1. Introduction
Breast cancer is a heterogeneous disease which can
be classified into four major subtypes: luminal A (ER and/or PR-
positive, HER2-negative), luminal B (ER and/or PR-positive,
HER2-positive), HER2-amplified (ER/PR-negative, HER2- positive),
and triple-negative breast cancer (ER/PR-negative, HER2-negative)
(1). Triple-negative breast cancer
is the most aggressive and poorly understood subtype due to the
absence of well-defined molecular targets. It has been considered
to be associated with invasive behavior and worse prognosis. In
addition, the intra-tumor heterogeneity challenges the appropriate
targeted therapies in TNBC. MicroRNAs are a class of small
non-coding RNAs which can reduce the abundance and transcriptional
efficiency of mRNAs by targeting the genes. Altered miRNAs
expression is common in various human malignancies and associated
with tumor initiation, progression and metastasis (2).
Recent studies have shown that a number of miRNAs
are correlated with the hormone receptor status in breast cancer.
Lowery et al (3) identified
three classes of miRNA signatures corresponding with ER (miR-342,
miR-299, miR-217, miR-190, miR-135b and miR-218), PR (miR-520g,
miR-377, miR-527-518a and miR-520f-520c) and HER2 (miR-520d,
miR-181c, miR-302c, miR-376b and miR-30e). Furthermore, TNBC can be
subdivided into core basal (CB) and five negative (5NP) subgroups,
based on a 4-miRNA signature given by miR-155, miR-493, miR-30e and
miR-27a expression levels, and deregulation of the four miRNAs has
been demonstrated to significantly influence prognosis of TNBC
patients (4). Indeed, the
prognostic miRNA signatures are distinct according to ER status.
For instance, miR-342 and miR-150 are associated with good
prognosis and miR-27b with poor prognosis in TNBC, whereas a
totally different cluster of miRNAs (miR-135a, miR-767-3p, miR-128
and miR-769-3p) are proven to be the prognostic markers of
ER-positive breast cancers (5).
Therefore, miRNAs not only play a pivotal role in breast cancer
differentiation, but also contribute to the biological process in
TNBC. Novel miRNAs are increasingly being investigated, and
evidence shows that numerous miRNAs are involved in a variety of
processes contributing to tumorigenesis and metastasis in TNBC.
However, the complex regulatory network formed by these miRNAs is
rarely well-established due to the continuously increasing members.
In the present study, we included the differentially expressed
miRNAs between TNBC and non-TNBC, and the deregulated ones involved
in cell proliferation, cell cycle, apoptosis, EMT, metastasis and
others. Moreover, we confirmed certain miRNAs of previous studies,
as well as discovered several novel miRNAs. The results from the
present study provide additional insight into the miRNA signatures
in TNBC.
2. Identification of microRNAs dysregulated
in TNBC
miRNAs control gene expression by targeting mRNAs,
and are identified to be correlated with specific clinicobiological
features. A set of miRNAs were discovered to be differentially
expressed between breast cancer and normal breast tissue as the
first miRNA signature characteristic of breast carcinoma (6). Since then, more studies have focused
on the miRNA expression patterns among subgroups in breast cancer.
Making use of miRNA microarray profiling, Cochrane et al
(7) found 53 miRNAs differentially
expressed in luminal A vs. TNBC cell lines. Notably, the miRNAs
associated with aggressiveness in lymph node-negative human breast
cancer were inconsistent between ER-positive and ER-negative cases.
The study further revealed the intrinsic miRNA signatures in these
two types of stromal cell lines, and actually the MDA-MB-435 and
MDA-MD-231 in ER-negative group are always considered to be
triple-negative (8). In a more
recent study, 34 miRNAs were observed to be significantly
differentially expressed among the luminal A, HER2-amplified and
triple negative cell lines (9),
and the biologic profiling of miRNA signatures for different
subtypes were also extensively explored in breast cancer tissue
samples (10–13).
The results from each independent study are
consistent for only a few miRNAs, and we summarized the
differentially expressed miRNAs observed in the comparison among
three independent studies with the Venn diagrams in Fig. 1. Comparing to non-TNBC, six miRNAs
(miR-146a, miR-100, miR-125b, miR-29a, miR-222 and miR-221) are
upregulated for TNBC in all three studies, while five miRNAs
(miR-200a, miR-200c, miR-141, miR-375 and miR-203) are
downregulated. Importantly, there are several miRNAs in the
overlapping area between each two of the studies (Table I), indicating the reliability of
the findings. The inconsistence may be caused by the different cell
lines and the different fold-changes in expression in each
study.
 | Table IDysregulated microRNAs in TNBC vs.
non-TNBC cell lines. |
Table I
Dysregulated microRNAs in TNBC vs.
non-TNBC cell lines.
| References | Upregulated
microRNAs | Downregulated
microRNAs |
|---|
| L1 + L2 (7,8) | miR-221, miR-222,
miR-29a, miR-29b, miR-100, miR-125b, miR-146a | miR-345, miR-200c,
miR-141, miR-375, miR-203, miR-200a, miR-93, miR-205, miR-183,
miR-182 |
| L1 + L3 (7,9) | miR-221, miR-222,
miR-224, miR-29a, miR-100, miR-125b, miR-138, miR-146a,
miR-23a, miR-503 | miR-200a, miR-200b,
miR-196a, miR-375, miR-148a, miR-203, miR-200c, miR-141,
miR-24a |
| L2 + L3 (8,9) | miR-146a, miR-100,
miR-125b, miR-29a, miR-222, miR-221 | miR-200c, miR-141,
miR-375, miR-203, miR-200a |
| L1 + L2 + L3
(7–9) | miR-146a, miR-100,
miR-125b, miR-29a, miR-222, miR-221 | miR-200a, miR-200c,
miR-141, miR-375, miR-203 |
To further identify differentially expressed miRNAs
between TNBC and non-TNBC cell lines, we profiled miRNA expression
in luminal A (MCF-7:
ER+/PR+/HER2−), HER2-amplified
(SK-BR-3: ER−/PR−/HER2+) and
triple-negative (MDA-MB-231 and Hs578T:
ER−/PR−/HER2−) molecular subtypes,
and only twelve miRNAs exhibited a 1.5-fold differential expression
between these two groups (Fig. 2).
Of these twelve miRNAs, four (miR-138-5p, miR-4324, miR-4800-3p and
miR-6836-3p) were observed to be upregulated in TNBC group, and the
other eight (miR-363-5p, miR-182-5p, miR-141-3p, miR-339-5p,
miR-4655-3p, miR-4784, miR-664b-5p and miR-6787-5p) were
downregulated. Among these, however, some miRNAs could be newly
discovered ones, which have not been identified previously,
including miR-4324, miR-4800-3p, miR-6836-3p, miR-4655-3p, miR-4784
and miR-6787-5p. The other miRNAs were validated earlier in other
forms, for instance, miR-138-5p was previously identified as
miR-138 in the overlap of L1 and L3, and miR-182-5p referred to
miR-182 in the overlap of L1 and L2. Likewise, miR-141-3p and
miR-363-5p, formerly known as miR-141 and miR-363, were
downregulated in L1-3 or only in L3 separately. In the deregulated
miRNA set of L1, miR-339-3p was regarded to be downregulated,
inconsistent with the miR-339-5p here, and they are the two types
of mature sequences from the same stem-loop sequence miR-339.
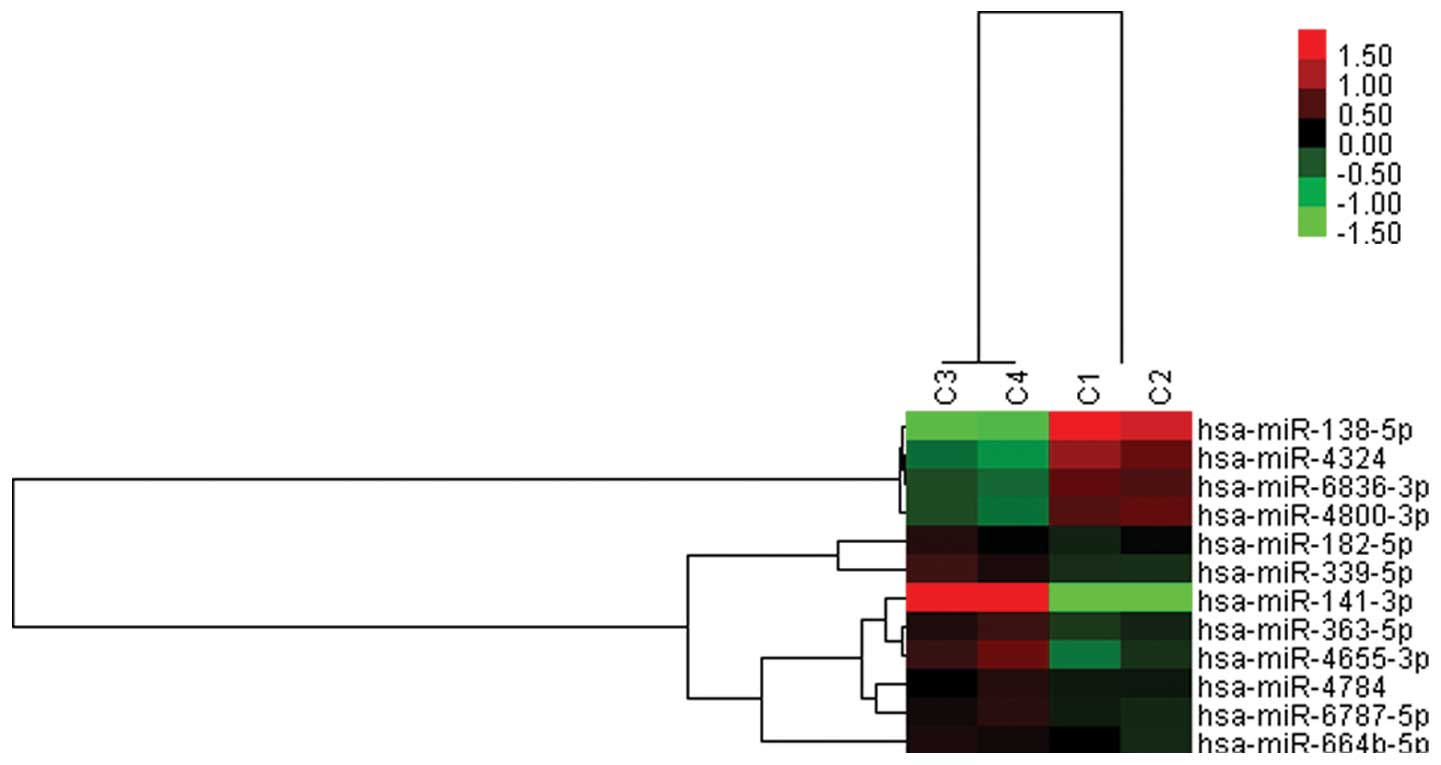 | Figure 2miRNA differential expression in TNBC
vs. non-TNBC cell lines. Hierarchical clustering of twelve miRNAs
exhibiting a 1.5-fold different expression. Rows, individual
miRNAs; colums, individual breast cancer cell lines (C1,
MDA-MB-231; C2, Hs578T; C3, MCF-7; C4, SK-BR-3). Pseudo-colors
represent transcript levels above, equal to, and below the mean
(red, black and green, respectively). The scale represents the
intensity of miRNA expression. |
Given the development in sequencing, our results are
partly consistent with previous findings. Moreover, the TNBC cell
line Hs578T was derived from carcinoma, while the MDA-MB-231 cells
from adenocarcinoma. The different histogenesis of the cell lines
may narrow the miRNA profile outcome, contributing to the fewer
miRNAs in our results comparing to other researches. Taken
together, we validated some previously discovered miRNAs in a
precision sequence, providing us additional insight into the miRNA
signatures in TNBC. It should be tenable to consider these miRNAs
as solid ones being functional in TNBC. However, the observations
need to be further confirmed in more cell lines and tissue
samples.
3. Functional evidence of microRNAs in
TNBC
In general, the miRNAs targeting mRNAs encoding
tumor suppressor proteins are defined as oncogenic miRNAs
(oncomiRs), while the miRNAs targeting oncogenes exhibiting tumor
suppressor properties are defined as oncosuppressor miRNAs. An
increasing body of evidence reveals the pivotal role of miRNAs in
all stages of TNBC. For instance, miR-221 is one of the best known
miRNAs in breast cancer field. Recently, miR-221 was shown to
regulate cell cycle progression by targeting p27 in TNBC,
and to modulate cell migration as well as EMT by decreasing the
E-cadherin level (14). Another
important oncomiR is miR-21 which can promote cell proliferation.
In the study by Sharma et al (15) miR-21 and miR-206 co-targeted
RASA1 and SPRED1, two repressors of RAS-ERK
signaling, resulting in resistance to cell death in MDA-MB-231
cells. In addition, miR-21 was proven to regulate PTEN by
targeting the mRNA 3′UTR, leading to an anti-apoptosis effect in
TNBC (16). Specifically, miR-21
exhibits a higher expression level in TNBC than non-TNBC, and it is
positively correlated with a poor clinical outcome. In contrast to
the miRNAs afore-mentioned, miR-203 is an oncosuppressor miRNA
which plays a specific metastic suppressor role by targeting
LASP1 in TNBC, it can significantly inhibit cell
proliferation by regulating BIRC5 (17). Thus, the miRNAs controlling
expression of quite a few key genes are likely to affect tumor
behavior and progression in TNBC.
microRNA regulation of EMT
An increasing number of miRNAs are demonstrated to
be involved in EMT (Table II).
Reportedly, miR-181a expression was markedly unregulated in TNBC,
and promoted EMT by suppressing the expression of proapoptotic
molecule Bim (18). The miR-200
family of miRNAs is comprised of miR-200a, miR-200b, miR-200c,
miR-141 and miR-429, and the expression of all the miR-200 family
members were absent in invasive breast cancer cell lines with
mesenchymal phenotype, cooperatively preventing TGF-β induced EMT
by attenuating the expression of ZEB1 and ZEB2
(19). Howe et al (20) demonstrated that in MDA-MB-231 or
BT549 cell lines, miR-200c maintained the epithelial phenotype by
targeting FN1 and MSN, encoding proteins normally
expressed in mesenchymal or neuronal origin. In our research,
miR-141, another member from miR-200 family, was identified
down-modulated in TNBC vs. non-TNBC cell lines, which highlights,
therefore, the positive role of miR-141 in epithelial phenotype
maintenance in breast cancer. Contrary to our expectation, however,
all the protective miR-200 family members were shown upregulated in
TNBC compared with normal breast tissues (21,22),
exhibiting a putative risky role in TNBC initiation. One possible
explanation for this phenomenon is that the miR-200 family may act
as an entirely inverse effect in tumorigenic TNBC vs. EMT.
Nevertheless, the actual role of miR-200 family in TNBC deserved to
be further verified by well-designed experiments.
 | Table IIMicroRNAs associated with EMT in
TNBC. |
Table II
MicroRNAs associated with EMT in
TNBC.
| MicroRNAs | Validated
targets | Ref. |
|---|
| Oncosuppressor
miRNAs |
| miR-200b, miR-107,
miR-15b, miR-145, miR-128b | SUZ12, ZEB1/2,
KLF4, BMI1 | (37) |
| miR-200a,
miR-200b, miR-200c | ZEB1/2, FN1, MSN,
N-cadherin, vimentin | (20,38,39) |
| Oncogenic
miRNAs |
| miR-181a | Bim | (18) |
| miR-155 | C/EBPβ | (40) |
| miR-221 | E-cadherin | (14) |
microRNA regulation of metastasis
In addition to regulation of EMT, emerging evidence
has shown the vital role of miRNAs in cell migration, invasion and
metastasis (Table III).
Expression of miR-206 was significantly suppressed in TNBC, and the
forced expression could markedly inhibit cell migration by
targeting 3′UTR of CORO1C (23). Similarly, restoration of miR-145 in
TNBC resulted in a dramatic decrease in ARF6, a known
regulator of breast cancer cell invasion (24). Additionally, another study found
that miR-200b could suppress TNBC cell migration and tumor
metastasis by directly targeting protein kinase Cα (PKCα) (25). Another miR-200 family member
miR-200a, is able to inhibit invasion and metastasis in TNBC by
reducing the ubiquitin-associated and SH3 domain-containing B
(UBASH3B) mRNA and protein expression along with ZEB1
and ZEB2 (26). Besides the
miRNAs mentioned above, miR-139-5p was also validated to repress
metastasis in MDA-MB-231 cells, by modulating a network of genes
underlying cellular processes involved in metastasis, including
HRAS, NFKB1, PIK3CA, RAF and
RHOT1.
 | Table IIIMicroRNAs associated with metastasis
in TNBC. |
Table III
MicroRNAs associated with metastasis
in TNBC.
| MicroRNAs | Validated
targets | Ref. |
|---|
| Oncosuppressor
miRNAs |
| let-7 | RAS, c-Myc,
ERK | (41) |
| miR-200a/c | UBASH3B,
ZEB1/2 | (26) |
| miR-638 | BRCA1 | (35) |
| miR-206 | CORO1C | (23) |
| miR-31 | LOC554202 | (42) |
| miR-145 | ARF6 | (24) |
| miR-200b | PKCα | (25) |
| miR-34a | AXL | (9) |
| miR-203 | LASP1 | (17) |
| miR-139-5p | HRAS, NFKB1,
PIK3CA, RAF, RHOT1 | (43) |
| Oncogenic
miRNAs |
| miR-181a | Bim | (18) |
| miR-17,
miR-20a | TIMP2/3 | (27) |
| miR-221 | E-cadherin | (14) |
| miR-182 | PFN1 | (28) |
In contrast, the miR-17-92 cluster is correlated
with a more aggressive behavior in breast cancer, and two members
from this cluster, miR-17 and miR-20a, could induce metastasis
partially by targeting the extracellular matrix (ECM) proteins
TIMP2 and TIMP3 (27). Comparing
with normal tissue adjacent to TNBC, miR-182 expression was
obviously increased in tumor tissues, which promotes cell invasion
by negatively regulating profilin 1 (PFN1) (28) and correspondently, miR-182-5p, a
mature loop from miR-182, was expressed relatively higher in TNBC
tissue vs. normal (12). Whereas,
miR-182 or miR-182-5p were found to be specifically expressed in
non-TNBC vs. TNBC cell lines based on L1, L2 or our result
(Table I and Fig. 2). Taken together, it would be
reasonable to treat miR-182 and miR-182-5p as potential therapeutic
targets for both TNBC and non-TNBC.
4. MicroRNAs as prognostic markers in
TNBC
With the improvement of tumor biology understanding,
an increasing number of known miRNAs have been identified to be
prognostic in TNBC. Inverse correlation between miR-27b and distant
relapse-free survival (DRFS) was found by performing miRNA
expression profiles, while miR-342 and miR-150 were correlated with
better prognosis (5). A set of
seven miRNAs were found to be associated with TNBC clinical
prognosis (11), and upregulation
of miR-16-2* and miR-766 was correlated with favorable distant
metastases-free survival (DMFS), while miR-381 and miR-409-5p
correlated with poor DMFS, and miR-409-5p was also negatively
associated with breast cancer specific survival (BCSS) along with
miR-376b, miR-410 and miR-193a-3p. Cascione et al (21) identified further miRNA signatures
(miR-16, miR-155, miR-125b, miR-374a and miR-16, miR-125b,
miR-374a, miR-374b, miR-421, miR-655 and miR-497) eligible for
predicting overall survival (OS) and distant disease-free survival
(DDFS), respectively. There are some other miRNAs, such as miR-210
(29), miR-155 (30), miR-27b-3p (31), miR-34b (32) and miR-21 (16), that could to be poor prognosis
indicators in TNBC. However, the prognostic roles of miR-155 in two
independent studies were completely the opposite, implying the
complexity in miRNA regulation.
It is worth mentioning that the protective miR-497,
was identified downregulated in TNBC vs. both normal tissue and
non-TNBC cells (9,21,22),
indicating its putative positive effect against aggressive
malignant phenotype in TNBC. The miR-210, was up-modulated in TNBC
vs. both normal and non-TNBC tissues (12,22,33),
thereby, providing a potential therapeutic target for TNBC.
5. Conclusion
TNBC is a heterogeneous subgroup in breast cancer
characterized by lack of effective targeted therapies, showing an
aggressive morphology and poor prognosis, and miRNA profiling
studies have shown an additional complexity of signal networks
driving the biology of this subtype. There is a need to shed light
on the miRNA related pathways to identify biomarkers able to
predict prognosis and response to therapy, even probably identify
novel therapeutic targets in TNBC. Although no therapeutics
targeting specific miRNAs have made into clinic, a few of miRNAs
have been implicated in altering sensitivity to treatment. A recent
study co-encapsulated miR-34a with doxorubicin into hyaluronic acid
(HA)-chitosan (CS) nanoparticles (NPs), and then co-delivered it
into TNBC cells or tissues. Surprisingly, the co-delivery of
miR-34a enhanced antitumor effects of doxorubicin by suppressing
the expression of Bcl-2 and blocking Notch-1 signaling
(34). Furthermore, miR-638
overexpression was associated with increased sensitivity to DNA
damaging agents, ultraviolet (UV) and cisplatin in TNBC cells
(35), while miR-181a and miR-181b
were regarded as negative regulators of the DNA damage response and
decreased the sensitivity to poly-ADP-ribose-polymerase 1 (PARP1)
inhibition in TNBC cells (36).
The present review has provided a new scenario of
the miRNA expression differences between TNBC and normal tissue or
TNBC and non-TNBC cells by summarizing previous profiling studies.
In our research, we verified some previously proven miRNAs and
identified several new ones as well, providing new evidence for
investigating the functional miRNAs in TNBC. Such studies will be
valuable for gaining better understanding of the role of miRNAs on
the pathogenesis of TNBC, and identifying novel biomarkers for
diagnosis, treatment and prognosis. However, although quite a few
miRNAs showed differences, demonstration of the functions of most
molecules has not been validated, and some miRNAs even show the
opposite effect among independent studies. Consequently, further
studies are required to manifest the underlying mechanism of miRNAs
in TNBC progression, including cell proliferation, cell cycle,
apoptosis, EMT and metastasis, before taking miRNAs as therapeutic
targets in TNBC.
Acknowledgements
The present review was supported by grants from the
National Natural Science Foundation of China (no. 81272252) and a
Foundation for Clinical Medicine Science and Technology Special
Project of the Jiangsu Province, China (no. BL2014071) (to
X.G).
Abbreviations:
|
TNBC
|
triple-negative breast cancer
|
|
miRNA
|
microRNA
|
|
EMT
|
epithelial-mesenchymal transition
|
|
ER
|
estrogen receptor
|
|
3′UTR
|
3′ untranslated region
|
References
|
1
|
Sorlie T, Perou CM, Tibshirani R, et al:
Gene expression patterns of breast carcinomas distinguish tumor
subclasses with clinical implications. Proc Natl Acad Sci USA.
98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lowery AJ, Miller N, Devaney A, et al:
MicroRNA signatures predict oestrogen receptor, progesterone
receptor and HER2/ neu receptor status in breast cancer. Breast
Cancer Res. 11:R272009. View
Article : Google Scholar
|
|
4
|
Gasparini P, Cascione L, Fassan M, et al:
microRNA expression profiling identifies a four microRNA signature
as a novel diagnostic and prognostic biomarker in triple negative
breast cancers. Oncotarget. 5:1174–1184. 2014.PubMed/NCBI
|
|
5
|
Buffa FM, Camps C, Winchester L, et al:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, et al:
MicroRNA gene expression deregulation in human breast cancer.
Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cochrane DR, Cittelly DM, Howe EN, et al:
MicroRNAs link estrogen receptor alpha status and Dicer levels in
breast cancer. Horm Cancer. 1:306–319. 2010. View Article : Google Scholar
|
|
8
|
Foekens JA, Sieuwerts AM, Smid M, et al:
Four miRNAs associated with aggressiveness of lymph node-negative,
estrogen receptor-positive human breast cancer. Proc Natl Acad Sci
USA. 105:13021–13026. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mackiewicz M, Huppi K, Pitt JJ, Dorsey TH,
Ambs S and Caplen NJ: Identification of the receptor tyrosine
kinase AXL in breast cancer as a target for the human miR-34a
microRNA. Breast Cancer Res Treat. 130:663–679. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Janssen EA, Slewa A, Gudlaugsson E, et al:
Biologic profiling of lymph node negative breast cancers by means
of microRNA expression. Mod Pathol. 23:1567–1576. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Rinaldis E, Gazinska P, Mera A, et al:
Integrated genomic analysis of triple-negative breast cancers
reveals novel microRNAs associated with clinical and molecular
phenotypes and sheds light on the pathways they control. BMC
Genomics. 14:6432013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calvano Filho CM, Calvano-Mendes DC,
Carvalho KC, et al: Triple-negative and luminal breast tumors:
differential expression of miR-18a-5p, miR-17-5p, and miR-20a-5p.
Tumour Biol. 35:7733–7741. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Farazi TA, Ten Hoeve JJ, Brown M, et al:
Identification of distinct miRNA target regulation between breast
cancer molecular subtypes using AGO2-PAR-CLIP and patient datasets.
Genome Biol. 15:R92014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nassirpour R, Mehta PP, Baxi SM and Yin
MJ: miR-221 promotes tumorigenesis in human triple negative breast
cancer cells. PLoS One. 8:e621702013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sharma SB, Lin CC, Farrugia MK, et al:
microRNAs-206 and -21 cooperate to promote RAS-ERK signaling by
suppressing the translation of RASA1 and SPRED1. Mol Cell Biol.
34:4143–4164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong G, Liang X, Wang D, et al: High
expression of miR-21 in triple-negative breast cancers was
correlated with a poor prognosis and promoted tumor cell in vitro
proliferation. Med Oncol. 31:572014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang C, Zheng X, Shen C and Shi Y:
MicroRNA-203 suppresses cell proliferation and migration by
targeting BIRC5 and LASP1 in human triple-negative breast cancer
cells. J Exp Clin Cancer Res. 31:582012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taylor MA, Sossey-Alaoui K, Thompson CL,
Danielpour D and Schiemann WP: TGF-beta upregulates miR-181a
expression to promote breast cancer metastasis. J Clin Invest.
123:150–163. 2013. View
Article : Google Scholar :
|
|
19
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Howe EN, Cochrane DR and Richer JK:
Targets of miR-200c mediate suppression of cell motility and
anoikis resistance. Breast Cancer Res. 13:R452011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cascione L, Gasparini P, Lovat F, et al:
Integrated microRNA and mRNA signatures associated with survival in
triple negative breast cancer. PLoS One. 8:e559102013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Avery-Kiejda KA, Braye SG, Mathe A, Forbes
JF and Scott RJ: Decreased expression of key tumour suppressor
microRNAs is associated with lymph node metastases in triple
negative breast cancer. BMC Cancer. 14:512014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang J, Tsouko E, Jonsson P, et al:
miR-206 inhibits cell migration through direct targeting of the
actin-binding protein Coronin 1C in triple-negative breast cancer.
Mol Oncol. 8:1690–1702. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Eades G, Wolfson B, Zhang Y, Li Q, Yao Y
and Zhou Q: lincRNA-RoR and miR-145 regulate invasion in
triple-negative breast cancer via targeting ARF6. Mol Cancer Res.
Sep 24–2014.(Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Humphries B, Wang Z, Oom AL, et al:
MicroRNA-200b targets protein kinase Calpha and suppresses
triple-negative breast cancer metastasis. Carcinogenesis.
35:2254–2263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lee ST, Feng M, Wei Y, et al: Protein
tyrosine phosphatase UBASH3B is overexpressed in triple-negative
breast cancer and promotes invasion and metastasis. Proc Natl Acad
Sci USA. 110:11121–11126. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jin L, Lim M, Zhao S, et al: The
metastatic potential of triple-negative breast cancer is decreased
via caloric restriction-mediated reduction of the miR-17~92
cluster. Breast Cancer Res Treat. 146:41–50. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu H, Wang Y, Li X, et al: Expression and
regulatory function of miRNA-182 in triple-negative breast cancer
cells through its targeting of profilin 1. Tumour Biol.
34:1713–1722. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Toyama T, Kondo N, Endo Y, et al: High
expression of microRNA-210 is an independent factor indicating a
poor prognosis in Japanese triple-negative breast cancer patients.
Jpn J Clin Oncol. 42:256–263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kong W, He L, Richards EJ, et al:
Upregulation of miRNA-155 promotes tumour angiogenesis by targeting
VHL and is associated with poor prognosis and triple-negative
breast cancer. Oncogene. 33:679–689. 2014. View Article : Google Scholar :
|
|
31
|
Shen S, Sun Q, Liang Z, et al: A
prognostic model of triple-negative breast cancer based on
miR-27b-3p and node status. PLoS One. 9:e1006642014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Svoboda M, Sana J, Redova M, et al:
MiR-34b is associated with clinical outcome in triple-negative
breast cancer patients. Diagn Pathol. 7:312012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Radojicic J, Zaravinos A, Vrekoussis T,
Kafousi M, Spandidos DA and Stathopoulos EN: MicroRNA expression
analysis in triple-negative (ER, PR and Her2/neu) breast cancer.
Cell Cycle. 10:507–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deng X, Cao M, Zhang J, et al: Hyaluronic
acid-chitosan nanoparticles for co-delivery of miR-34a and
doxorubicin in therapy against triple negative breast cancer.
Biomaterials. 35:4333–4344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Tan X, Peng J, Fu Y, et al: miR-638
mediated regulation of BRCA1 affects DNA repair and sensitivity to
UV and cisplatin in triple negative breast cancer. Breast Cancer
Res. 16:4352014. View Article : Google Scholar
|
|
36
|
Bisso A, Faleschini M, Zampa F, et al:
Oncogenic miR-181a/b affect the DNA damage response in aggressive
breast cancer. Cell Cycle. 12:1679–1687. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Polytarchou C, Iliopoulos D and Struhl K:
An integrated transcriptional regulatory circuit that reinforces
the breast cancer stem cell state. Proc Natl Acad Sci USA.
109:14470–14475. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ahmad A, Sarkar SH, Bitar B, et al:
Garcinol regulates EMT and Wnt signaling pathways in vitro and in
vivo, leading to anti-cancer activity against breast cancer cells.
Mol Cancer Ther. 11:2193–2201. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Si L, Jiang F, Li Y, et al: Induction of
the mesenchymal to epithelial transition by demethylation-activated
microRNA-200c is involved in the anti-migration/invasion effects of
arsenic trioxide on human breast cancer cells. Mol Carcinog. Apr
14–2014.(Epub ahead of print). View
Article : Google Scholar
|
|
40
|
Johansson J, Berg T, Kurzejamska E, et al:
MiR-155-mediated loss of C/EBPbeta shifts the TGF-beta response
from growth inhibition to epithelial-mesenchymal transition,
invasion and metastasis in breast cancer. Oncogene. 32:5614–5624.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Aceto N, Sausgruber N, Brinkhaus H, et al:
Tyrosine phosphatase SHP2 promotes breast cancer progression and
maintains tumor-initiating cells via activation of key
transcription factors and a positive feedback signaling loop. Nat
Med. 18:529–537. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
42
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krishnan K, Steptoe AL, Martin HC, et al:
miR-139-5p is a regulator of metastatic pathways in breast cancer.
RNA. 19:1767–1780. 2013. View Article : Google Scholar : PubMed/NCBI
|
















