylase inhibitors (10), we
investigated the effect of BCE on histone deacetylase inhibition.
We demonstrated that stimulation of both TGCT cell lines with HDAC
inhibitors valproic acid and trichostatin A (TSA) leads to a
significant decrease of protein expression of the stem cell genes
and the decrease is accompanied by a hyperacetylation of histone
protein H4. In contrast, the phytoestrogen-induced inhibition of
NANOG and POU5F1 protein expression is independent of acetylation
of histone protein H4 (Fig.
4).
Discussion
In this study, we showed that BCE and tectorigenin
inhibit the proliferation of TGCT cells in a time- and
concentration-dependent manner. The anti-proliferative potential of
BCE and the isolated isoflavone thereof tectorigenin has been
evaluated on different cancer types and cell lines (14,22).
Showing time-and concentration-dependent effects on proliferation
in human promyelocytic leukemia HL-60 cells, Lee et al also
described induction of differentiation in these cells. They
reasoned that the anti-proliferative effect of tectorigenin was
ascribed to induction of differentiation and apoptosis (14). Recent studies refuted this because
reduction of cells was exclusively associated with inhibition of
proliferation (changes in G1, S and G2M phase) but not with
increasing amount of apoptosis. The conclusion was that the
inhibitory effect on cell viability is based on cell cycle arrest
(22).
At first sight tectorigenin seems more capable in
reduction of cell viability than BCE. Focusing the used
concentrations of BCE (62.5–1,000 μg/ml) and tectorigenin
(100–500 μM) shows that a direct comparison is not feasible.
Morrissey et al described that 100 μg/ml BCE comprise
17 μM tectorigenin (22)
and Thelen et al also specified the content of tectorigenin
in BCE of about 5%. BCE and tectorigenin were directly compared
before with consistent results (15). Later a potential synergistic effect
from the combination of phytochemicals in BCE was hypothesized
because tumor cell proliferation and androgen receptor expression
were more affected by BCE than by the pure isoflavone tectorigenin
alone (16). But there are also
different views for the variable potential of tectorigenin and BCE.
For example the 5-hydroxyl group of the isoflavone structure of
several phytoestrogens plays a leading role for cytotoxic effects
(14). Equally to other
phytoestrogens tectorigenin comprises this structure (22,23).
BCE also includes other compounds containing this structure as well
as isoflavone glycoside (25)
which is poorly permeable for cell membranes (14).
Furthermore, we investigated whether treatment of
TGCT cells with phytoestrogens influence the expression of stem
cell factors involved in self-renewal and proliferation of poorly
differentiated tumors (10,20).
BCE is capable of downregulating the expression of the stem cell
factors NANOG and POU5F1 in TGCT cells, whereas expression of SOX2
remained unchanged. The detection of SOX2 in seminoma cells TCam-2
is inconsistent with other authors (26), though it was described later that
this cell line also features non-seminomatous characteristics
(4). Previous studies on
HDACi-treated stem cells and embryonal carcinoma cells showed a
connection between downregulation of NANOG and inhibition of
proliferation, however, the exact relation between cell cycle
arrest and NANOG suppression is not yet known. Furthermore You
et al described dependency of the transcription factors
POU5F1 and SOX2 from NANOG in NANOG-siRNA experiments and
stimulation with high dose of apicidin. The conclusion was that
additionally to the known circuitries, other mechanisms are in
involved in the downregulation of POU5F1 and SOX2, independent of
NANOG (10). Previous studies
exhibited incoherent effects on the dependency of these three stem
cell factors (27), being later
interpreted as incomplete knockdown (10). Also, results from estrogen-related
receptor β-knockdown showed that NANOG is depleted followed by
reduction of POU5F1 expression ascribed as POU5F1-dependent
inhibition of NANOG (28).
However, further investigations are needed to explore the
circuitries as well as additional factors.
Effects of phytoestrogens on stem cell factors have
been described before by analyzing genistein. The isoflavonoid
genistein contained in soy induces downregulation of NANOG and
decreased protein levels of POU5F1 and NANOG. It was concluded that
the observed decrease in the transcript level of NANOG is a
downstream effect of genistein-induced depletion of POU5F1 protein
(29). Furthermore it should be
noted, that in our experiments NANOG and POU5F1 mRNA expression are
just mildly repressed compared with the protein expression where
both stem cell factors were barely detectable after stimulation
with BCE and tectorigenin, respectively. One explanation may be
that protein analyses do not provide quantitative results, or that
other mechanisms such as post-transcriptional gene silencing are
involved.
Gene expression analyses of TCam-2 and NTera-2 cells
revealed several genes differentially expressed after BCE
treatment. Using DAVID we focused on genes involved in
differentiation or associated with malignancies. Genes with
repressed expression in stimulated NTera-2 cells: GDF3, which is
also expressed in primordial germ cells, is overexpressed in TGCT
compared with normal testis, just as NANOG and POU5F1 (30). DAZL is only expressed in IGCNU but
not for example in breast cancer cells and is therefore regarded as
germ cell origin of these cells (30). CALCA stimulates growth and motility
of prostate cancer cells and also has essential functions in
angiogenesis (31). In addition
EGFL6 abets, probably through paracrine mechanisms, angiogenesis
and promotes migration of endothelia cells (32). GLI1-knockout experiments showed
that suppression of this transcription factor compromises
proliferation, invasion and migration of cancer cells (33). Similar effects are known for ASCL2.
Liver metastases from colorectal cancer exhibit an ASCL2-related
stem cell signature which likely influences the metastatic activity
of tumor cells (34).
Induced genes in stimulated NTera-2 cells: HAND1 is
known to be involved in morphogenesis and embryogenesis (35). Additionally it is induced in
GADD45G-overexpressed NCCIT cells, as well as in POU5F1-knockout
cells and therefore is involved in cell cycle arrest and
differentiation (36). Besides
being involved in morphogenesis (37) HMX2, like HAND1, is repressed in
cancer and induction seems to be involved in inhibition of
proliferation (38).
Repressively expressed genes in stimulated TCam-2
cells: GDF3, CALCA and EGFL6 are inhibited as well in TCam-2 as in
NTera-2 cells. For information on their function in differentiation
see above. Furthermore, AKT1-expression was reduced, being known
for central roles in proliferation and survival pathways in cancer
(39). Similar mechanisms have
been reported for ANGPTL4, which supports tumor progression through
metastasis and vasculogenesis (40).
Induced gene expression in stimulated TCam-2 cells:
MAFB induces differentiation (41), coexistently it is known as oncogene
in multiple myeloma (42).
Altogether the maf protein can play antagonistic functions in
oncogenesis and plays a dual role as oncogene and tumor
suppressor-like protein (43).
NEUROG3 has important functions in specialization of organs
(44,45). GADD45B expression is associated
with the level of differentiation of tumors being clearly expressed
at lower levels in poorly differentiated compared to well
differentiated tumors. Hence it was proposed as a marker for the
state of differentiation of tumors (46). In summary, BCE-stimulated TGCT cell
lines revealed repression of oncogenes and induction of genes
central for differentiation indicating anti-cancerogenic activities
of this substance.
Assuming a connection between the repression of
NANOG and POU5F1 and hyperacetylation of histone proteins by
genistein (29,47,48)
we investigated whether BCE acts similarly by also affecting
hyperacetylation. HDACi showed, comparable with BCE/tectorigenin,
inhibition of the stem cell signature and inhibition of
proliferation, probably in relation with cell cycle arrest
(10). No hyperacetylation of
histone H4 by BCE highlights the different mode of action of
phytoestrogens despite the comparable effects on the stem cell
signature through these two agents.
Attempting to identify the mode of action of BCE on
TGCT cell lines we utilized cmap. BCE-induced gene signature in
NTera-2 cells revealed high connections to the HDAC inhibitors
vorinostat, CP-690334-01 and trichostatin A. Given that BCE does
not cause hyperacetylation of histone H4 we assume that the
observed commonalities are based on histone-independent mechanisms
like direct hyperacetylation of various transcription factors
(49). Genistein, showing almost
no congruence to BCE, seems to act in a different way and that the
observed similarities are just exceptions. Additionally it should
be noted that we used high dose of BCE compared to genistein in the
cmap (1,000 μg/ml BCE for 72 h vs. 10 μM genistein)
presumably provoking toxic effects.
TCam-2 cells showed just one high ranged HDACi,
CP-690334-01. Strongest positive connection was obtained for
lisuride, which is used in treatment of prolactinoma. Beside
inhibition of prolactin secretion it is also known for reduction of
tumor cell mass and inhibition of transcription (50), furthermore necrotic effects have
been detected (51). By viewing
the ‘permuted results’, genistein had hardly any connection to BCE
in TCam-2 cells. In contrast, on the ‘detailed results’ table BCE
revealed the two highest connections with genistein (used
concentrations see above).
Overall BCE acts differently in seminoma and
non-semi-noma cells. To seize again the suggestion that mechanisms
similar to HDACi may be involved, it has been described that the
effects are partially dependent of the cell type (49) and that different target structures
of HDACi are differentially expressed in TGCT. For example the DNA
methyltransferase DNMT1 is not expressed in seminoma, however, in
embryonal carcinoma it is induced (52).
In summary, we demonstrated that the phytoestrogens
BCE and tectorigenin are capable of inhibiting the proliferation of
TGCT cell lines and lead to a downregulation of the stem cell genes
NANOG and POU5F1. Furthermore, various kinds of genes involved in
differentiation and carcinogenesis were differentially regulated by
phytoestrogens in TGCT cells. In addition cmap reveals high
positive connections to histone deacetylase inhibitors (HDACi) but
BCE stimulation had no effect on histone deacetylase inhibition
thus histone-independent mechanisms such as direct hyperacetylation
of transcription factors are possible. Further investigations are
needed to clarify the molecular mechanism(s) of phytoestrogens
being beneficial in the treatment of TGCT.
Acknowledgements
We thank Anke Klages, Nicole Kerl,
Marion Striepe, Christian Alfen, Lennart Opitz, Gabriela
Salinas-Riester and Perri Hartenstein for excellent technical
assistance and support.
References
|
1.
|
Hayes-Lattin B and Nichols CR: Testicular
cancer: a prototypic tumor of young adults. Semin Oncol.
36:432–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Mannuel HD and Hussain A: Update on
testicular germ cell tumors. Curr Opin Oncol. 22:236–241. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Rajpert-De Meyts E: Developmental model
for the pathogenesis of testicular carcinoma in situ: genetic and
environmental aspects. Hum Reprod Update. 12:303–323.
2006.PubMed/NCBI
|
|
4.
|
Looijenga LHJ: Human testicular
(non)seminomatous germ cell tumours: the clinical implications of
recent pathobiological insights. J Pathol. 218:146–162. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Nichols J, Zevnik B, Anastassiadis K, Niwa
H, Klewe-Nebenius D, Chambers I, Schöler H and Smith A: Formation
of pluripotent stem cells in the mammalian embryo depends on the
POU transcription factor Oct4. Cell. 95:379–391. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Kehler J, Tolkunova E, Koschorz B, Pesce
M, Gentile L, Boiani M, Lomelí H, Nagy A, McLaughlin KJ, Schöler HR
and Tomilin A: Oct4 is required for primordial germ cell survival.
EMBO Rep. 5:1078–1083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Abate-Shen C: Homeobox genes and cancer:
new OCTaves for an old tune. Cancer Cell. 4:329–330. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liu N, Lu M, Tian X and Han Z: Molecular
mechanisms involved in self-renewal and pluripotency of embryonic
stem cells. J Cell Physiol. 211:279–286. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Jeter CR, Badeaux M, Choy G, Chandra D,
Patrawala L, Liu C, Calhoun-Davis T, Zaehres H, Daley GQ and Tang
DG: Functional evidence that the self-renewal gene NANOG regulates
human tumor development. Stem Cells. 27:993–1005. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
You JS, Kang JK, Seo D-W, Park JH, Park
JW, Lee JC, Jeon YJ, Cho EJ and Han JW: Depletion of embryonic stem
cell signature by histone deacetylase inhibitor in NCCIT cells:
involvement of Nanog suppression. Cancer Res. 69:5716–5725. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Adlercreutz H: Phytoestrogens:
epidemiology and a possible role in cancer protection. Environ
Health Perspect. 103:103–112. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Glazier MG and Bowman MA: A review of the
evidence for the use of phytoestrogens as a replacement for
traditional estrogen replacement therapy. Arch Intern Med.
161:1161–1172. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Wagner H, Bauer R, Xiao P, Chen J and
Nenninger A: Rhizoma belamcandae sinensis (Shegan). Chinese Drug
Monographs and Analysis. 2:Verlag fur Ganzheitliche Medizin Dr.
Erich Wuhr, Kotzing. 1–13. 1999.
|
|
14.
|
Lee KT, Sohn IC, Kim YK, Choi JH, Choi JW,
Park HJ, Itoh Y and Mikayamoto K: Tectorigenin, an isoflavone of
Pueraria thunbergiana Benth, induces differentiation and
apoptosis in human promyelocytic leukemia HL-60 cells. Biol Pharm
Bull. 24:1117–1121. 2001.PubMed/NCBI
|
|
15.
|
Thelen P, Scharf J-G, Burfeind P,
Hemmerlein B, Wuttke W, Spengler B, Christoffel V, Ringert R-H and
Seidlova-Wuttke D: Tectorigenin and other phytochemicals extracted
from leopard lily Belamcanda chinensis affect new and
established targets for therapies in prostate cancer.
Carcinogenesis. 26:1360–1367. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Thelen P, Peter T, Hünermund A, Kaulfuss
S, Seidlová-Wuttke D, Wuttke W, Ringert R-H and Seseke F:
Phytoestrogens from Belamcanda chinensis regulate the
expression of steroid receptors and related cofactors in LNCaP
prostate cancer cells. BJU Int. 100:199–203. 2007.PubMed/NCBI
|
|
17.
|
Venkataramani V, Rossner C, Iffland L,
Schweyer S, Tamboli IY, Walter J, Wirths O and Bayer TA: Histone
deacetylase inhibitor valproic acid inhibits cancer cell
proliferation via down-regulation of the alzheimer amyloid
precursor protein. J Biol Chem. 285:10678–10689. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Smyth GK: Linear models and empirical
Bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article 3. 2004.PubMed/NCBI
|
|
19.
|
Gentleman RC, Carey VJ, Bates DM, Bolstad
B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik
K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C,
Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L,
Yang JYH and Zhang J: Bioconductor: open software development for
computational biology and bioinformatics. Genome Biol. 5:R802004.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Ben-Porath I, Thomson MW, Carey VJ, Ge R,
Bell GW, Regev A and Weinberg RA: An embryonic stem cell-like gene
expression signature in poorly differentiated aggressive human
tumors. Nat Genet. 40:499–507. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Lamb J, Crawford ED, Peck D, Modell JW,
Blat IC, Wrobel MJ, Lerner J, Brunet J-P, Subramanian A, Ross KN,
Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons
PA, Wei R, Carr SA, Lander ES and Golub TR: The connectivity map:
using gene-expression signatures to connect small molecules, genes,
and disease. Science. 313:1929–1935. 2006. View Article : Google Scholar
|
|
22.
|
Morrissey C, Bektic J, Spengler B, Galvin
D, Christoffel V, Klocker H, Fitzpatrick JM and Watson WG:
Phytoestrogens derived from Belamcanda chinensis have an
antiproliferative effect on prostate cancer cells in vitro. J Urol.
172:2426–2433. 2004.
|
|
23.
|
Yamaki M, Kato T, Kashihara M and Takagi
S: Isoflavones of Belamcanda chinensis. Planta Med.
56:3351990.
|
|
24.
|
Ito H, Onoue S and Yoshida T:
Isoflavonoids from Belamcanda chinensis. Chem Pharm Bull
(Tokyo). 49:1229–1231. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zhang YY, Wang Q, Qi LW, Qin XY and Qin
MJ: Characterization and determination of the major constituents in
Belamcandae Rhizoma by HPLC-DAD-ESI-MS(n). J Pharm Biomed
Anal. 56:304–314. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
De Jong J, Stoop H, Gillis AJM, Hersmus R,
van Gurp RJ, van de Geijn GJM, van Drunen E, Beverloo HB, Schneider
DT, Sherlock JK, Baeten J, Kitazawa S, van Zoelen EJ, van
Roozendaal K, Oosterhuis W and Looijenga LHJ: Further
characterization of the first seminoma cell line TCam-2. Genes
Chromosomes Cancer. 47:185–196. 2008.PubMed/NCBI
|
|
27.
|
Greber B, Lehrach H and Adjaye J:
Silencing of core transcription factors in human EC cells
highlights the importance of autocrine FGF signaling for
self-renewal. BMC Dev Biol. 7:462007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Van den Berg DLC, Zhang W, Yates A,
Engelen E, Takacs K, Bezstarosti K, Demmers J, Chambers I and Poot
RA: Estrogen-related receptor beta interacts with Oct4 to
positively regulate Nanog gene expression. Mol Cell Biol.
28:5986–5995. 2008.PubMed/NCBI
|
|
29.
|
Regenbrecht CR, Jung M, Lehrach H and
Adjaye J: The molecular basis of genistein-induced mitotic arrest
and exit of self-renewal in embryonal carcinoma and primary cancer
cell lines. BMC Med Genomics. 1:492008. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ezeh UI, Turek PJ, Reijo RA and Clark AT:
Human embryonic stem cell genes OCT4, NANOG, STELLAR, and GDF3 are
expressed in both seminoma and breast carcinoma. Cancer.
104:2255–2265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Dong Y-L, Reddy DM, Green KE, Chauhan MS,
Wang HQ, Nagamani M, Hankins GDV and Yallampalli C: Calcitonin
gene-related peptide (CALCA) is a proangiogenic growth factor in
the human placental development. Biol Reprod. 76:892–899. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Chim SM, Qin A, Tickner J, Pavlos N, Davey
T, Wang H, Guo Y, Zheng MH and Xu J: EGFL6 promotes endothelial
cell migration and angiogenesis through the activation of
extracellular signal-regulated kinase. J Biol Chem.
286:22035–22046. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Das S, Harris LG, Metge BJ, Liu S, Riker
AI, Samant RS and Shevde LA: The hedgehog pathway transcription
factor GLI1 promotes malignant behavior of cancer cells by
up-regulating osteopontin. J Biol Chem. 284:22888–22897. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Stange DE, Engel F, Longerich T, Koo BK,
Koch M, Delhomme N, Aigner M, Toedt G, Schirmacher P, Lichter P,
Weitz J and Radlwimmer B: Expression of an ASCL2 related stem cell
signature and IGF2 in colorectal cancer liver metastases with
11p15.5 gain. Gut. 59:1236–1244. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Riley P, Anson-Cartwright L and Cross JC:
The Hand1 bHLH transcription factor is essential for placentation
and cardiac morphogenesis. Nat Genet. 18:271–275. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Jung M, Peterson H, Chavez L, Kahlem P,
Lehrach H, Vilo J and Adjaye J: A data integration approach to
mapping OCT4 gene regulatory networks operative in embryonic stem
cells and embryonal carcinoma cells. PLoS One. 5:e107092010.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Wang W, Chan EK, Baron S, Van de Water T
and Lufkin T: Hmx2 homeobox gene control of murine vestibular
morphogenesis. Development. 128:5017–5029. 2001.PubMed/NCBI
|
|
38.
|
Jin B, Yao B, Li J-L, Fields CR, Delmas
AL, Liu C and Robertson KD: DNMT1 and DNMT3B modulate distinct
polycomb-mediated histone modifications in colon cancer. Cancer
Res. 69:7412–7421. 2009. View Article : Google Scholar
|
|
39.
|
Carpten JD, Faber AL, Horn C, Donoho GP,
Briggs SL, Robbins CM, Hostetter G, Boguslawski S, Moses TY, Savage
S, Uhlik M, Lin A, Du J, Qian Y-W, Zeckner DJ, Tucker-Kellogg G,
Touchman J, Patel K, Mousses S, Bittner M, Schevitz R, Lai MHT,
Blanchard KL and Thomas JE: A transforming mutation in the
pleckstrin homology domain of AKT1 in cancer. Nature. 448:439–444.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Nakayama T, Hirakawa H, Shibata K, Nazneen
A, Abe K, Nagayasu T and Taguchi T: Expression of angiopoietin-like
4 (ANGPTL4) in human colorectal cancer: ANGPTL4 promotes venous
invasion and distant metastasis. Oncol Rep. 25:929–935. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Kelly LM, Englmeier U, Lafon I, Sieweke MH
and Graf T: MafB is an inducer of monocytic differentiation. EMBO
J. 19:1987–1997. 2000. View Article : Google Scholar
|
|
42.
|
Barillé-Nion S, Barlogie B, Bataille R,
Bergsagel PL, Epstein J, Fenton RG, Jacobson J, Kuehl WM,
Shaughnessy J and Tricot G: Advances in biology and therapy of
multiple myeloma. Hematology Am Soc Hematol Educ Program. 248–278.
2003.PubMed/NCBI
|
|
43.
|
Pouponnot C, Sii-Felice K, Hmitou I,
Rocques N, Lecoin L, Druillennec S, Felder-Schmittbuhl M-P and
Eychène A: Cell context reveals a dual role for Maf in oncogenesis.
Oncogene. 25:1299–1310. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Zhang YQ, Mashima H and Kojima I: Changes
in the expression of transcription factors in pancreatic AR42J
cells during differentiation into insulin-producing cells.
Diabetes. 50:S10–S14. 2001. View Article : Google Scholar
|
|
45.
|
Gasa R, Mrejen C, Lynn FC, Skewes-Cox P,
Sanchez L, Yang KY, Lin CH, Gomis R and German MS: Induction of
pancreatic islet cell differentiation by the neurogenin-neuroD
cascade. Differ Res Biol Divers. 76:381–391. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Zenmyo M, Tanimoto A, Sakakima H, Yokouchi
M, Nagano S, Yamamoto T, Ishido Y, Komiya S and Ijiri K: Gadd45β
expression in chondrosarcoma: a pilot study for diagnostic and
biological implications in histological grading. Diagn Pathol.
5:692010.
|
|
47.
|
Kikuno N, Shiina H, Urakami S, Kawamoto K,
Hirata H, Tanaka Y, Majid S, Igawa M and Dahiya R: Genistein
mediated histone acetylation and demethylation activates tumor
suppressor genes in prostate cancer cells. Int J Cancer.
123:552–560. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Majid S, Dar AA, Shahryari V, Hirata H,
Ahmad A, Saini S, Tanaka Y, Dahiya AV and Dahiya R: Genistein
reverses hypermethylation and induces active histone modifications
in tumor suppressor gene B-cell translocation gene 3 in prostate
cancer. Cancer. 116:66–76. 2010.PubMed/NCBI
|
|
49.
|
Bolden JE, Peart MJ and Johnstone RW:
Anticancer activities of histone deacetylase inhibitors. Nat Rev
Drug Discov. 5:769–784. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Iván G, Szigeti-Csúcs N, Oláh M, Nagy GM
and Góth MI: Treatment of pituitary tumors: dopamine agonists.
Endocrine. 28:101–110. 2005.PubMed/NCBI
|
|
51.
|
Ferrari C and Crosignani PG: Medical
treatment of hyperprolactinaemic disorders. Hum Reprod. 1:507–514.
1986.
|
|
52.
|
Omisanjo OA, Biermann K, Hartmann S,
Heukamp LC, Sonnack V, Hild A, Brehm R, Bergmann M, Weidner W and
Stege K: DNMT1 and HDAC1 gene expression in impaired
spermatogenesis and testicular cancer. Histochem Cell Biol.
127:175–181. 2007. View Article : Google Scholar : PubMed/NCBI
|