Introduction
In the last few decades, there has been an
exponential rise of the available biological data including human
genomic sequences, gene expression profiles, protein-protein
interaction networks, and metabolomic data of physiologically
active compounds. This vast amount of information provides us with
a challenging opportunity to develop computational approaches for
systematic analysis of various disorders, including cancers, in
order to utilize the biomedical data for prediction of patient
prognosis, disease modeling and biological systems analysis
(1). The genetic diversity in
cancer is vast, given cancers can arise from most of the cells in
the body (~6×1013 cells), and that tumors themselves
also harbor heterogeneous cellular components (~1010
cells), including a subpopulation of cancer initiating cells
(~108 cells) (2,3). The
gene expression between cancer cells is also highly variable, this
variability being generated through numerous mechanisms, including
chromosomal translocations, genetic mutations, epigenetic chromatin
remodeling of the 3×109 base paired DNA and alteration
of histone structures (4–6). For example, sequencing analysis of
liver cancer revealed that there are 11×103 genetic
mutations, of which 10–100 are driver mutations, as well as 21
chromosomal structural abnormalities (7). Genetic heterogeneity occurs in many
tumors (http://cancergenome.nih.gov),
indicating that mathematical and statistical analysis is
indispensable to distinguish between driver and passenger mutations
in order to identify druggable targets.
In 2008 and 2009, two independent cancer sequencing
projects reported high frequencies of mutations of the gene
encoding the cytosolic enzyme IDH1, a critical component of the
tricarboxylic acid (TCA) cycle, in glioblastoma multiforme (GBM)
(8) and acute myeloid leukemia
(AML) (9). Further study reported
that IDH2, the gene encoding the homologous enzyme in the
mitochondria was also frequently mutated in GBM patients (10). Studies indicated that >75% of
grade 2 and 3 GBM and 20% of AML harbor mutations of IDH1 at R132,
or IDH2 at the homologous R172 residue (11). These oncogenic mutations in IDH1
and IDH2 enzymes reduce their native activity, and generate
neomorphic activity that converts α-ketoglutarate (α-KG; also known
as 2-oxyoglutarate) to D-2-hydroxygutarate (D2HG) (12,13).
D2HG is an oncome-tabolite that can cause changes to the epigenetic
landscape by inhibiting the activities of iron (II), α-KG-dependent
dioxygenases, including: prolyl-hydroxylase-domains (PHDs)
resulting in pseudo-hypoxic environment and activation of
hypoxia-inducible-factor (HIF) pathway leading to aberrant cellular
proliferation; ten-eleven translocation (TET)-family demethylases,
AlkB-family dioxygenases, which protect nucleotides against
methylating reactions by directly dealkylating bases (1mA, 3mC, 1mG
and 3mT); and histone lysine demethylases (KDMs) (14).
Other oncometabolites that have been identified are
succinate and fumarate, which are generated by mutations of
succinate dehydrogenase (SDH) and fumarate hydratase (FH),
respectively. These also competitively inhibit α-KG-dependent
dioxygenases (14), and moreover
provide a strong rationale that TCA enzymes as a family may be
important in other malignancies.
Here we applied computational analysis to study the
association between gene expression profiles of colorectal cancer
(CRC) patients and their prognosis. Using this approach, we
identified that the imbalances between the expressions of IDH1 and
IHD2 was associated with poorer prognosis of patients with CRC.
Subsequent unsupervised analysis of targeted genes identified low
expression of hydroxyl-CoA dehydratase (HCDH) 4 to amplify the
effect of imbalanced IDH1 and IDH2 expressions on the survival of
CRC patients. HCDH4 is an enzyme that metabolizes D2HG in the
β-oxygenation pathway, and is encoded by a gene that is located at
chromosome 9p21.3, which is frequently deleted in cancers alongside
tumor suppressor INK4A and microRNA-31 (miR-31). The present
computational analysis linked the association combining expression
of IDH1 and IDH2, rather than mutations, with β-oxidization pathway
in the prognosis of CRC patients.
Materials and methods
Gene Expression database of colorectal
cancer patients
We used the published GSE17536 database (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE17536)
(15) from the Gene Expression
Omnibus in NCBI to analyze the effects of gene expression on
disease-free survival (DFS) and overall survival (OS) of CRC
patients. This database contains the microarray data of 177
patients, generated using the Affymetrix Human Genome U133 Plus 2.0
Array. The microarray data were generated with multiple probes for
one gene in some cases; if this was the case, the probe that
demonstrated the widest variance of genetic expression amongst the
cohort was selected for statistical analysis. Each selected gene
was divided into low and high expression groups at the median point
of expression. Additionally, to verify the relevance of our results
with GSE17536 database, we used another reference dataset for CRC
patients (16). The 91 patients
with both CRC stage and DFS time information were selected from
this reference dataset and used in validation analysis.
Analysis of the expression of genes of
the oxidative phosphorylation pathway and their effect on the
survival of colorectal cancer patients
Genes of the oxidative phosphorylation (OxPhos)
pathway that are already known to be involved in neoplastic
diseases were selected for initial analysis: TET 1–3 proteins,
IDH1, IDH2, SDH, FH, MYC, glutaminase (GLS), and the mitochondrial
alcohol dehydrogenase iron-containing 1 (ADHFE) (14). Genes in the above group were
analyzed in all possible combinations as pairs in order to
ascertain whether there were any associations between combined gene
expressions and DFS or OS (Fig.
1). These analyses were conducted through the generation of
Kaplan-Meier curves. Thirty-two patients were excluded from the
analysis concerning the DFS given these were stated as being
zero.
Screening for genes affecting the
survival of colorectal cancer patients in a manner dependent on the
expression of oxidative phosphorylation genes
Other genes that are associated with DFS and OS of
CRC patients in a manner that is dependent on the selected genes of
the OxPhos pathway were screened. The rationale for adopting this
approach was to identify a biological pathway important in CRC,
instead of emphasizing isolated gene expression. Only CRC patients
with stages II or III were selected for this stage of analysis.
Unbiased screening was performed for ‘the third genes’, searching
for genes that do not have independent association with DFS or OS,
but were nevertheless able to accentuate the effect of the selected
OxPhos pathway genes on the DFS and OS (Fig. 1). Statistical analysis was
performed; with statistical significance being defined when the
p-value was <0.05. Kaplan-Meier curves were generated using the
survival package (16) on R
version 3.0.2 (17).
Results
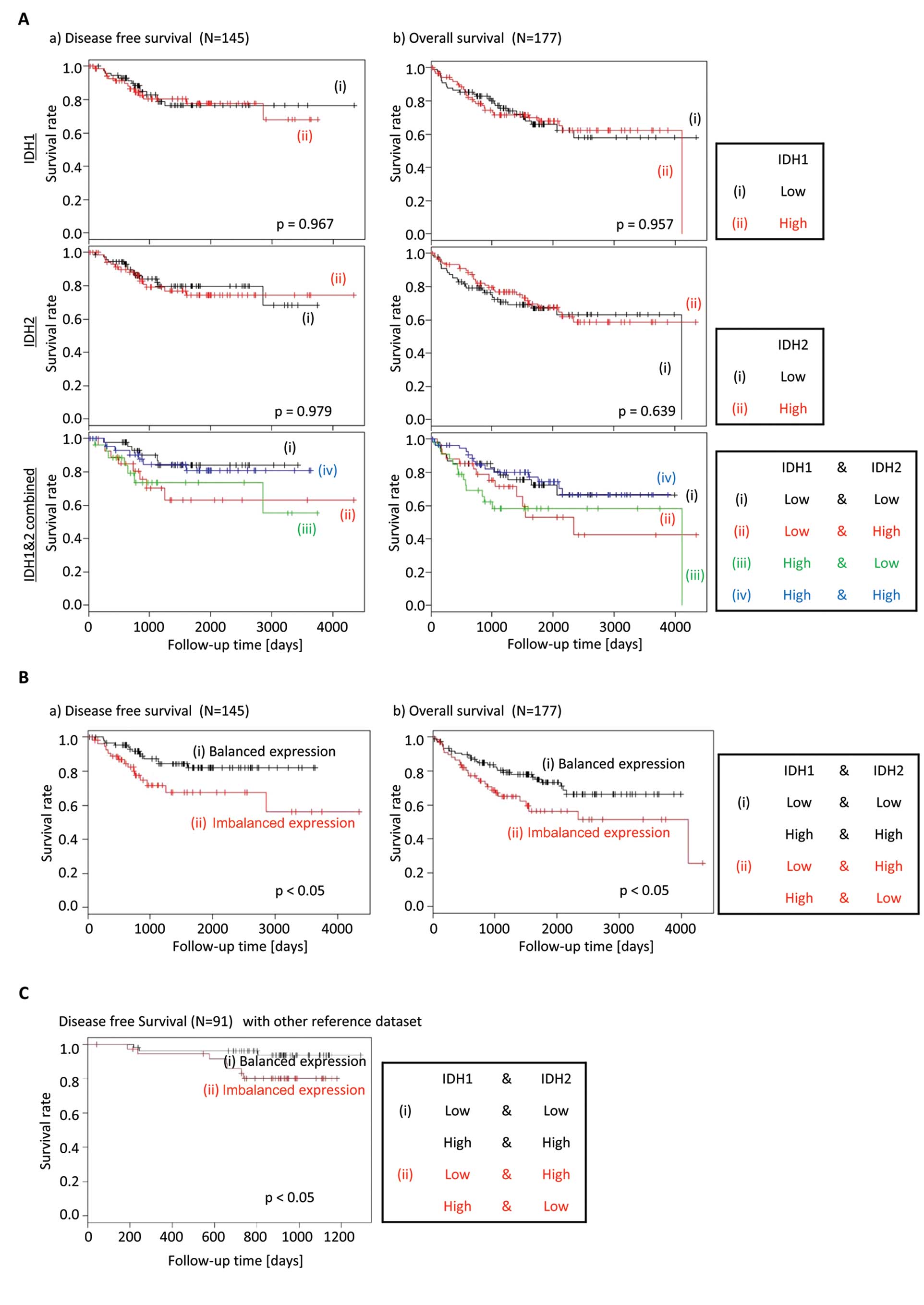
Imbalance between the IDH1 and IDH2
expression is associated with decreased survival of colorectal
cancer patients
The TCA cycle in mitochondria plays a critical role
in OxPhos, which produces ATP via the electron transport chain
(14). Given mutations of several
of the OxPhos-related genes, such as families of TET proteins, IDH,
SDH, and FH, are observed in human malignancies, we investigated
whether the expression of these genes in combination as pairs would
be associated with patient survival in CRC. Alterations of families
of TETs, IDHs, SDHs, and FHs are expected to result in attenuation
of α-KG-dependent dioxygenases, such as demethylases, which
characterize cancers (14). There
were no differences in the Kaplan-Meier curves for DFS or OS, when
IDH1 or IDH2 expressions were analyzed in isolation. However, when
the gene expression of IDH1 and IDH2 was analyzed together, the
patients with an imbalance of IDH1 and IDH2 gene expression (i.e.,
IDH1high;IDH2low, or
IDH1low;IDH2high), had a shorter DFS and OS
compared to patients with a ‘balanced’ IDH gene expression (i.e.,
IDH1high;IDH2high, or
IDH1low;IDH2low) (Fig. 2A and B).
We performed the same prognosis analysis for cancer
recurrence with another reference dataset (16) to confirm that this biological
behavior is not unique to the GSE17536 database. Also the
validation results showed the same tendency that the patients with
imbalanced expression of IDH1/2 had a shorter DFS compared to those
with balanced expression (Fig.
2C), although this reference dataset is a small one, which has
only 10 patients with relapsed cancer.
Reduced HCDH4 expression exacerbates the
poor prognosis in colorectal cancer patients with imbalanced IDH
expression
In order to identify novel partners of imbalanced
IDH1 and IDH2 expression, we performed non-biased comparison
against the total transcriptome data. Analysis was conducted in two
steps, the first being to identify candidate genes that could
separate the IDH1high:IDH2low and
IDH1low:IDH2high patients from
IDH1high:IDH2high and
IDH1low:IDH2low patients in terms of the DFS
and OS. Genes identified using this analysis were also checked for
their ability to independently predict DFS or OS depending on their
expression, and was excluded if this was the case. By using this
approach, we identified 43 and 44 genes that had a significant
effect on the DFS and OS, respectively (Table I). Of these ZNF91 (18,19)
and HCDH4 [protein tyrosine phosphatase-like a domain containing 2
(PTPLAD2)] (20), were found to
accentuate the reduction of both DFS and OS of CRC patients when
IDH1 and IDH2 expression was imbalanced.
 | Table IGenes that have a significant effect
on the DFS and OS, in a manner dependent on the imbalance between
the expressions of IDH1 and IDH2.a |
Table I
Genes that have a significant effect
on the DFS and OS, in a manner dependent on the imbalance between
the expressions of IDH1 and IDH2.a
| DFS time | OS time |
|---|
|
|
|---|
| Gene name | Stage 2 | Stage 3 | Gene name | Stage 2 | Stage 3 |
|---|
| AIM2 | 0.049 | 0.032 | ADH5 | 0.029 | 0.000 |
| ANKRD49 | 0.042 | 0.011 | AK7 | 0.023 | 0.002 |
| ARHGEF19 | 0.006 | 0.044 | ANKRD49 | 0.029 | 0.007 |
| C16orf73 | 0.042 | 0.000 | ANKS1B | 0.042 | 0.008 |
| C20orf72 | 0.021 | 0.028 | ANXA9 | 0.041 | 0.005 |
| C2orf73 | 0.021 | 0.012 | ATP5F1 | 0.029 | 0.014 |
| C7 | 0.006 | 0.024 | BMP2K | 0.028 | 0.008 |
| CA11 | 0.031 | 0.018 | CHCHD2 | 0.034 | 0.010 |
| CDH8 | 0.031 | 0.006 | EMX2 | 0.023 | 0.023 |
| CHEK2 | 0.021 | 0.039 | FAM118B | 0.016 | 0.011 |
| DIO2 | 0.042 | 0.049 | FAM124B | 0.008 | 0.033 |
| FAM54B | 0.012 | 0.025 | FAM54B | 0.008 | 0.007 |
| KIAA0355 | 0.042 | 0.028 | FDX1 | 0.010 | 0.003 |
| LELP1 | 0.031 | 0.049 | GNN | 0.023 | 0.037 |
| LOC100129335 | 0.042 | 0.045 | GOLT1A | 0.029 | 0.042 |
| LOC284379 | 0.031 | 0.005 | GPR87 | 0.013 | 0.004 |
| LOC388789 | 0.031 | 0.011 | LOC100132354 | 0.042 | 0.023 |
| LOC728804 | 0.014 | 0.009 | LOC100132356 | 0.023 | 0.000 |
| NCKAP5L | 0.021 | 0.048 | LOC100289086 | 0.034 | 0.046 |
| OSBPL9 | 0.031 | 0.012 | LOC389043 | 0.008 | 0.014 |
| PHLDB3 | 0.014 | 0.039 | LOC729420 | 0.034 | 0.037 |
| POP4 | 0.042 | 0.002 | LRRK1 | 0.029 | 0.025 |
| PTP4A3 | 0.012 | 0.007 | NINJ1 | 0.029 | 0.032 |
| HCDH4 | 0.031 | 0.000 | NR1H4 | 0.013 | 0.043 |
| RPF2 | 0.021 | 0.035 | OXA1L | 0.023 | 0.036 |
| SCN4B | 0.021 | 0.045 | PCMT1 | 0.029 | 0.015 |
| SEC13 | 0.031 | 0.046 | PEX16 | 0.029 | 0.033 |
| SLC16A2 | 0.049 | 0.025 | PRR13 | 0.029 | 0.003 |
| SLC9A1 | 0.031 | 0.041 | PTP4A3 | 0.023 | 0.041 |
| SNTA1 | 0.021 | 0.025 | HCDH4 | 0.029 | 0.002 |
| STK25 | 0.012 | 0.022 | RPS3 | 0.023 | 0.044 |
| TBC1D19 | 0.014 | 0.037 | SIX5 | 0.010 | 0.016 |
| TGFBRAP1 | 0.031 | 0.004 | SLC24A2 | 0.029 | 0.041 |
| TGM6 | 0.042 | 0.029 | SLITRK4 | 0.028 | 0.003 |
| TLR1 | 0.042 | 0.035 | SMARCAL1 | 0.034 | 0.029 |
| UCHL3 | 0.042 | 0.044 | TGFBRAP1 | 0.034 | 0.001 |
| ULBP2 | 0.021 | 0.044 | TMSB15B | 0.029 | 0.020 |
| WWP1 | 0.021 | 0.032 | TRIM48 | 0.029 | 0.022 |
| ZDHHC16 | 0.049 | 0.030 | TUFM | 0.029 | 0.029 |
| ZNF142 | 0.042 | 0.044 | UCHL3 | 0.034 | 0.016 |
| ZNF573 | 0.021 | 0.004 | WIPI1 | 0.029 | 0.033 |
| ZNF814 | 0.042 | 0.015 | YLPM1 | 0.018 | 0.004 |
| ZNF91 | 0.042 | 0.015 | ZNF362 | 0.010 | 0.040 |
| - | - | - | ZNF91 | 0.029 | 0.011 |
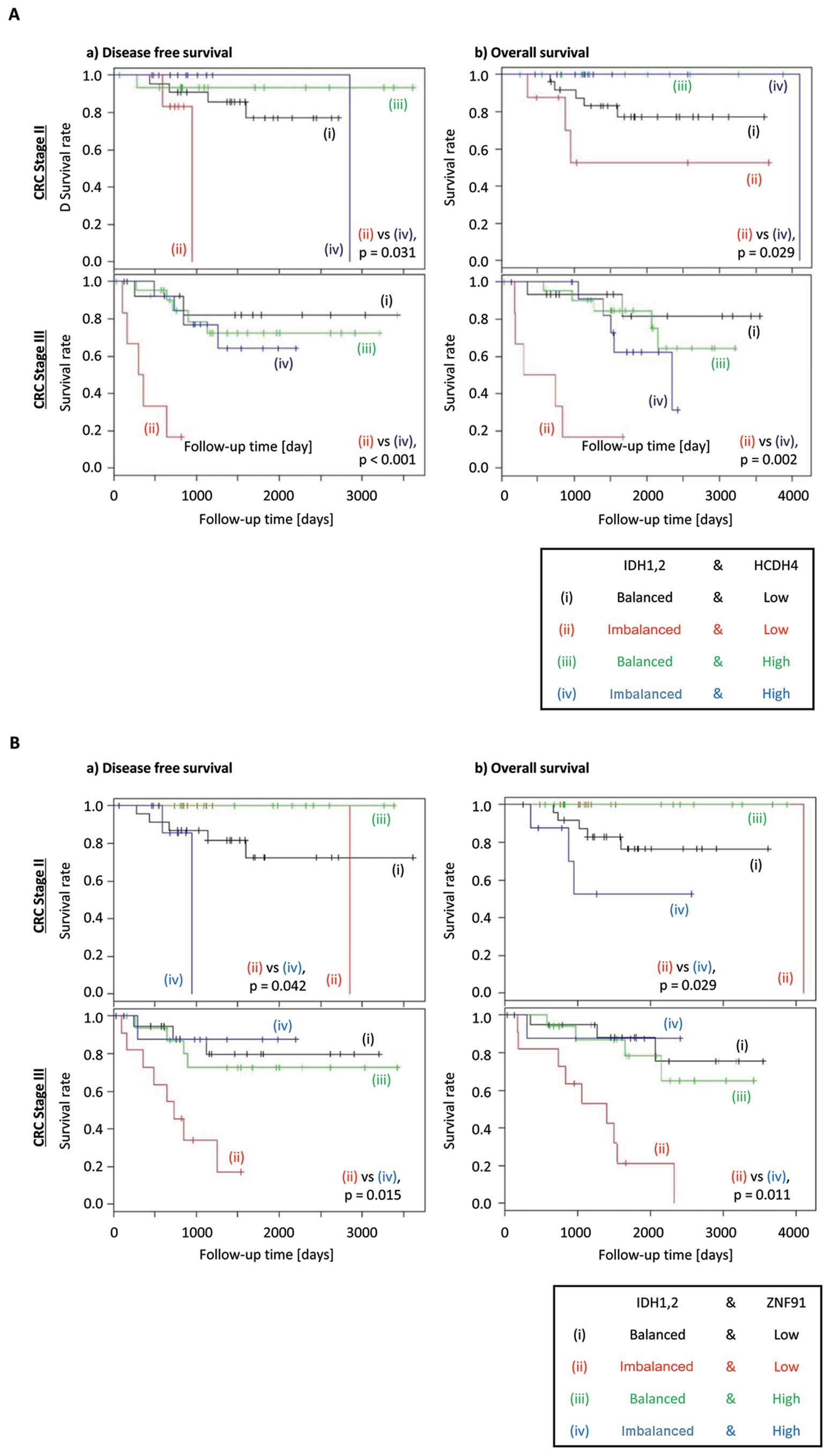
When the above analysis was conducted further for
individual stages of CRC, reduced expression of HCDH4 was shown to
exacerbate the poorer DFS and OS in CRC patients with stages II or
III disease (Fig. 3A). However,
ZNF91 expression showed discordant effect on DFS and OS between
different stages of CRC, with high expression associated with worse
prognosis in stage II disease, but low expression is associated
with worse prognosis in stage III disease (Fig. 3B). This result indicates that HCDH4
was more likely to be a suitable candidate for further analysis.
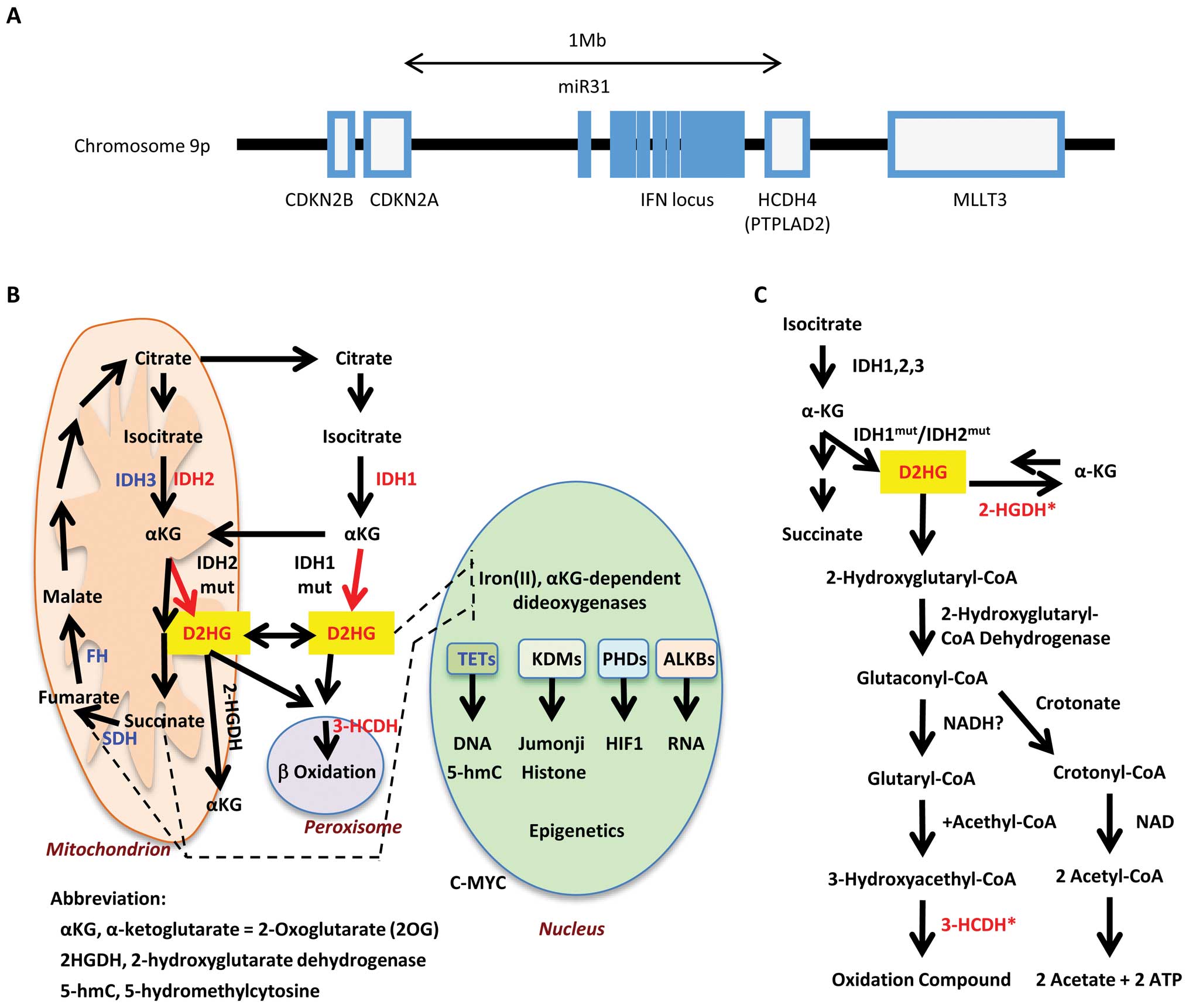
HCDH4 is an enzyme that metabolizes D2HG in the β-oxygenation
pathway. The genome data-base (http://www.ncbi.nlm.nih.gov/guide/genomes-maps/) shows
that the HCDH4 gene is located at chromosome 9p21.3, which is a
genomic region frequently deleted in cancer, also containing the
tumor suppressor gene p16/CDKN2A/ INK4A. p16/CDKN2A/INK4A is
inactivated by methylation of the gene promoter in early stages of
gastrointestinal cancer (21). The
gene locus containing p16/CDKN2A/INK4A and HCDH4 also harbors
interferon genes and cancer-associated miR-31 (22), suggesting the alterations of this
locus increases the susceptibility to cancer (Fig. 4A).
Prediction of a common pathway of IDH1,
IDH2 and HCDH4 genes
Although the IDH expression changes analyzed in this
study are likely to be associated with wild-type genes, we propose
that an imbalance between the expressions of IDH1 and IDH2 could
lead to the overproduction of D2HG, of which more will be explained
in Discussion. Increasing the levels of the oncometabolite D2HG is
potentially a common denominator of IDH1, IDH2 and HCDH4, with the
decreased expression of the latter enzyme leading to reduced
metabolism of D2HG. Therefore, D2HG provides an explanation for
poorer survival of patients with imbalanced IDH1 and IDH2 as well
as low HCDH4 expression. Previous studies have shown that HCDH is
involved with the metabolization of D2HG in the glutaconyl pathway
(http://www.genome.jp/kegg/) (Fig. 4). The direction of chemical
reaction generally depends on the ΔG value instead of the
activation energy. Given that the conversion from D2HG to α-KG is a
relatively small exothermic reaction of 0.791 (kcal/mol)
(calculated with MP2/6-31G level in Gaussian 09 package) (23), this suggests that D2HG is produced
under physiological conditions, with IDH1 and IDH2 expression
determining rate of production and D2HGDH and HCDH governing
metabolization (Figs. 4C and
6A).
 | Figure 6(A) At the state of equilibrium, the
abundance of substance A and B depends not on the activation energy
but the ΔG. (B) Schematic diagram of the relationship of IDH1, IDH2
and the other genes involved in the TCA cycle. Although, on a
routine basis, α-KG and D2HG transfer to and from between
mitochondria and cell cytoplasm keeping the same conditions, their
conditions are changed by imbalanced IDH1/2 expression. High MYC
expression makes improvements because of acceleration of TCA cycle.
(C) Schematic diagram explaining how an imbalance between the
expressions of IDH1 and IDH2 could lead to increased production of
D2HG. (a) IDH1low;IDH2low: (1) transfer to
and from caused by the same condition, (2) insensitive to transfer
to and from caused by each low concentration, (3) insensitive to
D2HG in low concentration of α-KG, (4) transfer to and from caused
by the same condition; (b) IDH1high;IDH2low:
(1) move to cell cytoplasm caused by high concentration in
mitochondria, (2 and 3) transfer from α-KG to D2HG caused by high
concentration of α-KG, (4) go and come keeping the same
concentration; (c) IDH1low;IDH2high: (1) move
to mitochondria caused by high concentration in the cell cytoplasm,
(2) a disproportionate emphasis on D2HG caused by high
concentration of α-KG and deacceleration of TCA cycle, (3) transfer
to D2HG caused by high concentration of α-KG, (4) go and come
transfer keeping the same concentration; (d)
IDH1high;IDH2high: (1) transfer to and from
caused by the same condition, (2) a disproportionate emphasis on
α-KG caused by the consume α-KG of along with acceleration of TCA
cycle, (3) insensitive to D2HG in low concentration of α-KG, (4)
transfer to and from caused by the same condition. |
MYC overexpression is not associated with
decreased survival of colorectal cancer patients
Recently, overexpression of the MYC oncogene
occurring in a sub-group of breast cancer patients was shown to be
associated with higher levels of D2HG and decreased survival. D2HG
production was demonstrated to be driven by MYC overexpression
in vitro, demonstrating a novel mechanism of D2HG production
given IDH1 or IDH2 mutations in breast cancers are rare (24). In this cohort of CRC patients, MYC
expression was not associated with decreased DFS or OS.
Interestingly, the Kaplan-Meier analysis showed that patients with
high MYC expression have better DFS or OS, regardless of the
expression status of FH, IDH2 and SDHB (Fig. 5), suggesting that the role of MYC
in CRC is distinct from that in breast cancers.
Discussion
In this study, an imbalance between the expression
of IDH1 and IDH2 was associated with decreased DFS and OS in CRC
patients; these reductions in DFS and OS were further accentuated
by reduced expression of HCDH4. While previous investigations
support the role of mutated forms of IDH and its importance in the
pathophysiology of cancer, the link with the relative levels of
expression between wild-type IDH1 and IDH2 in cancer survival is
unprecedented. Mutated IDH1 and IDH2 have been implicated in
causing cancer through the production of the oncometabolite D2HG
(11). A recent study of
hematopoietic malignancies show that the IDH2R140Q
mutation was necessary alongside overexpression of Hox9A and
Meis1a, or mutation of FMS-like tyrosine kinase 3 (FLT3), to
initiate and maintain acute leukemia, most likely through increased
levels of D2HG (25).
We postulate a role for D2HG in promoting CRC
progression based on our current results, along with other recent
published evidence. First, D2HG levels are raised in the tumors and
plasma of mice with azoxymethane-induced intestinal cancer
(26). Second,
5-hydroxymethylcytosine levels are decreased in human CRC,
indicating decreased TET2 activity, which could reflect inhibition
through high levels of D2HG (27).
We propose a model that explains how imbalanced IDH1
and IDH2 expression could lead to increased production of D2HG
(Fig. 6B and C). In this scheme,
we make several assumptions, including that both α-KG and D2HG are
able to permeate between mitochondria and cell cytoplasm and
equilibrate by osmosis. We also assume that the direction of
chemical reaction depends on the difference of free energy between
substances rather than the activation energy. When both IDH1 and
IDH2 expressions are low, the production of α-KG is also low,
leading to low levels of D2HG. When both IDH1 and IDH2 expressions
are high, α-KG levels will rise due to increased production, but
will either be metabolized effectively by the TCA cycle or be
converted back to isocitric acid by the IDH enzymes, and the D2HG
levels will remain low. When IDH1 expression is high and IDH2
expression is low, there will be increased production of α-KG in
the cytoplasm, which will diffuse into the mitochondria by osmosis.
In the absence of a high TCA cycle activity, the low level of IDH2
is not able to convert all of the α-KG into isocitric acid,
resulting in the production of D2HG through a relatively small
endothermic reaction. Similarly, when IDH2 expression is high and
IDH1 expression is low, mitochondrial α-KG will diffuse into the
cytoplasm, where the low levels of IDH1 is unable to convert all
the α-KG into isocitric acid, instead leading to the production of
D2HG.
We next focused on the relationship between MYC and
components of the TCA cycle, given MYC has previously been reported
to accelerate the TCA cycle. We proposed that the TCA cycle in an
accelerated state is able to more efficiently metabolize α-KG,
thereby preventing the production of D2HG and increasing the
survival of CRC patients. As expected, high expression of MYC
alongside high expression of the TCA cycle enzymes IDH2, FH and
SDHB were associated with increased DFS and OS. However, the
decreased DFS and OS when MYC and SDHA expression is high were
inconsistent with the above notion. We are therefore preparing
additional analysis concerning SDHA. The increased MYC expression
associated with improved survival in this study is consistent with
previous studies of CRC (28). In
breast cancer however, increased MYC signaling was responsible for
D2HG production and was associated with decreased survival
(24), suggesting that MYC plays a
different role to that in CRC.
HCDH4 is an enzyme that metabolizes D2HG in the
β-oxygenation pathway and its mutation has been reported to cause
hereditary peroxisomal disorders (20). Therefore the IDH enzymes together
with HCDH4 affecting the prognosis of patients with CRC adds
further support that D2HG is likely to play a role. Given that
HCDH4 gene is located in the frequently deleted genomic region with
tumor suppressor INK4A and miR-31 at chromosome 9p21.3, the
inactivation of HCDH4 may contribute to CRC progression together
with other oncogenic mutations.
The importance of D2HG in pathology is further
demonstrated in a rare autosomal recessive neurometabolic disorder
called D2HG aciduria (30). This
condition occurs when D-2-hydroxyglutarate dehydrogenase (D2-HGHD),
a mitochon-drial enzyme belonging to the FAD-binding
oxidoreductase/transferase type 4 family, is mutated. This enzyme,
which is most active in liver and kidney but also active in heart
and brain, converts D2HG to α-KG. The condition is characterized by
developmental delay, epilepsy, hypotonia, and dysmorphic features
(29). Given multiple mechanisms
are involved in metabolizing D2HG, this suggests the importance of
maintaining D2HG levels low. Biochemical studies indicated that
HCDH functions to inactivate D2HG via the glutaconyl pathway,
whereas D-2-hydroxyglutarate dehydrogenase (D2-HGHD) is involved in
the inactivation of physiological level of D2HG (http://www.genome.jp/kegg/) (Fig. 4B and C).
In order to be certain of the significance of IDH1
and IDH2 expression on the prognosis of colorectal cancer patients,
the data requires validation using other cohorts of CRC patients,
ideally with matched levels of D2HG. We are presently planning to
evaluate D2HG levels in CRC patients.
This study provides direction for future studies and
has demonstrated the following: i) the expressions of both IDH1 and
IDH2, critical enzymes involved in oxidative phosphorylation in
mitochondria, are associated with patient survival in CRC; ii) the
usefulness of computational analysis of high volume data to suggest
novel biological mechanisms and predict patient survival; iii) the
benefit of the gene expression microarray for identifying potential
novel therapeutic targets; iv) a link between IDH1, IDH2 and
D2HG-inactivating β-oxidization pathway in CRC.
Acknowledgements
We thank Dr R. Daniel Beauchamp and Dr Pengcheng Lu,
Vanderbilt Medical Center, TN, USA, for the critical review of our
manuscript. This study was supported in part by a Grant-in-Aid for
Scientific Research from the Ministry of Education, Culture,
Sports, Science, and Technology; a Grant-in-Aid from the Third
Comprehensive 10-year Strategy for Cancer Control, Ministry of
Health, Labor, and Welfare; a grant from the Kobayashi Cancer
Research Foundation; a grant from the Princess Takamatsu Cancer
Research Fund, Japan; a grant from the National Institute of
Biomedical Innovation; and a grant from the Osaka University Drug
Discovery Funds. Partial support was received from Taiho
Pharmaceutical Co., Ltd., EBMRCE, Chugai Co., Ltd., Yakult Honsha
Co., Ltd. (J.K., N.N., M.K., K.K. and H.I.), Merck Co., Ltd.,
Takeda Science Foundation and Takeda Medical Research Foundation
through institutional endowments.
References
|
1
|
Chena M and Hofestädtb R: A medical
bioinformatics approach for metabolic disorders: Biomedical data
prediction, modeling, and systematic analysis. J Biomed Inform.
39:147–159. 2006. View Article : Google Scholar
|
|
2
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer, and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Meacham CE and Morrison SJ: Tumour
heterogeneity and cancer cell plasticity. Nature. 501:328–337.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nowell PC and Croce CM: Chromosomes,
genes, and cancer. Am J Pathol. 125:8–15. 1986.
|
|
5
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yaniv M: Chromatin remodeling: from
transcription to cancer. Cancer Genet. Mar 21–2014.(Epub ahead of
print). View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fujimoto A, Totoki Y, Abe T, et al:
Whole-genome sequencing of liver cancers identifies etiological
influences on mutation patterns and recurrent mutations in
chromatin regulators. Nat Genet. 44:760–764. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parsons DW, Jones S, Zhang X, et al: An
integrated genomic analysis of human glioblastoma multiforme.
Science. 321:1807–1812. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mardis ER, Ding L, Dooling DJ, et al:
Recurring mutations found by sequencing an acute myeloid leukemia
genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yan H, Parsons DW, Jin G, et al: IDH1 and
IDH2 mutations in gliomas. N Engl J Med. 360:765–773. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cairns RA and Mak TW: Oncogenic isocitrate
dehydrogenase mutations: mechanisms, models, and clinical
opportunities. Cancer Discov. 3:730–741. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dang L, White DW, Gross S, et al:
Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ward PS, Patel J, Wise DR, et al: The
common feature of leukemia-associated IDH1 and IDH2 mutations is a
neomorphic enzyme activity converting alpha-ketoglutarate to
2-hydroxyglutarate. Cancer Cell. 17:225–234. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ponnaluri VK, Maciejewski JP and Mukherji
M: A mechanistic overview of TET-mediated 5-methylcytosine
oxidation. Biochem Biophys Res Commun. 436:115–120. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Smith JJ, Deane NG, Wu F, Merchant NB, et
al: Experimentally derived metastasis gene expression profile
predicts recurrence and death in patients with colon cancer.
Gastroenterology. 138:958–968. 2010. View Article : Google Scholar
|
|
16
|
Takatsuno Y, Mimori K, Yamamoto K, et al:
The rs6983267 SNP is associated with MYC transcription efficiency,
which promotes progression and worsens prognosis of colorectal
cancer. Ann Surg Oncol. 20:1395–1402. 2013. View Article : Google Scholar
|
|
17
|
Therneau TM and Grambsch PM: Modeling
Survival Data: Extending the Cox Model. Springer; NY: 2000
|
|
18
|
Team RC: A language and environment for
statistical computing. R Foundation for Statistical Computing;
Vienna: 2013, http://www.R-project.org/.
|
|
19
|
Unoki M, Okutsu J and Nakamura Y:
Identification of a novel human gene, ZFP91, involved in acute
myelogenous leukemia. Int J Oncol. 22:1217–1223. 2003.PubMed/NCBI
|
|
20
|
Micci F, Skotheim RI, Haugom L, et al:
Array-CGH analysis of microdissected chromosome 19 markers in
ovarian carcinoma identifies candidate target genes. Genes
Chromosomes Cancer. 49:1046–1053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Suzuki Y, Jiang LL, Souri M, et al:
D-3-hydroxyacyl-CoA dehydratase/D-3-hydroxyacyl-CoA dehydrogenase
bifunctional protein deficiency: a newly identified peroxisomal
disorder. Am J Hum Genet. 61:1153–1162. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bian YS, Osterheld MC, Fontolliet C,
Bosman FT and Benhattar J: p16 inactivation by methylation of the
CDKN2A promoter occurs early during neoplastic progression in
Barrett’s esophagus. Gastroenterology. 122:1113–1121. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Alder H, Taccioli C, Chen H, et al:
Dysregulation of miR-31 and miR-21 induced by zinc deficiency
promotes esophageal cancer. Carcinogenesis. 33:1736–1744. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frisch MJ, Trucks GW, Schlegel HB, et al:
Gaussian 09. Gaussian, Inc; Wallingford, CT: 2009
|
|
25
|
Terunuma A, Putluri N, Mishra P, et al:
MYC-driven accumulation of 2-hydroxyglutarate is associated with
breast cancer prognosis. J Clin Invest. 124:398–412. 2014.
View Article : Google Scholar :
|
|
26
|
Kats LM, Reschke M, Taulli R, et al:
Proto-oncogenic role of mutant IDH2 in leukemia initiation and
maintenance. Cell Stem Cell. 14:329–341. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Montrose DC, Zhou XK, Kopelovich L, et al:
Metabolic profiling, a noninvasive approach for the detection of
experimental colorectal neoplasia. Cancer Prev Res Phila.
5:1358–1367. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Haffner MC, Chaux A, Meeker AK, et al:
Global 5-hydroxy-methylcytosine content is significantly reduced in
tissue stem/ progenitor cell compartments and in human cancers.
Oncotarget. 2:627–637. 2011.PubMed/NCBI
|
|
29
|
Smith DR and Goh HS: Overexpression of the
c-myc proto-oncogene in colorectal carcinoma is associated with a
reduced mortality that is abrogated by point mutation of the p53
tumor suppressor gene. Clin Cancer Res. 2:1049–1053.
1996.PubMed/NCBI
|
|
30
|
Struys EA, Salomons GS, Achouri Y, et al:
Mutations in the D-2-hydroxyglutarate dehydrogenase gene cause
D-2-hydroxy-glutaric aciduria. Am J Hum Genet. 76:358–360. 2005.
View Article : Google Scholar :
|