Introduction
Osteosarcoma is one of the common primary malignant
bone tumor diagnosed in children and teenagers (1,2).
Current chemotherapy regimens of osteosarcoma are based on a
combination of doxorubicin, methotrexate (MTX) and cisplatin
(3–6). Only 50–60% of tumors are
chemosensitive, demonstrating the dismal outcome that occurs far
too often in osteosarcoma (7–9). One
potential strategy to overcome known chemotherapy agents in
osteosarcoma is to seek out alternative anticancer agents,
particularly those appearing from natural products or traditional
Chinese medicine (TCM) (10–13).
Curcumin is from the plant Curcuma longa
(tumeric) and has been used in traditional Chinese medicine for
thousands of years (14–18). Many pharmacological effects have
been reported including anti-amyloid, anti-bacterial,
anti-depressant, anti-inflammatory, anti-oxidant and anticancer
properties (19–24). Curcumin has also been proven to
affect multiple signaling pathways such as inhibiting cancer cell
proliferation, inducing apoptosis or autophagy (25–27),
blocking cell invasion and migration (28–31)
and suppressing inflammatory responses (19,32–34).
Phase II and III clinical trials with curcumin have advocated its
use for patients with colon and pancreatic cancers (35–41).
The low water solubility contributed to the poor bioavailability is
the primary limiting factor for the efficacy and safety of curcumin
(42–46). To improve the oral bioavailability
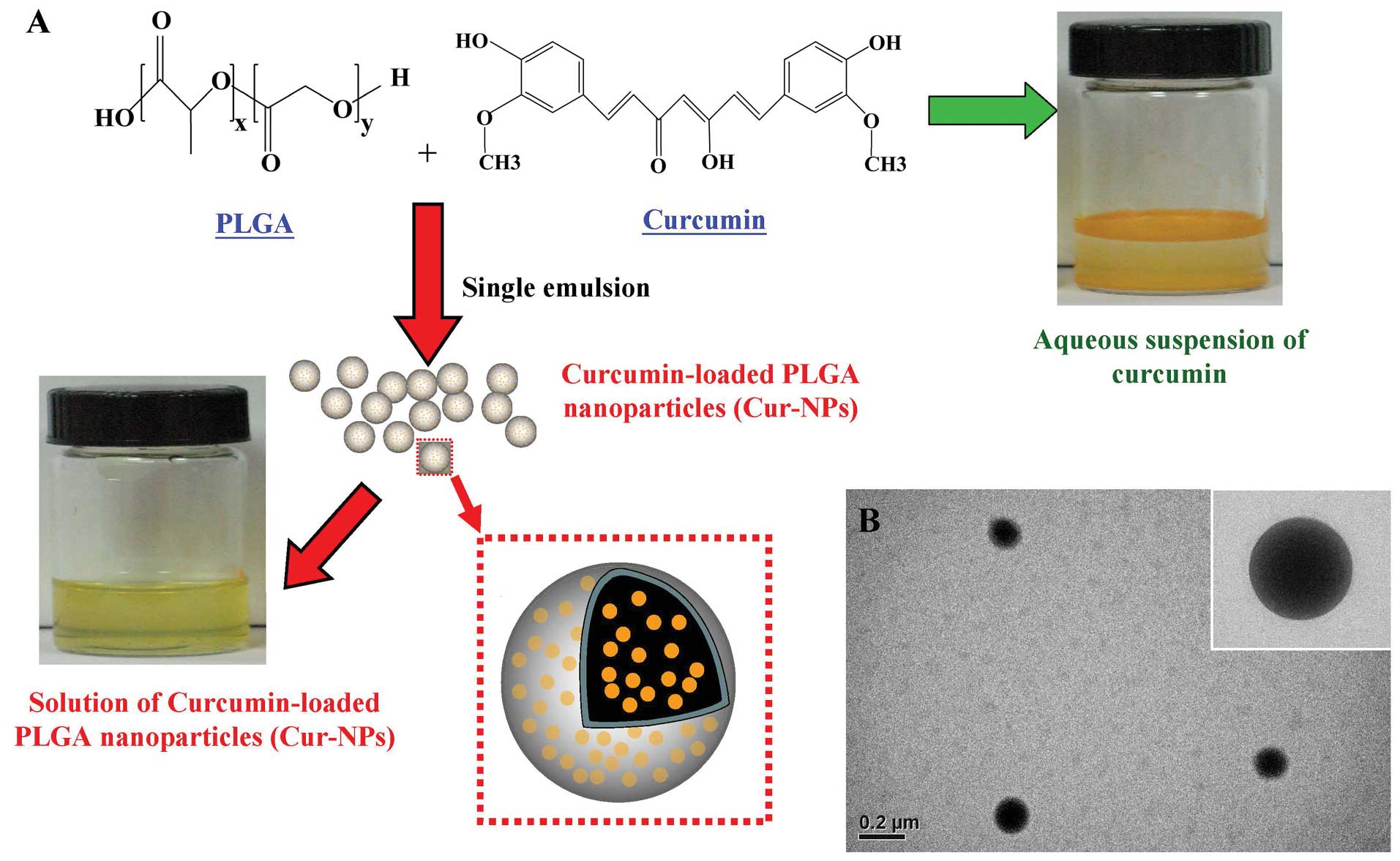
of curcumin, we designed and developed Cur-NPs (PLGA nanoparticles
loaded with curcumin) (Fig. 1A)
(42). The morphology of the
Cur-NPs was examined by transmission electron microscopy. The
produced Cur-NPs are spherical in shape with smooth surface
(Fig. 1B). Our previous study
showed that the Cur-NPs caused anti-proliferation effects on
cisplatin-resistant human oral cancer CAR cells, but little
cytotoxicity to the normal human gingival fibroblasts cells (HGF)
and normal human oral keratinocyte cells (OK) (42). The aims of this study were to
characterize the properties of Cur-NPs and to investigate the
molecular mechanisms triggered by Cur-NPs in human osteosarcoma
U2OS cells.
Materials and methods
Chemicals and reagents
Cisplatin,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
poly(D,L-lactide-co-glycolide) (PLGA, copolymer ratio 75:25,
molecular weight = 66,000–92,000), polyvinyl alcohol (PVA, average
molecular weight = 30,000–70,000) and curcumin were purchased from
Sigma-Aldrich Corp. (St. Louis, MO, USA). Fetal bovine serum (FBS),
L-glutamine, penicillin G and trypsin-EDTA were obtained from Life
Technologies (Carlsbad, CA, USA). Caspase-3/-7 and caspase-9
activity assay kits were purchased from R&D Systems Inc.
(Minneapolis, MN, USA). The primary antibodies against caspase-3,
caspase-9, Apaf-1, cytochrome c, AKT, p-AKT and BAD were
obtained from Cell Signaling Technology Inc. (Beverly, MA, USA).
All other antibodies used in this study and horseradish peroxidase
(HRP)-conjugated secondary antibodies against rabbit or mouse
immunoglobulin were purchased from Santa Cruz Biotechnology Inc.
(Santa Cruz, CA, USA). The enhanced chemiluminescence (ECL)
detection kit (Immobilon Western Chemiluminescent HRP Substrate)
was obtained from Merck Millipore (Billerica, MA, USA).
Cell culture
Human osteosarcoma U2OS cell line was obtained from
the Food Industry Research and Development Institute (Hsinchu,
Taiwan). Cells were maintained at 37°C under a humidified 5%
CO2 atmosphere in 90% McCoy’s 5a medium (Invitrogen Life
Technologist, Grand Island, NY, USA) containing 2 mM L-glutamine,
10% fetal bovine serum (Life Technologies) and 1%
penicillin-streptomycin (100 U/ml penicillin and 100 μg/ml
streptomycin) (Life Technologies) (47–50).
Curcumin loaded nanoparticles
Curcumin-loaded PLGA nanoparticles (Cur-NPs) were
prepared by using single emulsion solvent evaporation method.
Cucurmin (1 mg) and PLGA (10 mg) were dissolved in dichloromethane.
The curcumin and PLGA solution (1 ml) was added to 2 ml of 10%
(w/v) PVA surfactant solution to form an oil-in-water emulsion by
sonication. The emulsion was carried out by setting sonication at
55 W of energy output for 3 min over an ice bath. The formed
emulsion was dispersed by dropping into the 0.5% (w/v) PVA solution
and stirred for an additional 4 h at room temperature on a magnetic
stir plate to allow evaporation of organic solvent. Nanoparticles
were collected by centrifugation at 12,000 rpm for 30 min and
washed twice with double distilled water to remove PVA and
un-encapsulated curcumin. The prepared nanoparticles were collected
and lyophilized (42).
Size, polydispersity index (PDI) and
encapsulation efficiency
The size and the polydispersity of prepared
nanoparticles (PLGA-NPs and Cur-NPs) were measured using DLS
(Zetasizer Nano ZS, 3000HS, Malvern Instruments Ltd.,
Worcestershire, UK). To determine the encapsulation efficiency in
Cur-NPs before freeze-drying, the amount of non-encapsulated
curcumin was measured the absorption value of OD450 nm
by ELISA reader. The encapsulation efficiency was calculated by
[(Total amount of curcumin- non-encapsulated curcumin)/Total amount
of curcumin] × 100% (51).
Transmission electron microscopy (TEM)
observation
The morphology of test nanoparticles was examined by
TEM (Jeol, Tokyo, Japan). A dilute suspension of nanoparticles
(1/10 dilution) was prepared in double distilled water. One drop of
this solution was placed on the TEM grid for 10 min, washed twice
with double distilled water and allowed to dry overnight. The
images were observed and captured at an accelerating voltage of 120
kV under a microscope (42).
NMR and mass spectra
NMR spectra were obtained on a Bruker 500 MHz FT-NMR
(model: Avance III DPX-500) spectrometer in CDCl3. The following
abbreviations are used: s, singlet; d, doublet; m, multiplet. Mass
spectra were measured with a Finnigan/Thermo Quest MAT 95XL
instrument (52,53).
Cell viability and apoptotic
morphological features
The cell viability was assessed by the MTT assay.
The U2OS cells were cultured in a 96-well plate at the density of
1×104 cells per well and were incubated with 0, 0.25,
0.5, 1 and 2 μg/ml of Cur-NPs for 24 and 48 h. Then, culture
medium containing 500 μg/ml MTT was added to each well and
incubated at 37°C for 4 h. After incubation, the supernatant was
removed. The formed blue formazan crystals in viable U2OS cells
were dissolved with isopropanol/0.04 N HCl, followed by measurement
of the absorbance of each well at 570 nm with the ELISA reader. All
experiments were performed in triplicate. The morphological
examination in Cur-NPs-treated U2OS cells was determined under a
phase-contrast microscope (50).
Internalization of curcumin
To track the internalization of Cur-NPs, U2OS cells
(1×106) were seeded on 6-well plates and incubated
overnight. Subsequently, cells were treated with 1 μg/ml of
Cur-NPs for 24 h. Finally, cells were washed with PBS twice and the
internalized curcumin particles were observed under a fluorescence
microscope with the filter of 488-nm excitation wavelength and
520-nm emission (27).
DNA fragmentation assay
The U2OS cells (1×107) were exposed to 1
μg/ml Cur-NPs for 48 h. Cells were harvested, washed by PBS
and then lysed in 500 μl lysis buffer at 4°C. The lysed
cells were digested overnight with proteinase K (100 μg/ml)
at 50°C followed by incubation with 50 μg/ml RNase A at 37°C
for 1 h. DNA fragments were extracted twice with
phenol/chloroform/isopropanol (24:25:1; v/v/v) and precipitated
with 50% isopropanol with glycogen (20 μg/ml) before being
re-suspended in 100 μl Tris-EDTA (TE) buffer (Amresco Inc.,
Solon, OH, USA). Samples were electrophoresed on 1.8% (w/v) agarose
gel (Sigma-Aldrich Corp.) in 0.5X TBE buffer (Amresco Inc.) and DNA
was stained with 1 μg/ml ethidium bromide (Life
Technologies). The gel was observed and photographed under a UV
lamp (54,55).
Western blot analysis
The U2OS cells (1×107) were treated with
0, 0.5, 1 and 2 μg/ml of Cur-NPs for 12 or 48 h. Cells were
then harvested, lysed and the total proteins were collected by SDS
sample buffer. Approximately 50 μg of proteins from each
treatment were resolved on 10% SDS-polyacrylamide gel
electrophoresis (PAGE) and electro-transferred to the Immobilon-P
Transfer Membrane (Merck Millipore). The transferred membranes were
blocked with 5% non-fat dry milk in 20 mM Tris-buffered
saline/0.05% Tween-20 for 1 h at room temperature followed by
incubation with appropriate primary antibodies at 4°C overnight. At
the end of incubation, membranes were washed with Tris-buffered
saline/Tween-20 and incubated with secondary antibodies conjugated
with HRP. The blots were developed by the chemiluminescence kit and
then exposed to X-ray film. Each membrane was stripped and reprobed
with anti-β-actin antibody (Sigma-Aldrich Corp.) to ensure equal
protein loading during the experiments (47,56,57).
Assays for caspase-3/-7 and caspase-9
activities
The U2OS cells (1×107) were exposed to 1
μg/ml of Cur-NPs for 0, 12, 24, 36 and 48 h. Subsequently,
cells were harvested, and cell lysates were assessed in accordance
with the manufacturer’s instructions provided in the caspase-3/-7
and caspase-9 colorimetric assay kits (R&D System Inc.). Cell
lysate containing 50 μg proteins were then incubated for 1 h
at 37°C with specific caspase-3/-7 substrate (DEVD-pNA) or
caspase-9 substrate (LEHD-pNA) and determined by measuring
OD405 of the released pNA (58–60).
Statistical analysis
All the statistical results are performed as the
mean ± standard error of the mean (SEM) for the indicated numbers
of independent experiments. Statistical analyses of data were done
using one-way ANOVA followed by Student’s t-test and the levels of
P<0.001 was considered significant between the treated and
untreated group (61).
Results
Characterization of Cur-NPs
In order to improve the application of curcumin,
curcumin was encapsulated by PLGA to form curcumin-loaded PLGA
nanoparticles (Cur-NPs) by single emulsion method (Fig. 1A). The nanoparticle form of
curcumin exhibited good water-solubility and formed a transparent
solution while dissolving in water. TEM was used to examine the
morphology of the Cur-NPs. As shown, the Cur-NPs showed spherical
shape (Fig. 1B). The size
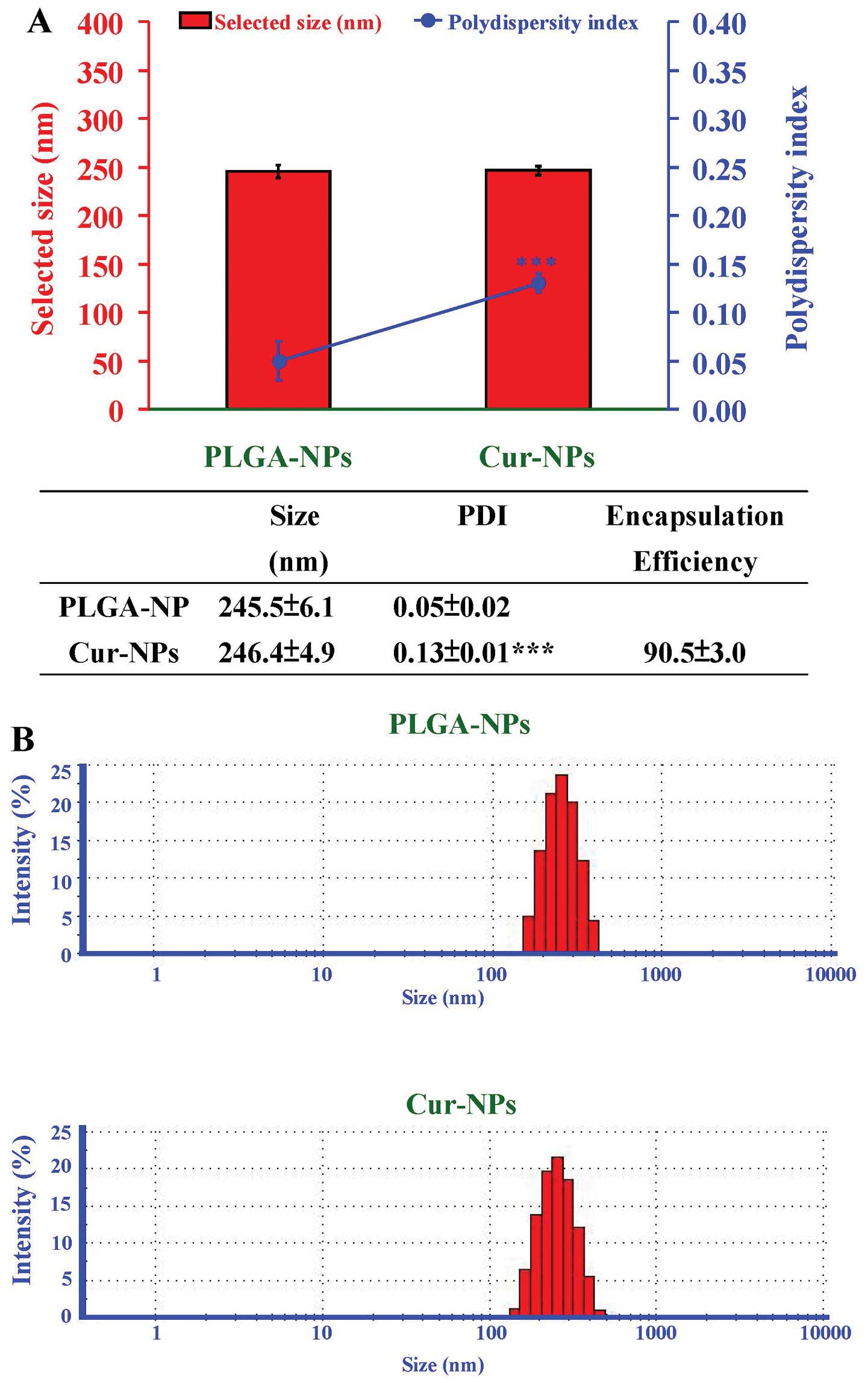
distribution of Cur-NPs in aqueous solution was measured by DLS. As
shown in Fig. 2A, the size of
Cur-NPs is ∼250 nm, similar to the curcumin-empty PLGA-NPs. Both
nanoparticles showed small polydispersity index (PDI) (Fig. 2A), indicating their homogeneous
size distribution (Fig. 2B). The
encapsulation efficiency of curcumin in Cur-NPs prepared by single
emulsion was 90.5±3.0% (Fig.
2A).
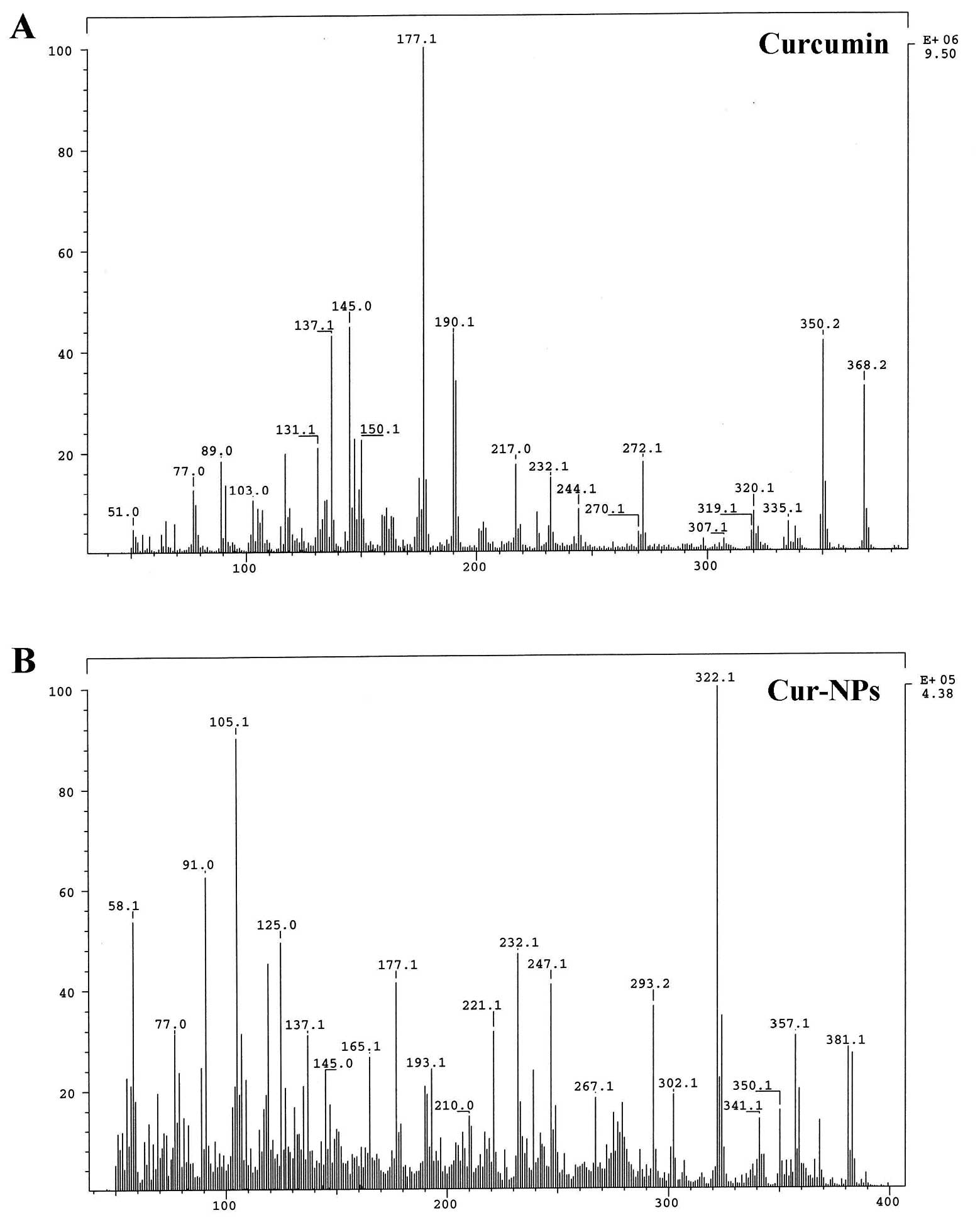
In order to confirm that curcumin was unaffected
after nano-technologization, mass (MS) spectroscopy of curcumin and
Cur-NPs were performed and we found the curcumin fragments on MS
spectrum of Cur-NPs (Fig. 3).
Furthermore, the proton nuclear magnetic resonance
(1H-NMR) spectroscopy was used to obtain the
1H-NMR spectra of PLGA-NPs [poly(lactic-co-glycolic
acid) nanoparticles], curcumin standard and Cur-NPs (Fig. 4). Comparing Fig. 4A and B, all the peaks of curcumin
standard (Fig. 4A) could be found
on the Cur-NPs spectrum (Fig. 4B)
with identical chemical shifts and integration. Besides the peaks
of curcumin, there were some other peaks, δ1.58–1.62 (354H, m),
4.66–4.93 (58H, m) and 5.17–5.27 (89H, m), on the Cur-NPs spectrum.
The chemical shifts and integration ratio (∼4:0.6:1) of these
additional peaks are identical with that of PLGA NPs, δ1.58–1.62
(4H, m), 4.63–4.93 (0.6H, m) and 5.17–5.27 (1H, m). From the
careful inspections of spectral data described above, curcumin was
constant and unaffected by nano-technologization.
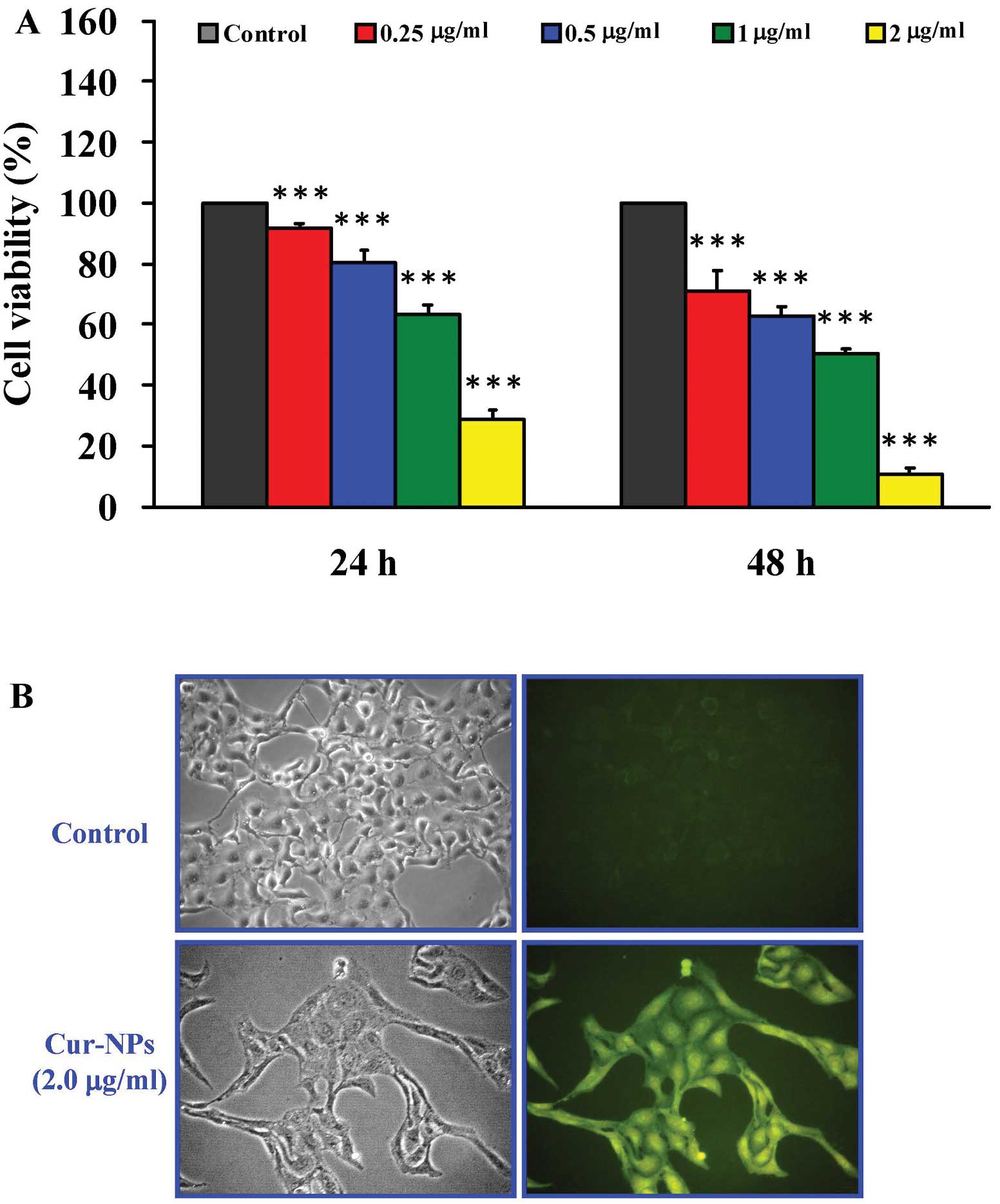
Cur-NPs reduce the viability of human
osteosarcoma U2OS cells
The U2OS cells were treated with Cur-NPs (0, 0.25,
0.5, 1 and 2 μg/ml) for 24 and 48 h. The cells were
collected and the cell viability was determined using MTT assay.
Our results showed that the concentrations of 0.25, 0.5, 1 and 2
μg/ml Cur-NPs significantly decreased cell viability in U2OS
cells concentration- and time-dependently (Fig. 5A). Cellular uptake of Cur-NPs was
visualized by green fluorescence of curcumin using fluorescence
microscopy (Fig. 5B). Intensified
fluorescence was detected in the cytoplasm and nucleus of cells
treated with Cur-NPs, suggesting the amount of curcumin
internalized into the cells. Our results demonstrated that Cur-NPs
display the anti-human osteosarcoma U2OS cells in vitro.
Cur-NPs induce apoptosis in human
osteosarcoma U2OS cells
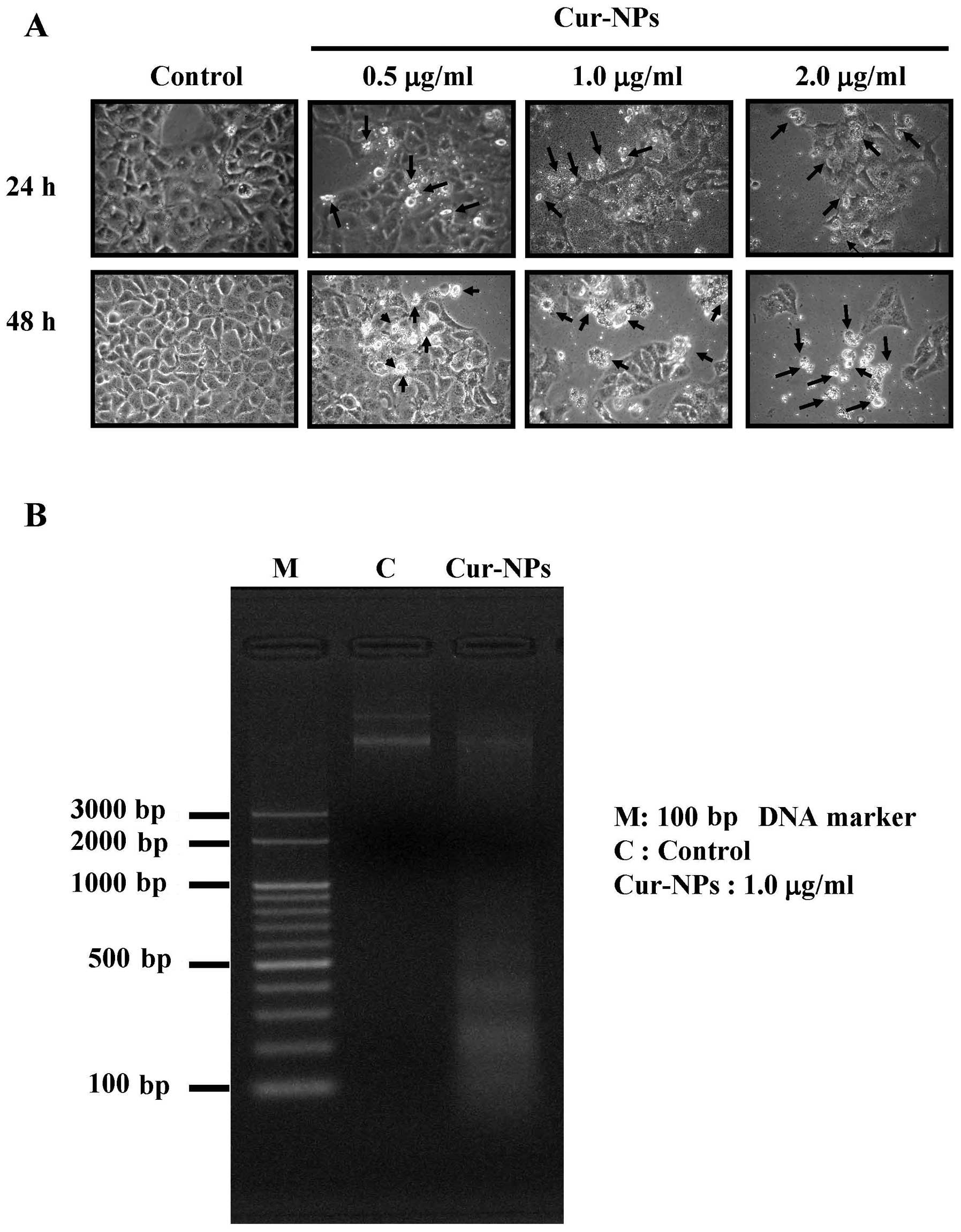
After treatment with 0.5, 1 and 2 μg/ml of
Cur-NPs for 48 h, Fig. 6A revealed
apoptotic bodies in Cur-NPs-treated U2OS cells and this effect is
concentration-dependent. Further results are demonstrated in
Fig. 6B, which indicated that
Cur-NPs induced DNA fragmentation in Cur-NPs-treated U2OS
cells.
Cur-NPs trigger mitochondria-dependent
apoptotic cell death in U2OS cells
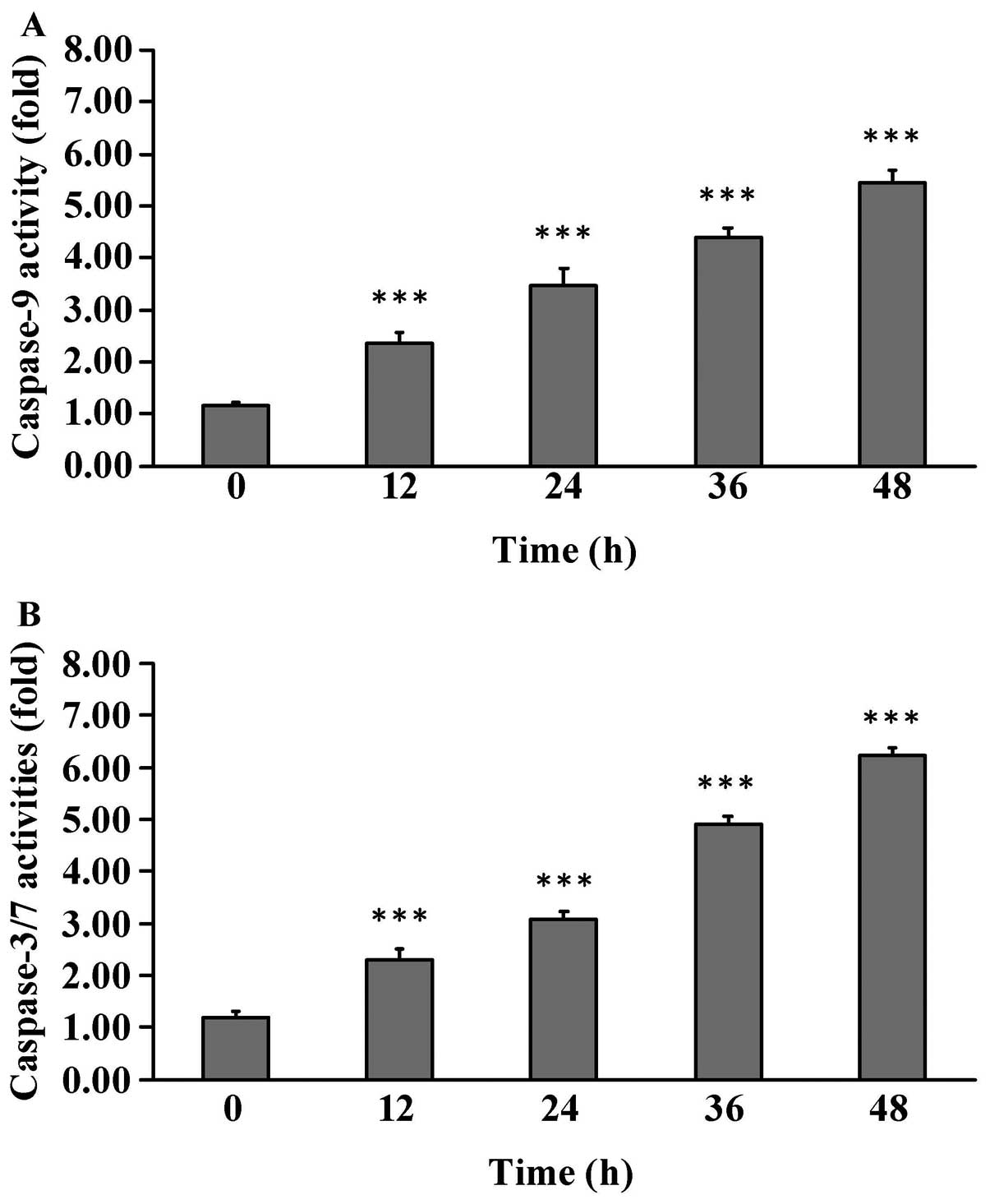
To examine whether Cur-NPs induces apoptosis in U2OS
cells, cells were treated with 1 μg/ml of Cur-NPs for 0, 12,
24, 36 and 48 h before subjected to caspase-3/-7 and caspase-9
activities. Cur-NPs stimulated caspase-9 (Fig. 7A) and caspase-3/-7 (Fig. 7B) activities in Cur-NPs-treated
U2OS cells and this effect is time-dependent. Based on these
findings, we provide evidence regarding the intrinsic caspase
contributing to Cur-NPs-induced apoptosis in U2OS cells.
Mitochondria-dependent and Akt-Bad
signaling pathways were involved in Cur-NPs-induced apoptosis in
U2OS cell apoptosis
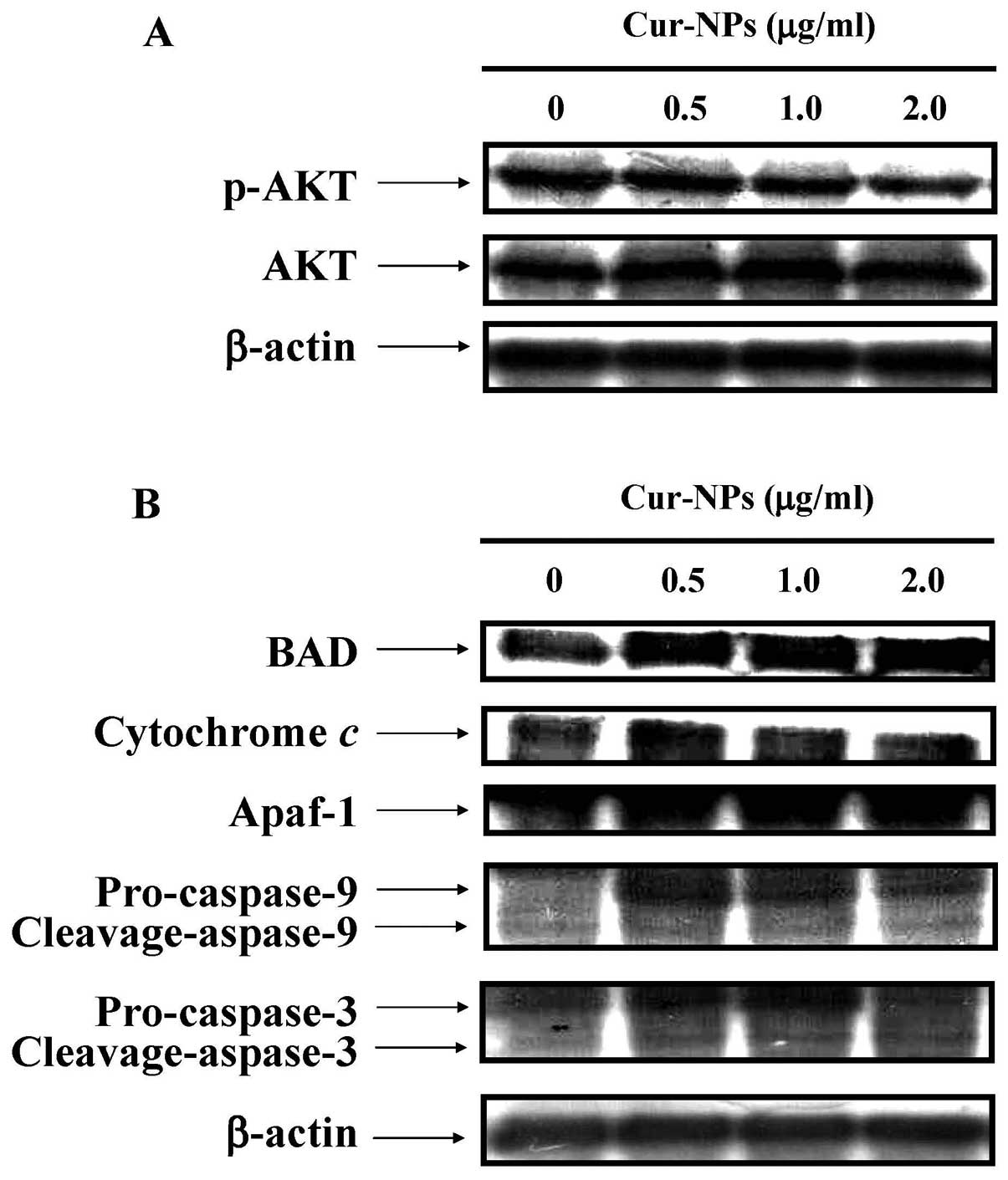
We examined the effects of Cur-NPs on
mitochondria-dependent and Akt-Bad signaling pathways in U2OS
cells. The immunoblotting results showed that the protein level of
p-Akt was decreased in Cur-NPs-treated U2OS cells (Fig. 8A). In contrast, the protein levels
of cleaved caspase-3, cleaved caspase-9, cytochrome c,
Apaf-1 and Bad were increased in Cur-NPs-treated U2OS cells
(Fig. 8B). In conclusion, our data
expand the current understanding of Cur-NPs treatment in U2OS cells
on causing cell death through the mitochondrial-dependent caspase
cascade and the Akt-Bad signaling in vitro.
Discussion
The study published by Yin et al demonstrated
that Cur-NPs are effective in inhibiting the growth of human lung
cancer and exhibited little toxicity to normal tissues in an
established A549 xenograft mouse model (42,62).
Our previous study also showed that the Cur-NPs used in our study
caused anti-proliferation effects on CAR cells in a dose- and
time-dependent manner but little cytotoxicity to the normal human
gingival fibroblasts cells (HGF) and normal human oral keratinocyte
cells (OK) (29). This is the
first study to investigate the anti-human osteosarcoma effects of
Cur-NPs on human osteosarcoma U2OS cells. Our results showed that
Cur-NPs inhibited U2OS cell proliferation (Fig. 5) and induced apoptotic cell death
(Fig. 6) in a concentration- and
time-dependent manner. The results in Fig. 7 show Cur-NPs enhanced cell
apoptosis through the activation of caspase-9 and caspase-3/-7 in
U2OS cells. Our results suggested that Cur-NPs-induced apoptosis
might be through the mitochondria-dependent signaling pathway,
which has a connection with the activation of caspase-9 and -3.
Previous research has shown that
mitochondrial-mediated apoptosis is regulated by the Bcl-2 family
proteins (63-65). The Bcl-2 family includes
pro-apoptotic proteins (Bax and Bad) and anti-apoptotic proteins
(Bcl-2 and Bcl-xL) (66-70). The ratio of pro-apoptotic and
anti-apoptotic proteins is thought to determine, at least in part,
the susceptibility of cells to a death signal (68,69,71).
Previous studies have shown that the apoptotic stimuli can
de-phosphorylate Bad and release Bad from the 14-3-3 protein
(72-75). Thus, Bad will compete with
Bcl-2/Bcl-xL in binding to Bax (76–79).
Previously, it was shown that functional Akt phosphorylated Bad at
Ser136 to promote the stabilization of the mitochondrial membrane
system and cell survival (78,80).
Our results demonstrated that the protein level of p-AKT was
decreased (Fig. 8A), while the
protein levels of cleaved caspase-3, cleaved caspase-9, cytochrome
c, Apaf-1 and BAD were increased in Cur-NPs-treated U2OS
cells (Fig. 8B). The results
suggest that Cur-NPs enhance apoptotic cell death of human
osteosarcoma U2OS cells through the Akt-Bad signaling pathway.
In conclusion, Cur-NPs induce cell apoptosis in
human osteosarcoma U2OS cells. The findings suggest that the major
pharmacologic action of Cur-NPs is to trigger apoptotic cell death
through activation of caspase-9 and caspase-3/-7 connected to
mitochondria-dependent and Akt-Bad signaling pathway in U2OS cells
(Fig. 9). Cur-NPs could be one of
the potential compounds to be developed as a novel medicine against
human osteosarcoma.
Acknowledgements
This study was supported by research
grant from the China Medical University (CMU101-N2-07) to Dr
Shu-Fen Peng.
References
|
1.
|
Ferrari S, Palmerini E, Fabbri N, et al:
Osteosarcoma of the pelvis: a monoinstitutional experience in
patients younger than 41 years. Tumori. 98:702–708. 2012.PubMed/NCBI
|
|
2.
|
Anninga JK, Picci P, Fiocco M, et al:
Osteosarcoma of the hands and feet: a distinct clinico-pathological
subgroup. Virchows Arch. 462:109–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Colomina J, Peiro A, Trullols L and Garcia
I: Telangiectatic osteosarcoma. J Orthop Surg. 21:96–99. 2013.
|
|
4.
|
Ebb D, Meyers P, Grier H, et al: Phase II
trial of trastuzumab in combination with cytotoxic chemotherapy for
treatment of metastatic osteosarcoma with human epidermal growth
factor receptor 2 overexpression: a report from the Children’s
Oncology Group. J Clin Oncol. 30:2545–2551. 2012.PubMed/NCBI
|
|
5.
|
Ritter J and Bielack SS: Osteosarcoma. Ann
Oncol. 21(Suppl 7): vii320–vii325. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Seki K, Yoshikawa H, Shiiki K, Hamada Y,
Akamatsu N and Tasaka K: Cisplatin (CDDP) specifically induces
apoptosis via sequential activation of caspase-8, -3 and -6 in
osteosarcoma. Cancer Chemother Pharmacol. 45:199–206. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Warzecha J, Gottig S, Chow KU, et al:
Inhibition of osteosarcoma cell proliferation by the
Hedgehog-inhibitor cyclopamine. J Chemother. 19:554–561. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Valteau-Couanet D and Minard V: Poor
prognosis childhood cancers. Rev Prat. 57:1087–1091. 2007.(In
French).
|
|
9.
|
Rouesse J and Le Chevalier J: Combination
of chemotherapy and surgery in pulmonary metastases of tumors
considered not very chemosensitive. Chirurgie. 111:538–541.
1985.(In French).
|
|
10.
|
Wang Y, Fu Q and Zhao W:
Tetramethylpyrazine inhibits osteosarcoma cell proliferation via
downregulation of NF-κB in vitro and in vivo. Mol Med
Rep. 8:984–988. 2013.PubMed/NCBI
|
|
11.
|
Shi QW, Li SG, Li J and Ling CQ:
Anti-tumor effects of triptolide on osteosarcoma cells in vitro and
in vivo: an experimental research. Zhongguo Zhong Xi Yi Jie He Za
Zhi. 33:659–663. 2013.(In Chinese).
|
|
12.
|
Yang JY, Cheng FW, Wong KC, et al: Initial
presentation and management of osteosarcoma, and its impact on
disease outcome. Hong Kong Med J. 15:434–439. 2009.PubMed/NCBI
|
|
13.
|
Zhang YK, Zhang XH, Li JM, Sun de S, Yang
Q and Diao DM: A proteomic study on a human osteosarcoma cell line
Saos-2 treated with diallyl trisulfide. Anticancer Drugs.
20:702–712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Yin H, Guo R, Xu Y, et al: Synergistic
antitumor efficiency of docetaxel and curcumin against lung cancer.
Acta Biochim Biophys Sin. 44:147–153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Tang N, Zhang J and Du Y: Curcumin
promoted the apoptosis of cisplain-resistant human lung carcinoma
cells A549/DDP through down-regulating miR-186*.
Zhongguo Fei Ai Za Zhi. 13:301–306. 2010.(In Chinese).
|
|
16.
|
Chen ZQ and Mo ZN: Curcumin in the
treatment of prostatic diseases. Zhonghua Nan Ke Xue. 14:67–70.
2008.(In Chinese).
|
|
17.
|
Sagar SM, Yance D and Wong RK: Natural
health products that inhibit angiogenesis: a potential source for
investigational new agents to treat cancer - Part 2. Curr Oncol.
13:99–107. 2006.
|
|
18.
|
Shishodia S, Sethi G and Aggarwal BB:
Curcumin: getting back to the roots. Ann NY Acad Sci. 1056:206–217.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Sareen R, Jain N and Pandit V: Curcumin: a
boon to colonic diseases. Curr Drug Targets. 14:1210–1218. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Patra D, Ahmadieh D and Aridi R: Study on
interaction of bile salts with curcumin and curcumin embedded in
dipalmitoylsn-glycero-3-phosphocholine liposome. Colloids Surf B
Biointerfaces. 110:296–304. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Mathew A, Fukuda T, Nagaoka Y, et al:
Curcumin loaded-PLGA nanoparticles conjugated with Tet-1 peptide
for potential use in Alzheimer’s disease. PLoS One.
7:e326162012.PubMed/NCBI
|
|
22.
|
Bolognesi ML, Bartolini M, Tarozzi A, et
al: Multitargeted drugs discovery: balancing anti-amyloid and
anticholinesterase capacity in a single chemical entity. Bioorg Med
Chem Lett. 21:2655–2658. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Khan S and Heikkila JJ: Curcumin-induced
inhibition of proteasomal activity, enhanced HSP accumulation and
the acquisition of thermotolerance in Xenopus laevis A6
cells. Comp Biochem Physiol A Mol Integr Physiol. 158:566–576.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Zhou H, Beevers CS and Huang S: The
targets of curcumin. Curr Drug Targets. 12:332–347. 2011.
View Article : Google Scholar
|
|
25.
|
Khan MA, Gahlot S and Majumdar S:
Oxidative stress induced by curcumin promotes the death of
cutaneous T-cell lymphoma (HuT-78) by disrupting the function of
several molecular targets. Mol Cancer Ther. 11:1873–1883. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Han J, Pan XY, Xu Y, et al: Curcumin
induces autophagy to protect vascular endothelial cell survival
from oxidative stress damage. Autophagy. 8:812–825. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
O’Sullivan-Coyne G, O’Sullivan GC,
O’Donovan TR, Piwocka K and McKenna SL: Curcumin induces
apoptosis-independent death in oesophageal cancer cells. Br J
Cancer. 101:1585–1595. 2009.
|
|
28.
|
Chen HW, Lee JY, Huang JY, et al: Curcumin
inhibits lung cancer cell invasion and metastasis through the tumor
suppressor HLJ1. Cancer Res. 68:7428–7438. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Wang WX, Sun SZ, Guo XL and Song Y: Effect
of curcumin on invasion and migration of tongue squamous cell
carcinoma cell line Tca8113. Zhonghua Kou Qiang Yi Xue Za Zhi.
43:101–104. 2008.PubMed/NCBI
|
|
30.
|
Lin CW, Hou WC, Shen SC, et al: Quercetin
inhibition of tumor invasion via suppressing PKC
delta/ERK/AP-1-dependent matrix metalloproteinase-9 activation in
breast carcinoma cells. Carcinogenesis. 29:1807–1815. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Philip S, Bulbule A and Kundu GC: Matrix
metalloproteinase-2: mechanism and regulation of NF-kappaB-mediated
activation and its role in cell motility and ECM-invasion.
Glycoconj J. 21:429–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Cho YJ, Yi CO, Jeon BT, et al: Curcumin
attenuates radiation-induced inflammation and fibrosis in rat
lungs. Korean J Physiol Pharmacol. 17:267–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Bowden RG, J JM, Deike E, et al: The use
of an anti-inflammatory supplement in patients with chronic kidney
disease. J Complement Integr Med. 10:1–10. 2013.PubMed/NCBI
|
|
34.
|
Asher GN and Spelman K: Clinical utility
of curcumin extract. Altern Ther Health Med. 19:20–22.
2013.PubMed/NCBI
|
|
35.
|
Shehzad A, Wahid F and Lee YS: Curcumin in
cancer chemo-prevention: molecular targets, pharmacokinetics,
bioavailability, and clinical trials. Arch Pharm. 343:489–499.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Kossler S, Nofziger C, Jakab M, Dossena S
and Paulmichl M: Curcumin affects cell survival and cell volume
regulation in human renal and intestinal cells. Toxicology.
292:123–135. 2012.PubMed/NCBI
|
|
37.
|
Ji JL, Huang XF and Zhu HL: Curcumin and
its formulations: potential anti-cancer agents. Anticancer Agents
Med Chem. 12:210–218. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Gulcubuk A, Altunatmaz K, Sonmez K, et al:
Effects of curcumin on tumour necrosis factor-alpha and
interleukin-6 in the late phase of experimental acute pancreatitis.
J Vet Med A Physiol Pathol Clin Med. 53:49–54. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Greenwald P, Milner JA, Anderson DE and
McDonald SS: Micronutrients in cancer chemoprevention. Cancer
Metastasis Rev. 21:217–230. 2002. View Article : Google Scholar
|
|
40.
|
Kelloff GJ, Boone CW, Crowell JA, et al:
New agents for cancer chemoprevention. J Cell Biochem (Suppl).
26:1–28. 1996. View Article : Google Scholar
|
|
41.
|
Kelloff GJ, Boone CW, Crowell JA, Steele
VE, Lubet R and Sigman CC: Chemopreventive drug development:
perspectives and progress. Cancer Epidemiol Biomarkers Prev.
3:85–98. 1994.PubMed/NCBI
|
|
42.
|
Chang PY, Peng SF, Lee CY, et al:
Curcumin-loaded nanoparticles induce apoptotic cell death through
regulation of the function of MDR1 and reactive oxygen species in
cisplatin-resistant CAR human oral cancer cells. Int J Oncol.
43:1141–1150. 2013.
|
|
43.
|
Manju S and Sreenivasan K: Gold
nanoparticles generated and stabilized by water soluble
curcumin-polymer conjugate: blood compatibility evaluation and
targeted drug delivery onto cancer cells. J Colloid Interface Sci.
368:144–151. 2012. View Article : Google Scholar
|
|
44.
|
Bhawana, Basniwal RK, Buttar HS, Jain VK
and Jain N: Curcumin nanoparticles: preparation, characterization,
and antimicrobial study. J Agric Food Chem. 59:2056–2061. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Bansal SS, Vadhanam MV and Gupta RC:
Development and in vitro-in vivo evaluation of polymeric implants
for continuous systemic delivery of curcumin. Pharm Res.
28:1121–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Mukerjee A and Vishwanatha JK:
Formulation, characterization and evaluation of curcumin-loaded
PLGA nanospheres for cancer therapy. Anticancer Res. 29:3867–3875.
2009.PubMed/NCBI
|
|
47.
|
Chen HJ, Lin CM, Lee CY, et al: Kaempferol
suppresses cell metastasis via inhibition of the ERK-p38-JNK and
AP-1 signaling pathways in U-2 OS human osteosarcoma cells. Oncol
Rep. 30:925–932. 2013.PubMed/NCBI
|
|
48.
|
Chen KT, Hour MJ, Tsai SC, et al: The
novel synthesized
6-fluoro-(3-fluorophenyl)-4-(3-methoxyanilino)quinazoline (LJJ-10)
compound exhibits anti-metastatic effects in human osteosarcoma U-2
OS cells through targeting insulin-like growth factor-I receptor.
Int J Oncol. 39:611–619. 2011.
|
|
49.
|
Hour MJ, Yang JS, Chen TL, et al: The
synthesized novel fluorinated compound (LJJ-10) induces death
receptor- and mitochondria-dependent apoptotic cell death in the
human osteogenic sarcoma U-2 OS cells. Eur J Med Chem.
46:2709–2721. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Chiu YJ, Hour MJ, Lu CC, et al: Novel
quinazoline HMJ-30 induces U-2 OS human osteogenic sarcoma cell
apoptosis through induction of oxidative stress and up-regulation
of ATM/p53 signaling pathway. J Orthop Res. 29:1448–1456. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Anton N, Gayet P, Benoit JP and Saulnier
P: Nano-emulsions and nanocapsules by the PIT method: an
investigation on the role of the temperature cycling on the
emulsion phase inversion. Int J Pharm. 344:44–52. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Witt E, Witt F, Trautwein N, et al:
Synthesis of lead chalcogenide nanocrystals and study of charge
transfer in blends of PbSe nano-crystals and
poly(3-hexylthiophene). Phys Chem Chem Phys. 14:11706–11714. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Dass A, Guo R, Tracy JB, Balasubramanian
R, Douglas AD and Murray RW: Gold nanoparticles with
perfluorothiolate ligands. Langmuir. 24:310–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Lin C, Tsai SC, Tseng MT, et al: AKT
serine/threonine protein kinase modulates baicalin-triggered
autophagy in human bladder cancer T24 cells. Int J Oncol.
42:993–1000. 2013.PubMed/NCBI
|
|
55.
|
Tsai SC, Yang JS, Peng SF, et al: Bufalin
increases sensitivity to AKT/mTOR-induced autophagic cell death in
SK-HEP-1 human hepatocellular carcinoma cells. Int J Oncol.
41:1431–1442. 2012.PubMed/NCBI
|
|
56.
|
Chen HJ, Lin CM, Lee CY, et al: Phenethyl
isothiocyanate suppresses EGF-stimulated SAS human oral squamous
carcinoma cell invasion by targeting EGF receptor signaling. Int J
Oncol. 43:629–637. 2013.PubMed/NCBI
|
|
57.
|
Tsai SC, Huang WW, Huang WC, et al:
ERK-modulated intrinsic signaling and G(2)/M phase arrest
contribute to the induction of apoptotic death by allyl
isothiocyanate in MDA-MB-468 human breast adenocarcinoma cells. Int
J Oncol. 41:2065–2072. 2012.
|
|
58.
|
Liao YR, Lu CC, Lai KC, et al: The novel
carboxamide analog ITR-284 induces caspase-dependent apoptotic cell
death in human hepatocellular and colorectal cancer cells. Mol Med
Rep. 7:1539–1544. 2013.PubMed/NCBI
|
|
59.
|
Liu CY, Yang JS, Huang SM, et al: Smh-3
induces G(2)/M arrest and apoptosis through calcium mediated
endoplasmic reticulum stress and mitochondrial signaling in human
hepatocellular carcinoma Hep3B cells. Oncol Rep. 29:751–762.
2013.
|
|
60.
|
Lee CY, Chien YS, Chiu TH, et al:
Apoptosis triggered by vitexin in U937 human leukemia cells via a
mitochondrial signaling pathway. Oncol Rep. 28:1883–1888.
2012.PubMed/NCBI
|
|
61.
|
Yang JS, Wu CC, Kuo CL, et al: Solanum
lyratum extracts induce extrinsic and intrinsic pathways of
apoptosis in WEHI-3 murine leukemia cells and inhibit allograft
tumor. Evid Based Complement Alternat Med. 2012:2549602012.
|
|
62.
|
Yin HT, Zhang DG, Wu XL, Huang XE and Chen
G: In vivo evaluation of curcumin-loaded nanoparticles in a A549
xenograft mice model. Asian Pac J Cancer Prev. 14:409–412. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Jiang L, Luo M, Liu D, et al: BAD
overexpression inhibits cell growth and induces apoptosis via
mitochondrial-dependent pathway in non-small cell lung cancer.
Cancer Cell Int. 13:532013. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Balogova L, Maslanakova M, Dzurova L,
Miskovsky P and Stroffekova K: Bcl-2 proapoptotic proteins
distribution in U-87 MG glioma cells before and after hypericin
photodynamic action. Gen Physiol Biophys. 32:179–187. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Plourde MB, Morchid A, Iranezereza L and
Berthoux L: The Bcl-2/Bcl-xL inhibitor BH3I-2′ affects the dynamics
and subcellular localization of sumoylated proteins. Int J Biochem
Cell Biol. 45:826–835. 2013.
|
|
66.
|
Kastelan M, Massari LP and Brajac I: The
role of bcl-2 family proteins in psoriasis. Lijec Vjesn. 132:31–33.
2010.(In Croatian).
|
|
67.
|
Danial NN: BAD: undertaker by night,
candyman by day. Oncogene. 27 (Suppl 1): S53–S70. 2008.PubMed/NCBI
|
|
68.
|
Levine B, Sinha S and Kroemer G: Bcl-2
family members: dual regulators of apoptosis and autophagy.
Autophagy. 4:600–606. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Stauffer SR: Small molecule inhibition of
the Bcl-X(L)-BH3 protein-protein interaction: proof-of-concept of
an in vivo chemopotentiator ABT-737. Curr Top Med Chem. 7:961–965.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Kuroda J and Taniwaki M: Involvement of
BH3-only proteins in hematologic malignancies. Crit Rev Oncol
Hematol. 71:89–101. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Li Y, Gu J, Liu Y, et al: iNOS
participates in apoptosis of spinal cord neurons via p-BAD
dephosphorylation following ischemia/reperfusion (I/R) injury in
rat spinal cord. Neurosci Lett. 545:117–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Hojabrpour P, Waissbluth I, Ghaffari M,
Cox ME and Duronio V: CaMKII-gamma mediates phosphorylation of BAD
at Ser170 to regulate cytokine-dependent survival and
proliferation. Biochem J. 442:139–149. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Kim W, Yang HJ, Youn H, Yun YJ, Seong KM
and Youn B: Myricetin inhibits Akt survival signaling and induces
Bad-mediated apoptosis in a low dose ultraviolet (UV)-B-irradiated
HaCaT human immortalized keratinocytes. J Radiat Res. 51:285–296.
2010. View Article : Google Scholar
|
|
75.
|
Yang J: Molecular modeling of human BAD, a
pro-apoptotic Bcl-2 family member, integrating glycolysis and
apoptosis. Protein Pept Lett. 17:206–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Bhat V, Olenick MB, Schuchardt BJ, et al:
Heat-induced fibrillation of BclXL apoptotic repressor. Biophys
Chem. 179:12–25. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Dorjgochoo T, Xiang YB, Long J, et al:
Association of genetic markers in the BCL-2 family of
apoptosis-related genes with endometrial cancer risk in a Chinese
population. PLoS One. 8:e609152013. View Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Zhang XH, Chen SY, Tang L, et al:
Myricetin induces apoptosis in Hepg2 cells through Akt/P70s6k/Bad
signaling and mitochondrial apoptotic pathway. Anticancer Agents
Med Chem. Feb 7–2013.(Epub ahead of print).
|
|
79.
|
Lim GE, Piske M and Johnson JD: 14-3-3
proteins are essential signalling hubs for beta cell survival.
Diabetologia. 56:825–837. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Riaz A, Zeller KS and Johansson S:
Receptor-specific mechanisms regulate phosphorylation of AKT at
Ser473: role of RICTOR in beta1 integrin-mediated cell survival.
PLoS One. 7:e320812012. View Article : Google Scholar : PubMed/NCBI
|