Introduction
Prostate cancer (PCa) is one of the most common
malignant tumors of male urinary and reproductive system.
Worldwide, PCa has the second highest incidence among malignant
tumors of the male. An estimated 220,800 new cases of PCa will
occur in the US during 2015. The incidence of PCa has surpassed
lung cancer, becoming the first health hazard of the men,
accounting for 26% of all male malignancies. With an estimated
27,540 deaths in 2015, PCa is the second-leading cause of cancer
death in men (1). PCa is the most
frequent cancer among males in Europe. In Europe in 2015, the
number of predicted PCa deaths is 72,600 (2). China is one of the nations which have
a lower incidence and mortality of PCa, but the incidence shows a
continued rapid growth trend in recent years. Risk factors for PCa
include age, race and heredity. Moreover, many molecular biological
changes which lead to alteration of gene expression and protein
functions have been found, including chromosomal aberration, gene
amplification and mutation. Similar to other human malignancies,
PCa is also the result of genetic and epigenetic factors working
together. However, for PCa, the exact molecular mechanism of tumor
occurrence and progression is still not clear.
NDRG family is a group of genes which have been
found in recent years, including NDRG1, NDRG2, NDRG3 and NDRG4.
Researches found that NDRG1 was related to tumor cell stress,
proliferation, differentiation and invasion. NDRG1 which is
conserved in evolution can interact with a variety of transcription
factors, acting as a transcriptional co-repressor factor. NDRG1 was
originally discovered with the mouse embryonic N-myc gene knockout
(3), located on human chromosome
8q 24.3, containing 16 exons and 15 introns. NDRG1 expression can
be upregulated by a variety of physiological conditions or external
stimulus, which could promote cell differentiation. NDRG1 has been
studied in breast (4,5), pancreatic (6–8),
gastric cancer (9), colon
(5,10), cervical (11), kidney cancer (12) and PCa (8,13–16),
but the findings are not consistent. Whether NDRG1 has a role in
tumor suppression or promotion remains controversial. The role may
be associated with specific tissues or tumor microenvironment.
Current research tends to consider NDRG1 as a tumor suppressor gene
(17) in PCa, inhibiting cell
proliferation, metastasis and invasion, however, the regulation
mechanism is unclear.
Studies have shown that DNA methylation may be
associated with the expression level of NDRG1 in breast cancer
(5). But in PCa, how the
expression level of NDRG1 changes and its epigenetic regulatory
mechanisms are not entirely clear. This research was conducted by a
preliminary study of the expression and function of NDRG1 in PCa,
and then by study of the methylation of NDRG1 promoter which could
be one of the epigenetic molecular mechanisms of the changes of
NDRG1 expression level.
Materials and methods
Ethics statement
The study was approved by the ethics board of the
Second Hospital of Tianjin Medical University. All samples were
obtained from patients who signed informed consent approving the
use of their tissues for research purposes after operation.
Prostate tissue samples
All of the PCa tissues were collected after radical
prostatectomy at the Department of Urology of the hospital. None of
the patients had received neoadjuvant hormone therapy before the
operation. Benign prostatic hyperplasia (BPH) tissues were
collected after suprapubic enucleation of the prostate. Fresh
prostate tissues were sampled directly after surgical removal of
the gland and were immediately frozen in liquid nitrogen. A
diagnostic H&E section was prepared to identify the tissues by
pathologists.
Cell culture
Human PCa cell lines (LNCap, PC-3, DU145 and 22Rv1)
and normal prostate epithelial cell line (RWPE-1) were obtained
from the American Type Culture Collection (ATCC; Manassas, VA, USA)
and maintained in our laboratory. Cells were cultured in RPMI-1640
(Gibco) supplemented with 10% fetal calf serum and penicillin (100
U/ml). Cultures were maintained under an atmosphere containing 5%
CO2.
RT-qPCR
Total RNA was extracted using TRIZol reagent
(Invitrogen, Carlsbad, CA, USA). cDNA was synthesized using M-MLV
MicroRNA reverse transcription kit (Promega, Madison, WI, USA).
RT-qPCR was performed with SYBR Premix Ex Taq™ (Takara,
Biotechnology Co., Ltd., Dalian, China). PCR primer for NDRG1 was
5′-CCGACAACCA CTACCTGA-3′ (forward) and 5′-CGTGAAGAATGTGCGAG AC-3′
(reverse). The expression level were normalized to GAPDH. PCR
primer for GAPDH was 5′-GGATTTGGTCG TATTGGG-3′ (forward) and
5′-GGAAGATGGTGATGGGA TT-3′ (reverse). PCR was performed under the
following conditions: 94°C for 4 min, followed by 40 cycles at 94°C
for 30 sec, 50°C for 30 sec and 72°C for 40 sec. Each sample was
run in triplicate.
Western blot analysis
All proteins were resolved on a 10% SDS-denatured
polyacrylamide gel and were then transferred onto a nitrocellulose
membrane. Membranes were incubated with blocking buffer for 60 min
at room temperature and then incubated with primary antibody
overnight at 4°C. The membranes were washed and incubated with a
horseradish peroxidase (HRP)-conjugated secondary antibody. Protein
expression was assessed by enhanced chemiluminescence and exposure
to chemiluminescent film. The LabWorks image acquisition and
analysis software (UVP, LLC Upland, CA, USA) was used to quantify
band intensities. All antibodies were purchased from Tianjin Saier
Biotechnology Co., Ltd. (Tianjin, China).
Immunohistochemistry staining (IHC)
Paraffin-embedded sections (4 μm) were
deparaffinized and hydrated in xylene, followed by graded alcohols
to water. Antigen retrieval was performed twice in 0.01 M citrate
10 min in a microwave oven, followed by a 60-min cool down. Slides
were then incubated with various primary antibodies followed by
EnVision-plus-labeled polymer-conjugated horseradish peroxidase and
DAB monitoring staining (Beijing Zhongshan Golden Bridge
Biotechnology Co., Ltd., Beijing). Then, slides were
counter-stained and dehydrated for viewing and imaging. The
antibody was anti-NDRG1 (GeneTech, Shanghai, China).
Plasmid vector construction and cell
transfection
Two types of plasmid vectors were constructed for
changing the expression level of NDRG1. PC-3 cells were divided
into several groups as follows according to different transfection
contents: i) for downregulation: a, PBS; b, pRNAT-U6.1/Neo (NC); c,
PSiHIV-U6/shRNA-1; d, PSiHIV-U6/shRNA-2; e, PSiHIV-U6/shRNA-3; f,
PSiHIV-U6/shRNA-4; ii) for upregulation: a, PBS; b, pReceiver-Lv103
(NC); c, pReceiver-Lv103-Expression. PC-3 cells were seeded in
triplicate in 96-well plates, allowed to settle for 24 h and then
co-transfected with different contents by using Lipofectamine 2000
(Invitrogen). After the transfection, using light microscope the
growth of the cells was observed.
MTT assay
PC-3 cells were plated on 96-well plates at
1×104 cells/well. Viable cells were measured on day 1,
2, 3 and 4 after plating. After incubation with
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),
the cells were lysed in 150 ml of 100% dimethylsulfoxide (DMSO) and
UV-visible absorbance was read at 490 nm. Each sample was run in
triplicate.
Flow cytometry
PC-3 cells were collected and fixed with 70% ethanol
for the detection of early apoptosis. The dyeing of cells was
performed according to the instructions of Annexin V-R-PE cell
apoptosis detection kit (SouthernBiotech, Birmingham, AL USA). Flow
cytometry (BD Biosciences, San Jose, CA, USA) was used to detect
the percentage of early apoptosis. The measurement was performed in
triplicate.
Transwell migration assay
PC-3 cells were collected. PC-3 (5×104)
cells were placed on the upper chamber of each insert coated with
50 ml of 2 mg/ml Matrigel (growth factor reduced BD Matrigel™
matrix), and 600 ml of RPMI-1640 with 20% FBS was added to the
lower part of the chamber. After incubating for 24 h, the chambers
were disassembled, and the membranes were stained with a 2% crystal
violet solution for 15 min and placed on a glass slide. Then, cells
that had migrated across the membrane were counted in five random
visual fields using a microplate reader at 490 nm. All assays were
performed three independent times in triplicate.
Bisulfite sequencing PCR (BSP) primer
design
NDRG1 promoter was predicted by NCBI (www.ncbi.nlm.nih.gov/gene) and Prosean (www-bimas.cit.nih.gov/molbio/Proscan/).
BSP Primer was designed by Promoter 2.0 (www.cbs.dtu.dk/services/Promoter/), BLAST (www.ncbi.nlm.nih.gov/BLAST/), methBLAST
(medgen.ugent.be/methBLAST/) and MethPrimer (www.urogene.org/methprimer/).
BSP
After genomic DNA treatment by bisulfite, all of the
unmethylated cytosines were converted to uracils, whereas
methylated cytosines were unchanged. BSP primers were designed for
PCR. Purified products were used for TA cloning. Positive clones
were selected from each of the TA clones. The methylation site
changes in CGIs of NDRG1 promoter were observed by comparing with
the original gene sequences (DNAMAN V6).
Statistical analysis
The data are presented as mean ± standard deviation
(± SD). The two-tailed Student's t-test was used to evaluate the
significance of the differences between two groups. P<0.05 was
considered significant.
Results
NDRG1 expression in normal prostate and
PCa
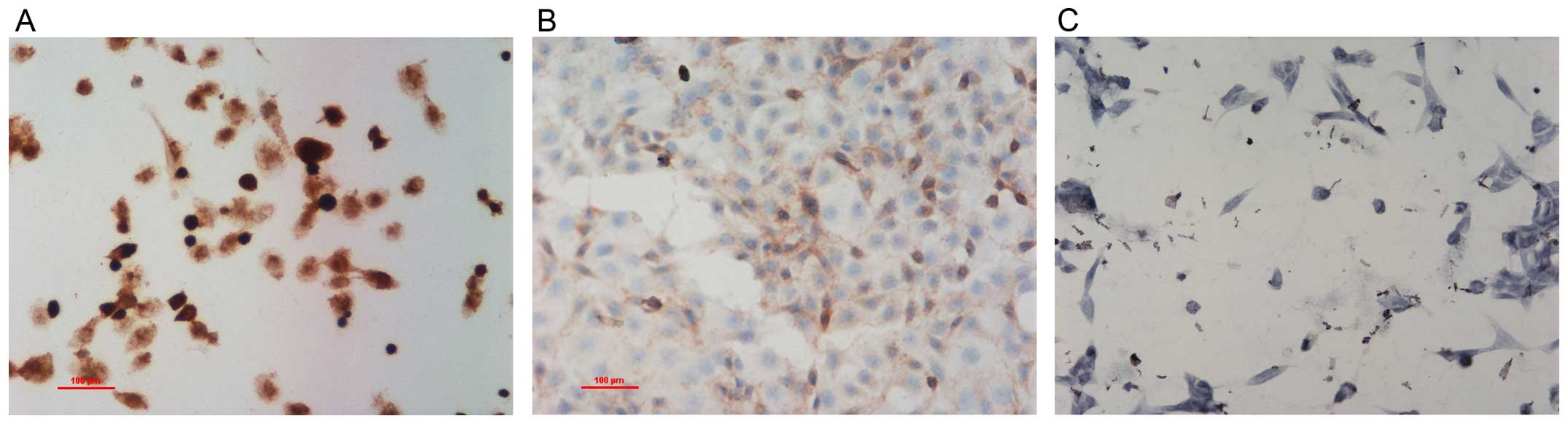
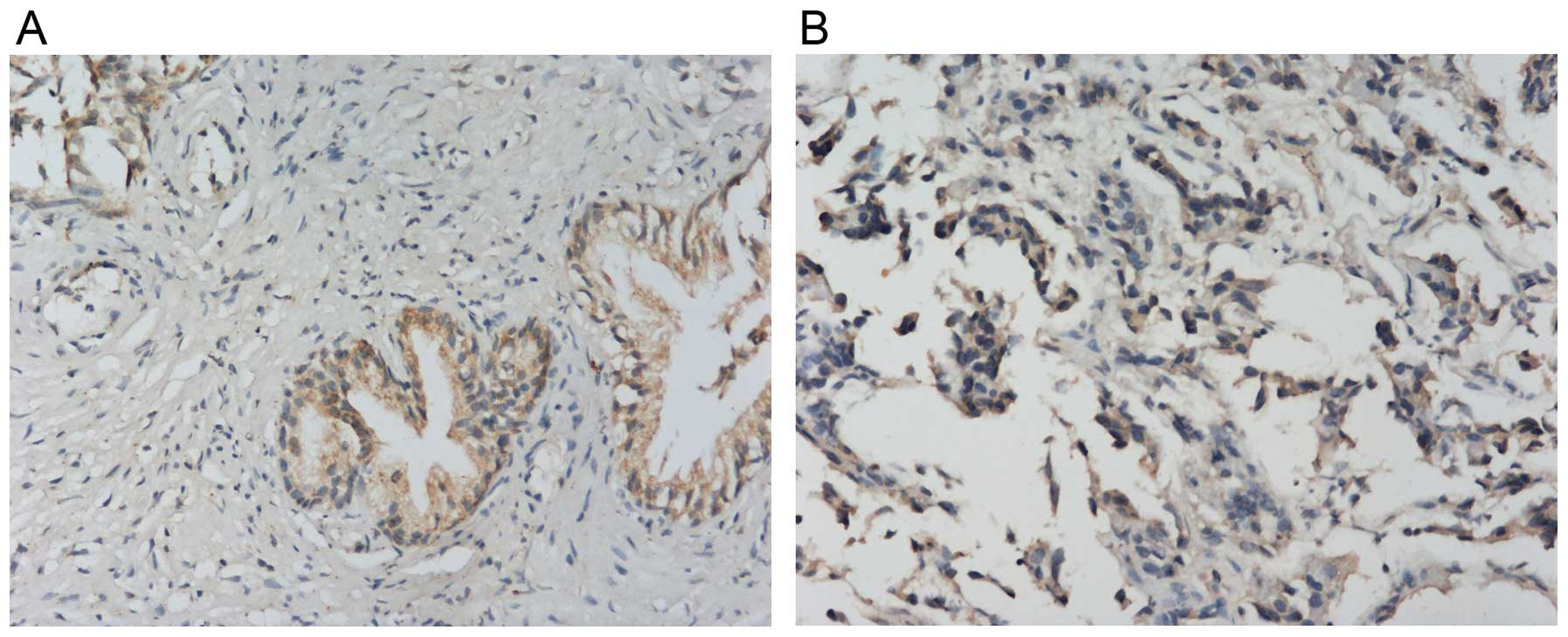
IHC showed that NDRG1 protein expressed both in the
cytoplasm and nucleus, but mainly in the cytoplasm (Fig. 1). NDRG1 expression in RWPE-1 cells
was higher than that in PC-3 and LNCap cells. NDRG1 expression in
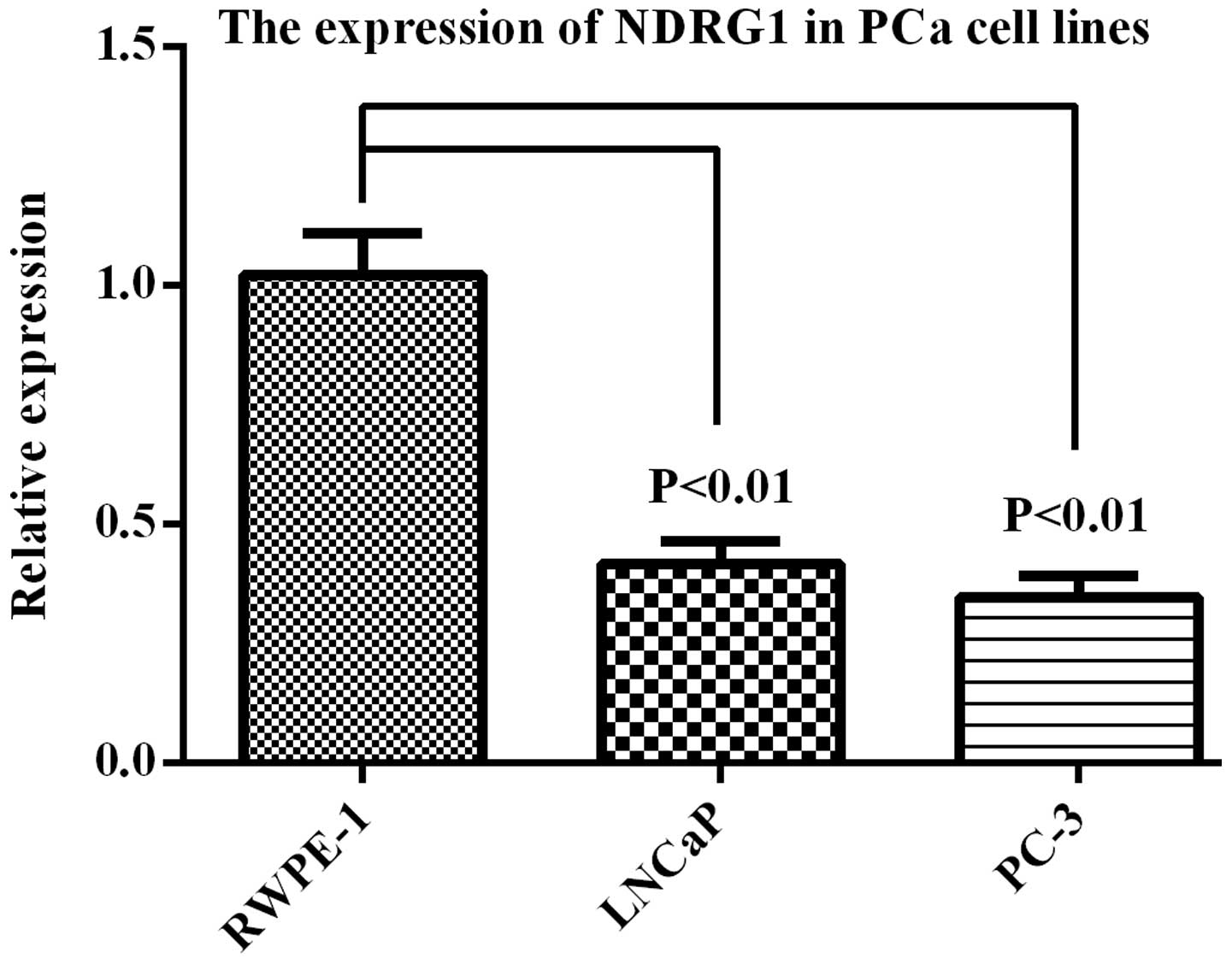
PCa tissues was lower than that in BPH tissues (Fig. 2). RT-qPCR showed that NDRG1 mRNA
expression was higher in RWPE-1 cells than that in LNCap and PC-3
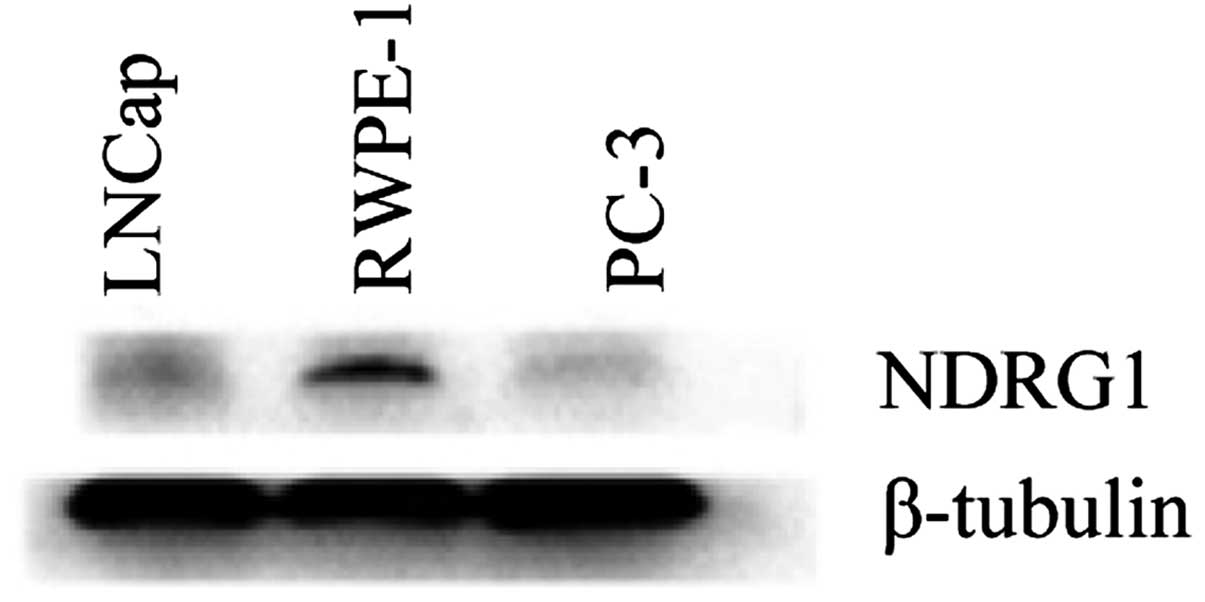
cells (Fig. 3). Western blot
analysis showed that NDRG1 protein expression in RWPE-1 cells was
higher than that in LNCap and PC-3 cells (Fig. 4).
NDRG1 expression is changed by
transfection with plasmid vectors
After PC-3 cell transfection with PSiHIV-U6/shRNA,
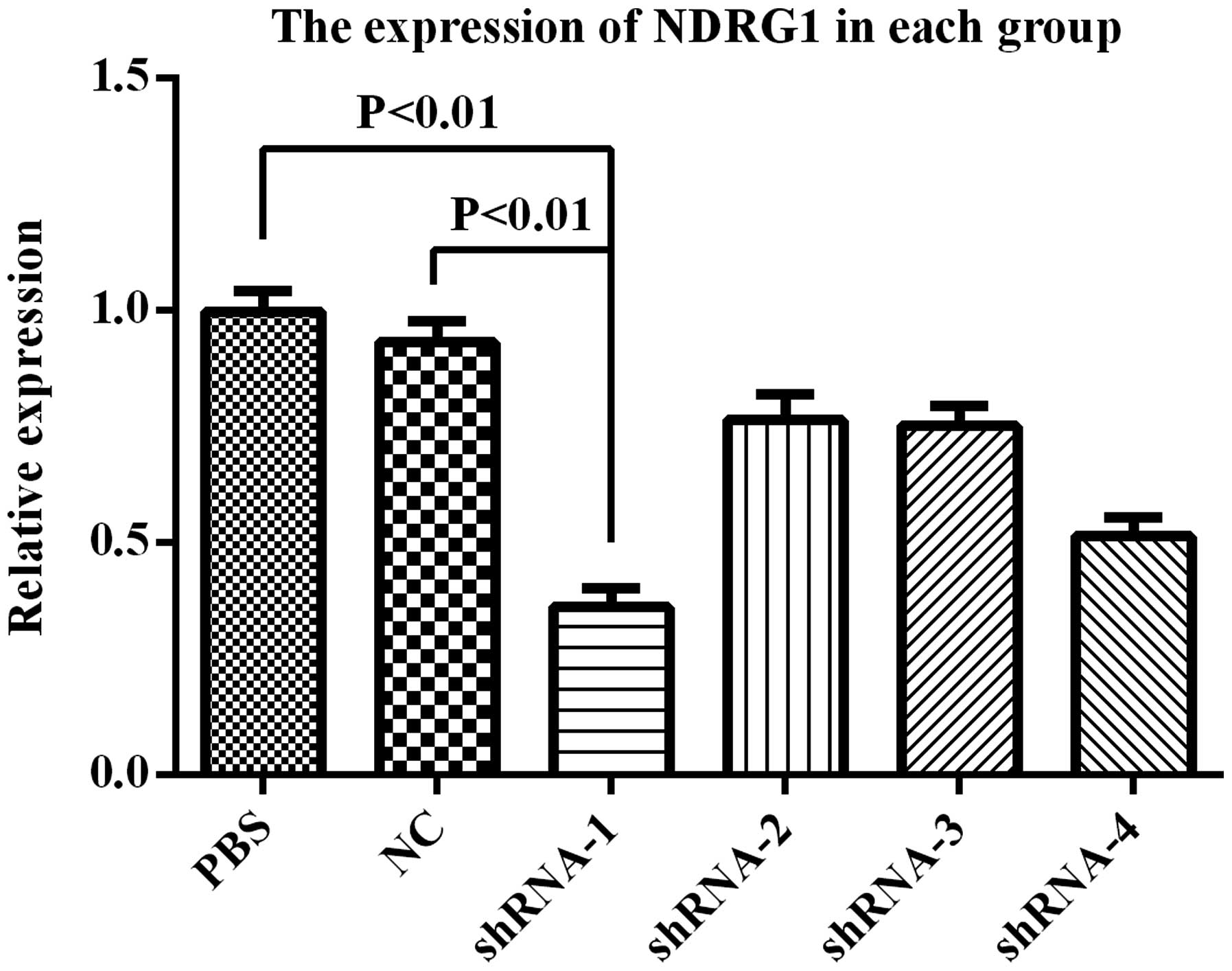
NDRG1 expression was downregulated. RT-qPCR showed that NDRG1 mRNA
expression was significantly decreased, and PSiHIV-U6/shRNA-1 had
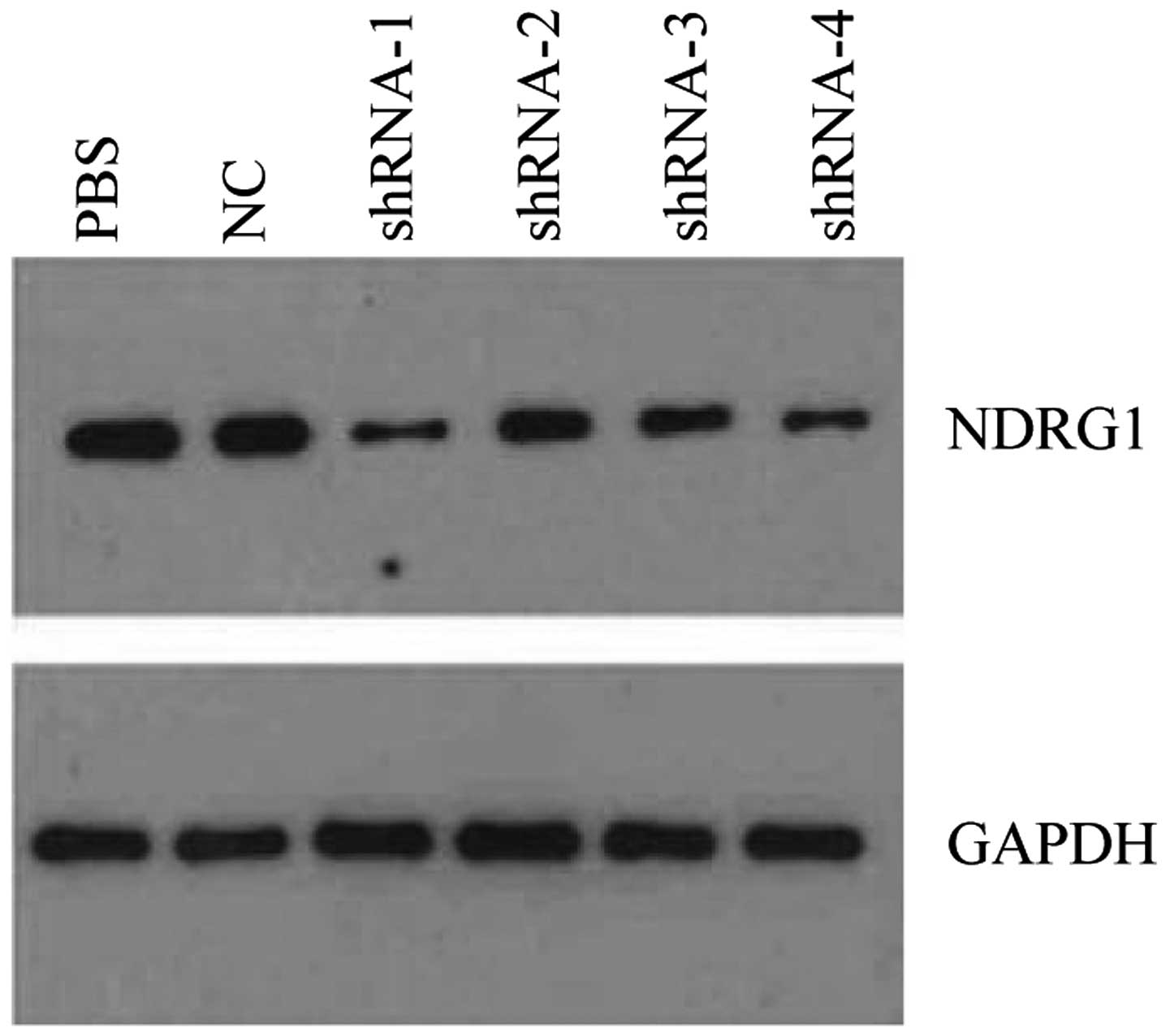
the most preferably interference effect (Fig. 5). Western blot analysis showed that
NDRG1 protein expression was significantly decreased, and
PSiHIV-U6/shRNA-1 had the most preferably interference effect
(Fig. 6). PSiHIV-U6/shRNA-1 was
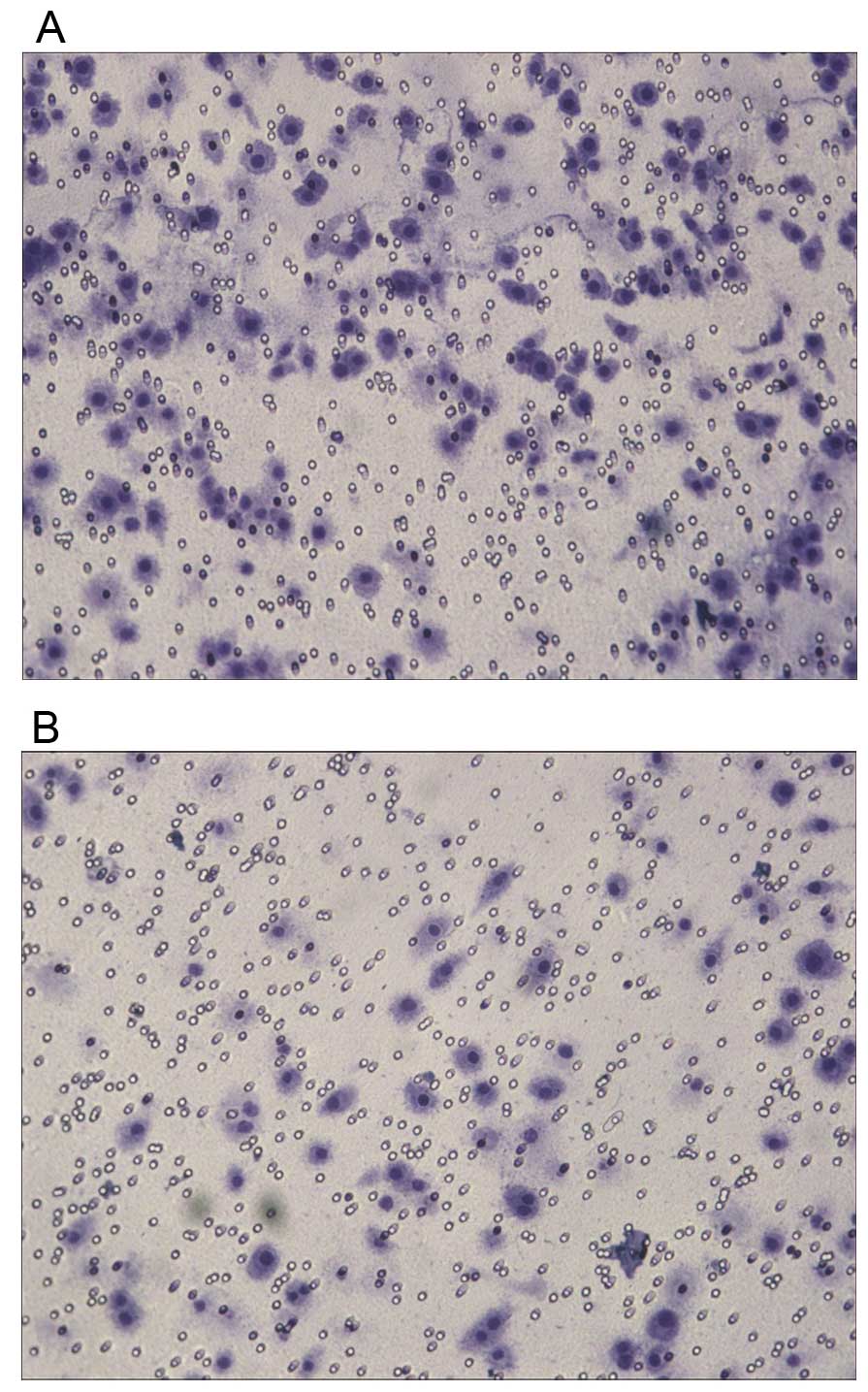
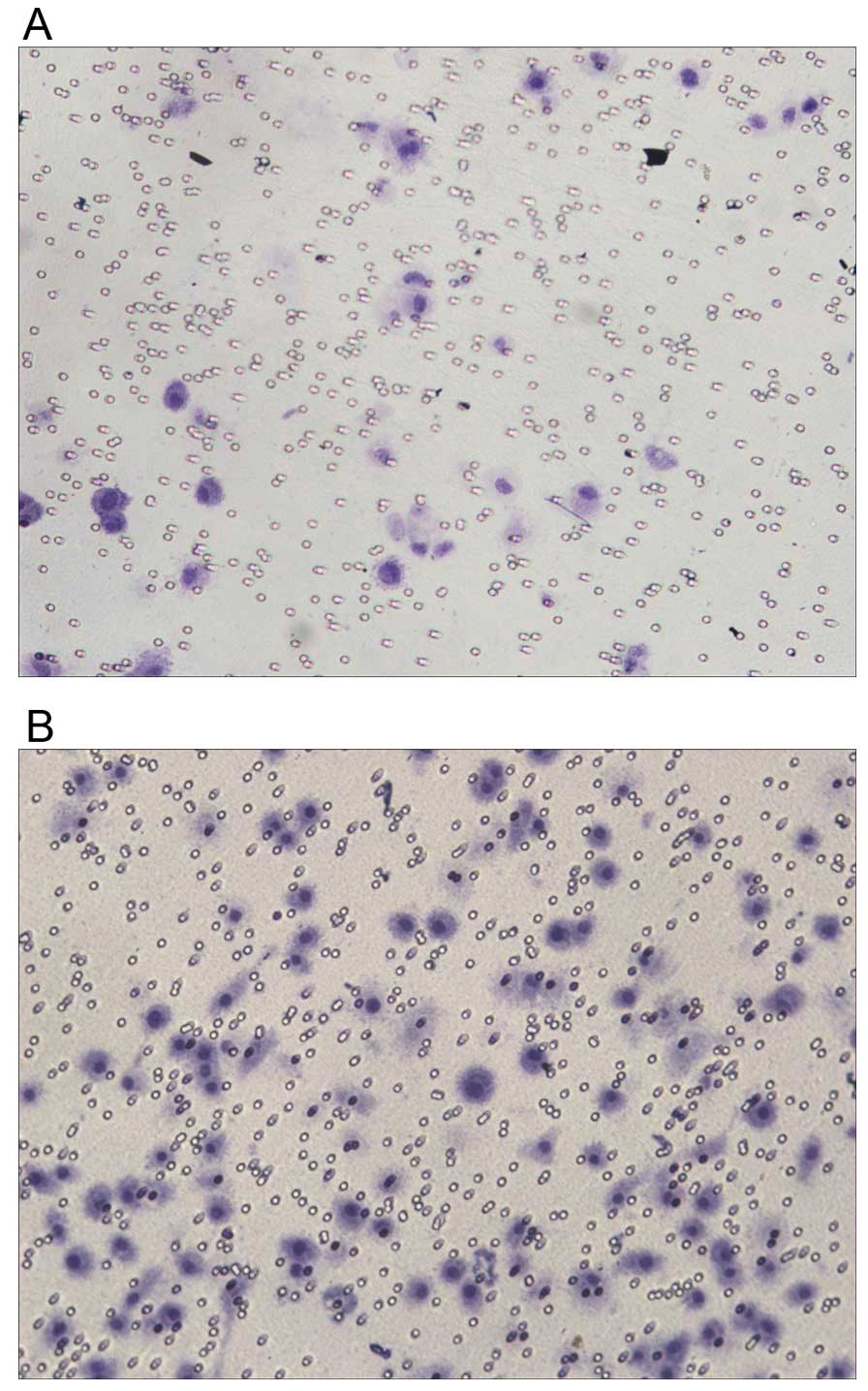
chosen for the following experiments. Transwell assay showed that
after the transfection, the invasive ability of PC-3 cells was
significantly increased (Table I),
suggesting that downregulation of NDRG1 expression had a
significant enhancing effect on the migration of PC-3 cells
(Fig. 7). Flow cytometry showed
that PSiHIV-U6/shRNA-1 group had the lowest early apoptosis rate at
0.02%. NC group had the highest early apoptosis rate at 1.34% and
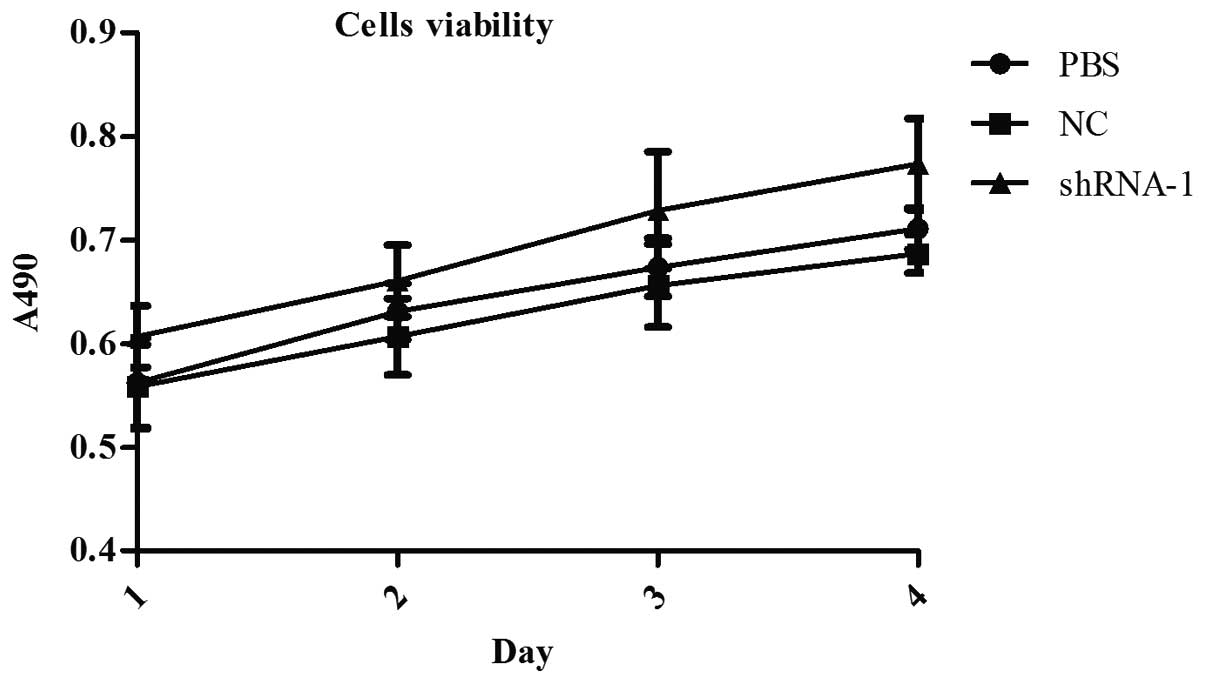
PBS group was 1.94%. MTT assay showed that the cell proliferation
rate of PSiHIV-U6/shRNA-1 group was significantly higher than the
others (Fig. 8). After
transfection for 24, 48, 72 and 96 h, the number of cells in
PSiHIV-U6/shRNA-1 group was always significantly higher than the
other groups (P<0.01). Howecer, at each time-point, the
difference between PBS group and NC group was no statistically
significant (P>0.05).
 | Table IResults of Transwell migration assay
after downregulation of NDRG1 expression (mean ± SD). |
Table I
Results of Transwell migration assay
after downregulation of NDRG1 expression (mean ± SD).
| Group | Fields counted | Invasion cells per
field | t | P-value |
|---|
|
PSiHIV-U6/shRNA-Expression-1 | 15 | 67±13 | 2.15 | <0.05 |
| PBS | 15 | 21±6 | | |
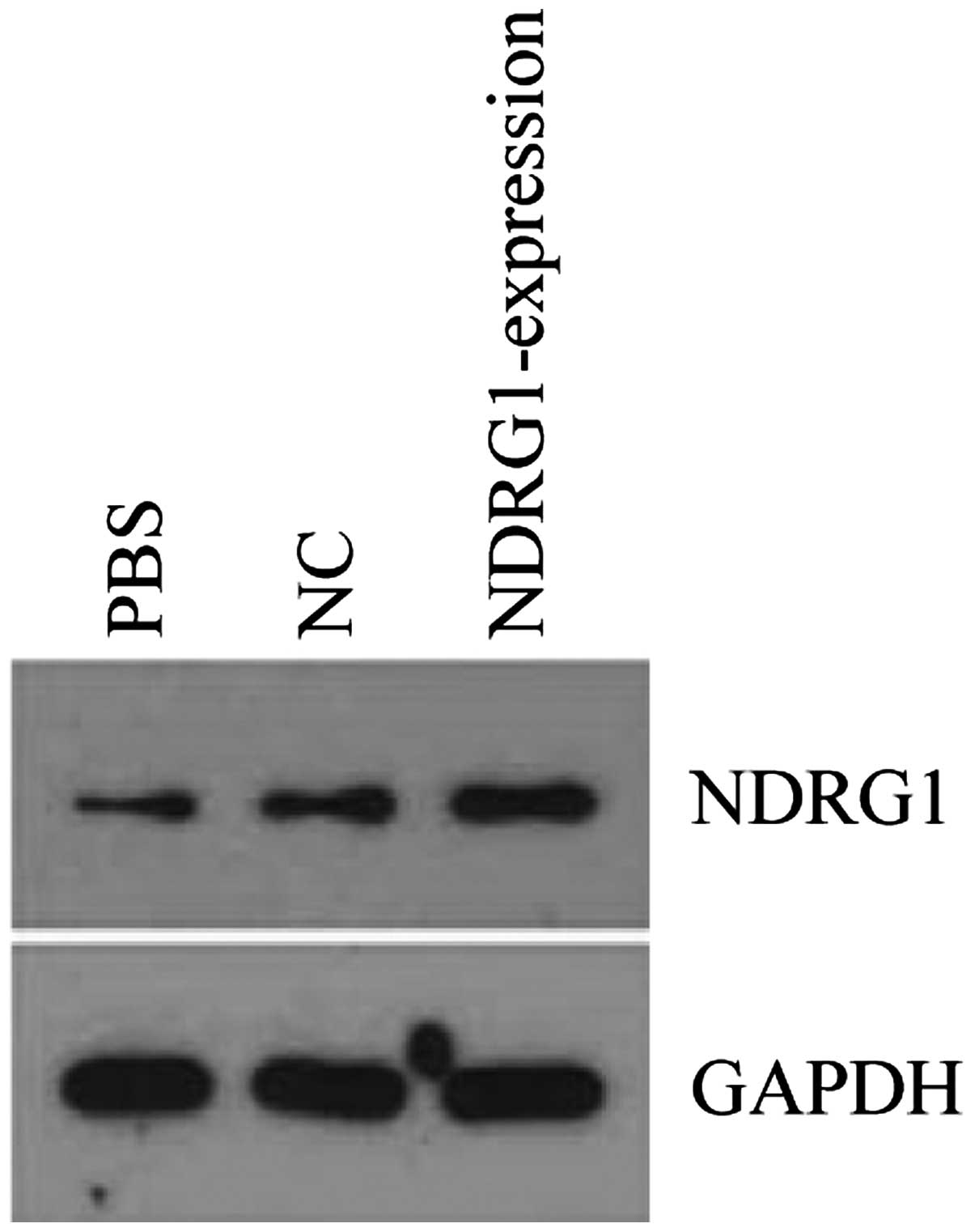
After PC-3 cell transfection with
pReceiver-Lv103-Expression, NDRG1 expression was upregulated.
Western blot analysis showed that NDRG1 protein expression was
higher in pReceiver-Lv103-expression group than that in control
group (Fig. 9). Transwell assay
showed that after the transfection, the invasive ability of PC-3
cells was significantly decreased (Table II), suggesting that upregulation
of NDRG1 expression did have a significant weakening effect on the
migration of PC-3 cells (Fig.
10). Flow cytometry showed that pReceiver-Lv103-Expression
group had the highest early apoptosis rate at 26.48%. NC group had
the lowest early apoptosis rate at 0.64% and PBS group was 4.97%.
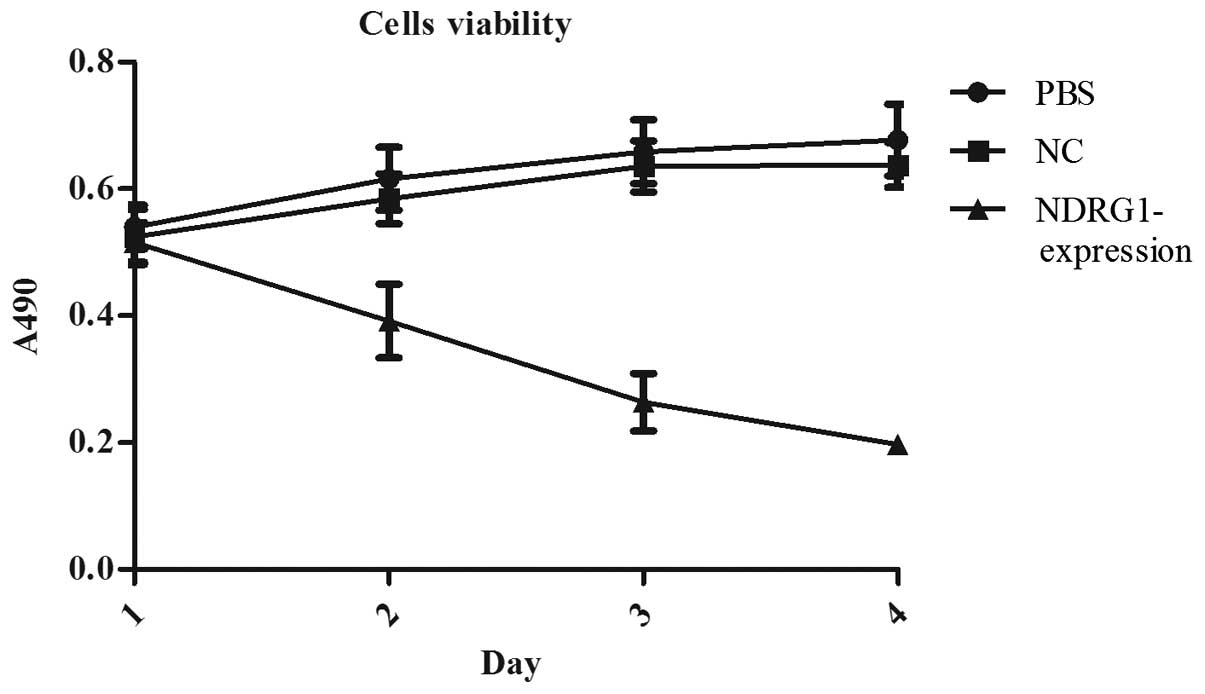
MTT assay showed that the cell proliferation rate of
pReceiver-Lv103-Expression group was significantly lower than the
others (Fig. 11). After
transfection for 24, 48, 72 and 96 h, the number of cells in
pReceiver-Lv103-Expression group was always significantly lower
than the other groups (P<0.01). At each time-point, the
difference between PBS group and NC group was no statistically
significant (P>0.05).
 | Table IIResults of Transwell migration assay
after upregulation of NDRG1 expression (mean ± SD). |
Table II
Results of Transwell migration assay
after upregulation of NDRG1 expression (mean ± SD).
| Group | Fields counted | Invasion cells per
field | t | P-value |
|---|
|
pReceiver-Lv103-Expression | 15 | 19±7 | 2.15 | <0.05 |
| PBS | 15 | 71±6 | | |
The methylation status of NDRG1 promoter
in prostate cells
There are four CGIs which may occur aberrant
methylation in the promoter region of human NDRG1 from −3000 bp to
+70 bp. The present study analyzed the first CGI from −213 bp to
+78 bp, containing 21 CpG sites (Table III). With BSP primer for NDRG1
promoter, using PCR amplified bisulfite modified DNA. The molecular
weight of PCR products were close to 291 bp. The rate of
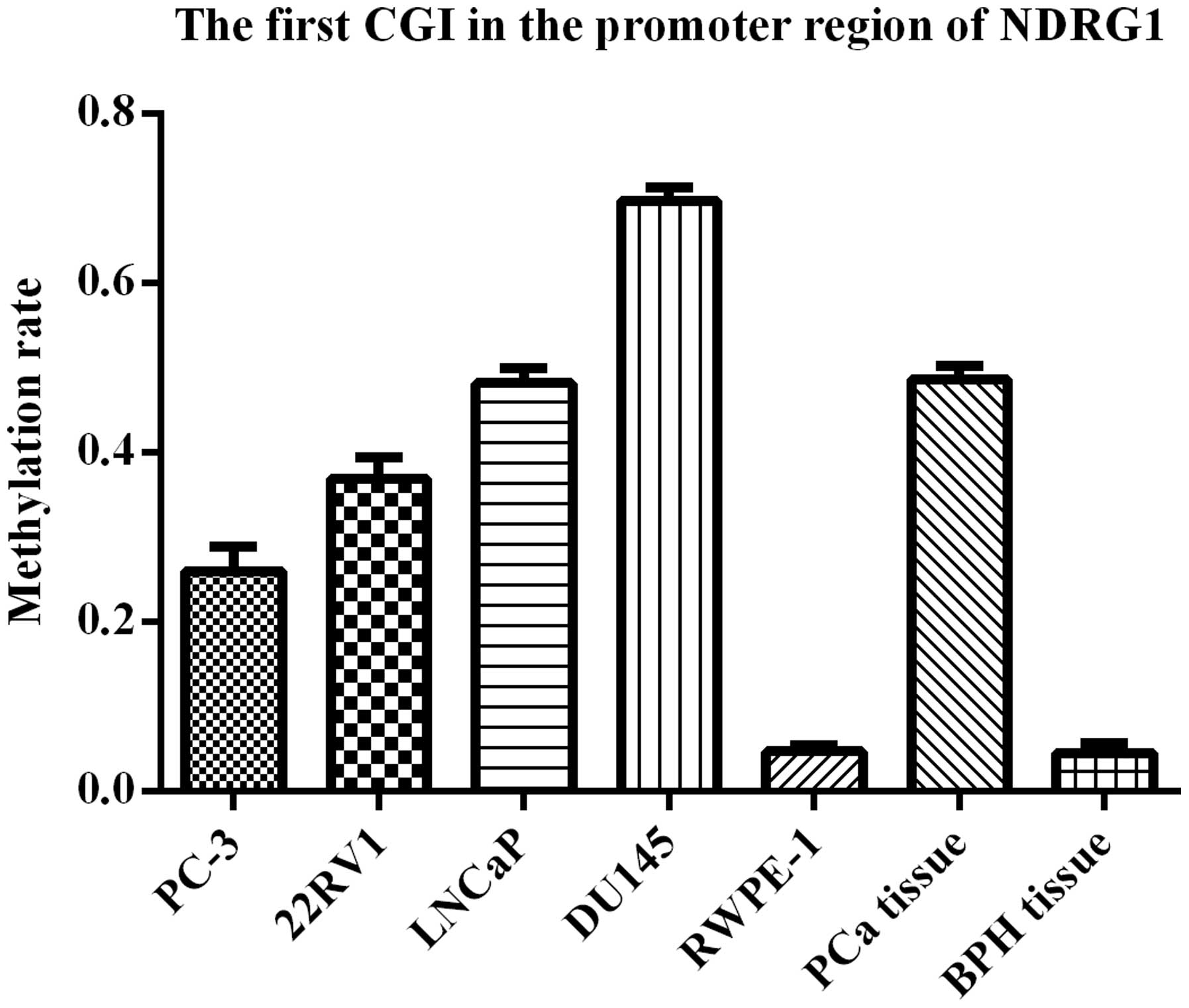
methylation of the first CGI was analyzed in different samples by
BIQ Analyzer (Fig. 12). The
results were 24.8% (PC-3 cells), 36.2% (22RV1 cells), 48.6% (LNCap
cells), 69.5% (DU145 cells), 4.8% (RWPE-1 cells), 48.6% (PCa
tissues) and 4.3% (BPH tissues). The rate of methylation in PCa
cells or tissues was significantly higher than that in normal
prostate cells or tissues (P<0.01). DU145 cells which had the
highest rate of methylation were chosen for the following
experiments. In PCa, the CpG site which was most likely to occur
aberrant methylation was the seventeenth CpG site (−127 bp) from
the 5′ end. Approximately 50% of methylation occurred at this
site.
 | Table IIIBSP primer sequences (designed by
methyl primer). |
Table III
BSP primer sequences (designed by
methyl primer).
| NDRG1 | Primer sequence
(5′-3′) |
|---|
| Methylated | F:
TTTAGTGGGTAAGGTTTAGTGAGTGT
R: CCTCAAAATTTCTTCTAAAAATCTC |
NDRG1 is demethylated by 5-Aza-CdR in
DU145 cells
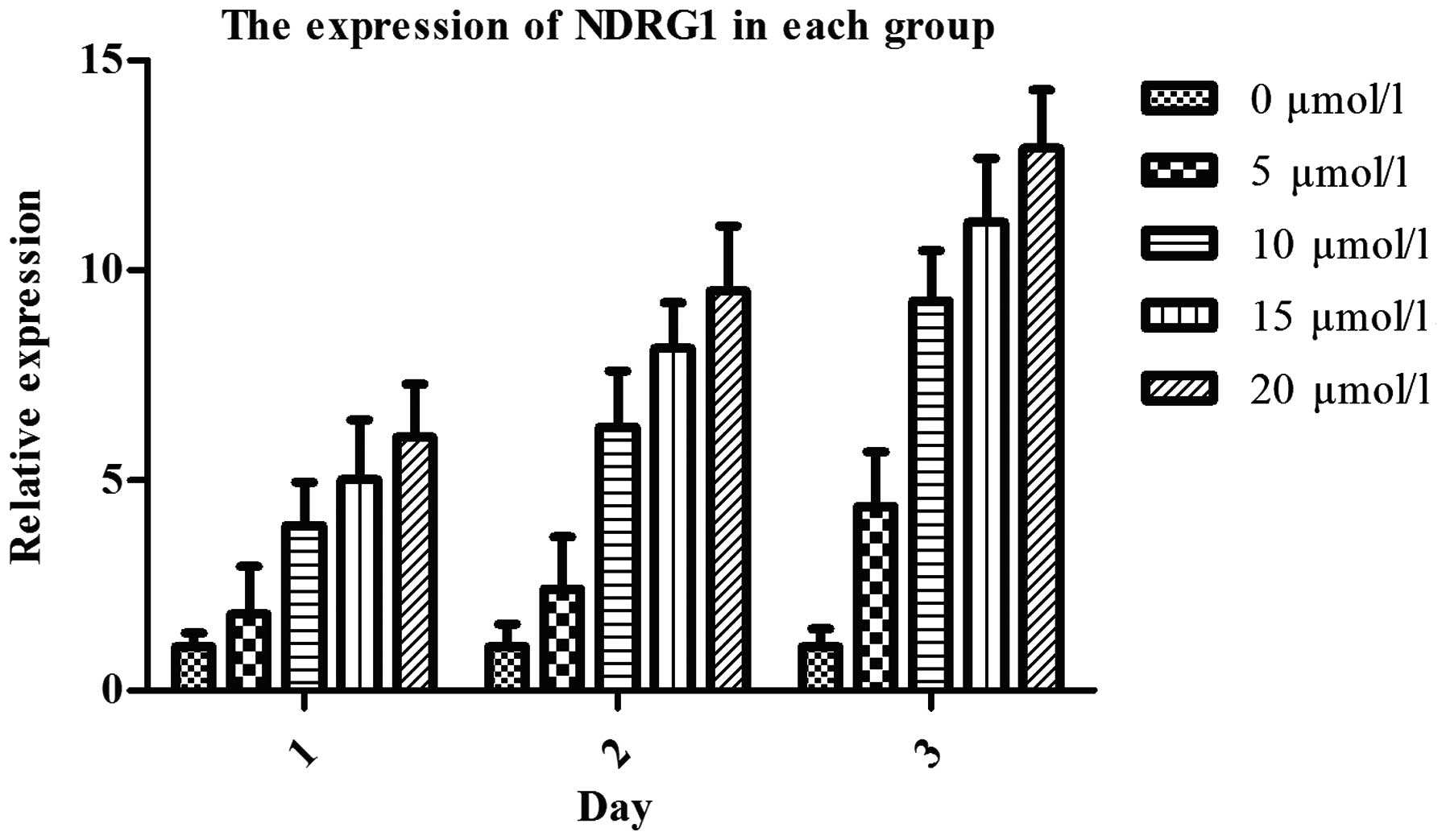
RT-qPCR showed that after treatment with 5-Aza-CdR
for 24, 48 and 72 h, NDRG1 mRNA expression in DU145 cells was
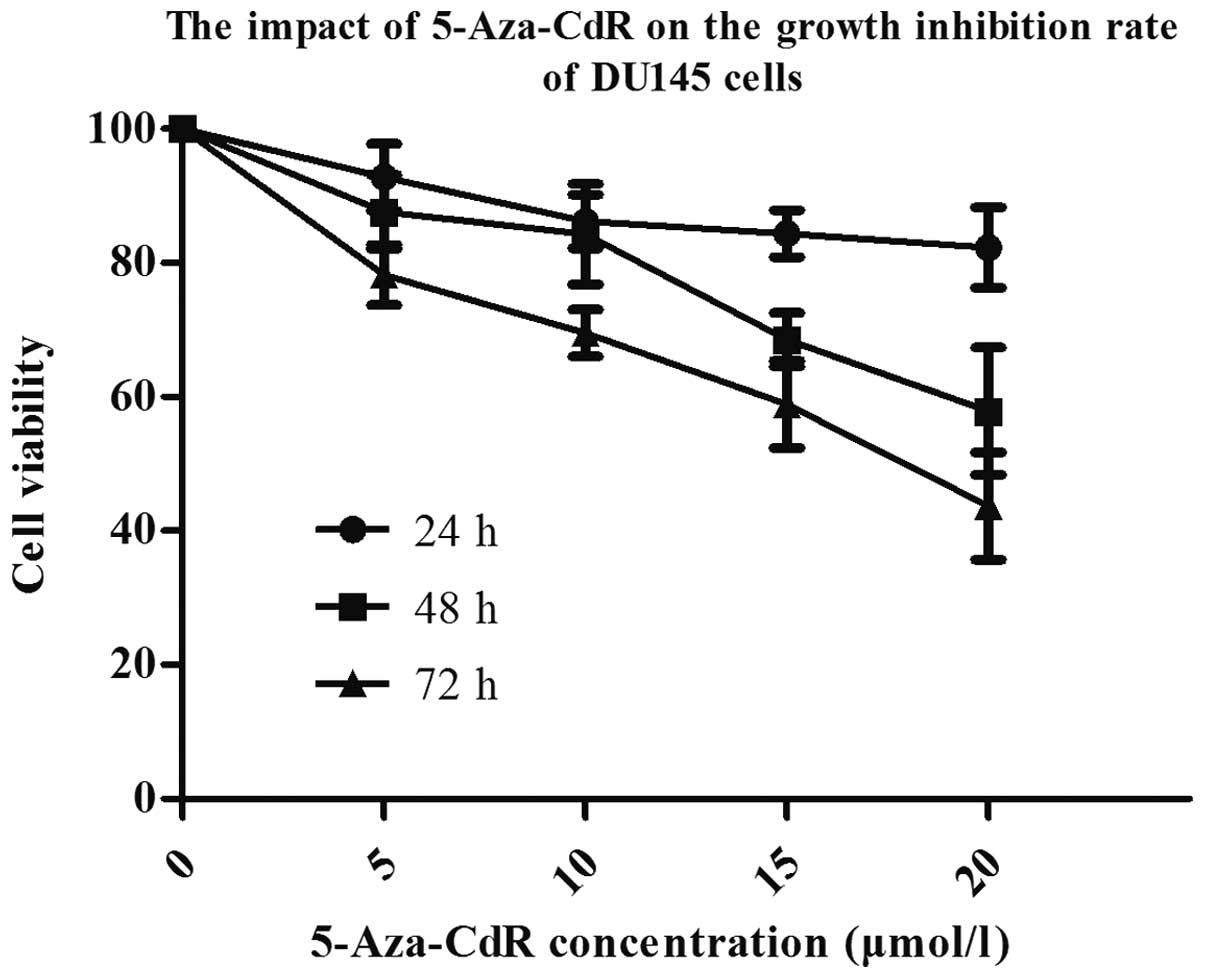
significantly upregulated (P<0.05), dose-dependently (Fig. 13). MTT assay showed that after
treatment with different concentrations of 5-Aza-CdR, the density
of cells was significantly reduced, and cells appeared shrunken and
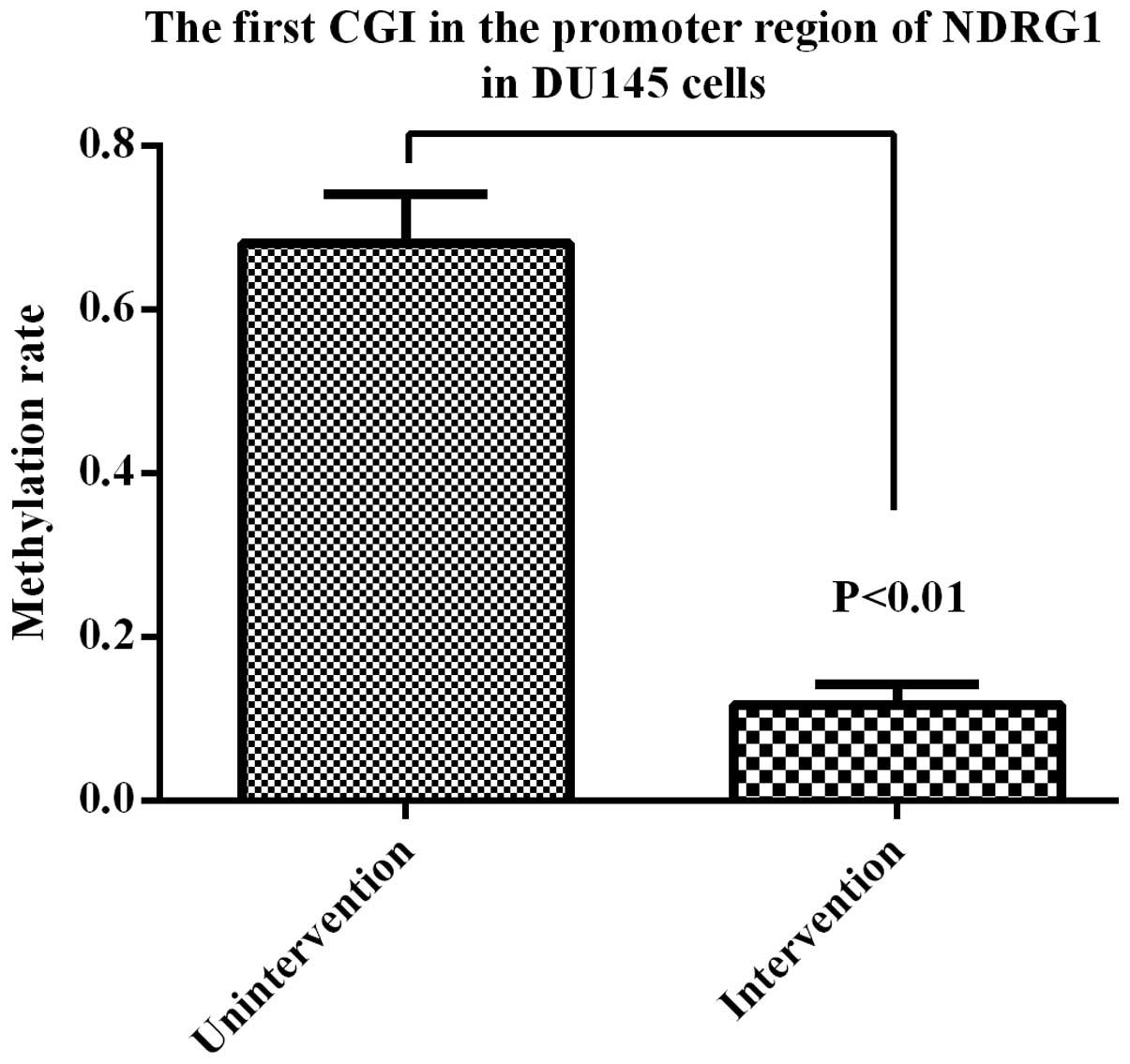
non-viable. The inhibiting effect was dose-dependent (Fig. 14). After 72 h, the methylation
rate of the first CGI located in the promoter region of NDRG1 was
11.4%, which was significant at <69.5% (Fig. 15). The results showed that
5-Aza-CdR was able to reverse the methylation status of NDRG1
promoter in DU145 cells.
Discussion
In the present study, we studied NDRG1 expression in
prostate tissues and cells. NDRG1 protein expressed in both the
cytoplasm and nucleus but mainly in the cytoplasm. NDRG1 expression
in PC-3 and LNCap cells was lower than that in RWPE-1 cells. With
these results, we confirmed that NDRG1 was associated with PCa.
For further research, we designed two types of
plasmid vectors, changing the expression level of NDRG1 in PC-3
cells. After downregulation of NDRG1 expression, the cell invasive
ability and proliferation rate in the experimental group was
significantly increased. The rate of early apoptosis significantly
decreased. After upregulation of NDRG1 expression, the cell
invasive ability and proliferation rate in the experimental group
was significantly decreased. The rate of early apoptosis
significantly increased. We can conclude that NDRG1 expression in
PCa is lower than that in normal prostate, and NDRG1 may have an
inhibitory effect on the progression and metastasis of PCa. NDRG1
is the downstream gene of p53 and may cooperate with p53, promoting
cell differentiation, maturation and apoptosis (18). Upregulation of NDRG1 reduced the
expression of vascular endothelial growth factor (VEGF) and
interleukin-8, inhibiting tumor angiogenesis and metastasis
(19). Moreover, it can change the
adhesion between tumor cells, inhibiting the extracellular matrix
degradation. NDRG1 expression was negatively correlated with the
activity of matrix metalloproteinase MMP-9, inhibiting the
extracellular matrix degradation and tumor metastasis (17).
Epigenetic (20)
changes are early molecular events occurring in the transcription
of tumor cells. DNA methylation is a common epigenetic change,
mostly occurring at 5′-CpG-3′ dinucleotide. The density of CpG
sequence is high in the genome regions named CGIs (21). CGIs usually locate in the gene
promoter region and may extend to the exon. The methylation of CGIs
was able to silence gene expression. In the human genome, the CpG
sites are usually methylated out of CGIs but stay unmethylated in
CGIs. This methylation pattern can be stably maintained during cell
division (22). Therefore, the
methylation of CGIs plays an important role in tumor cell
proliferation, DNA repair, cell cycle regulation and apoptosis
(23–25). DNA methylation may prevent the
combination of specific transcription factors and promoter
recognition, inhibiting gene expression. Kalaydjieva et al
(26) found a CGI located at the
5′ end of NDRG1 and considered that NDRG1 expression may be
regulated by DNA methylation. Guan et al (27) reported that NDRG1 expression was
regulated through multiple mechanisms, including DNA methylation
(27). Han et al (4) found the high methylation rate of
NDRG1 promoter in DNA promoter region was related to the occurrence
and progression of breast cancer. Chang et al (9) reported that the methylation of DNA
promoter may be related to the occurrence and progression of
gastric cancer. Gravina et al (28) found that increased levels of DNA
methyltransferases (DNMT) are associated with the tumorigenic
capacity of PCa cells. Gillio-Tos et al (29) reported that the DNMT3b may play an
important role in PCa progression. Therefore, we speculated that
the low expression of NDRG1 in PCa may be related to the
methylation status of CGIs in gene promoter region. Then, we
analyzed the methylation rate of the first CGI located in the
promoter region of NDRG1 in prostate cells and tissues. The results
showed that the methylation rate in PCa was higher than that in
BPH, which could explain the abnormal expression of NDRG1. Our
research suggested that the high methylation rate of NDRG1 promoted
the occurrence and progression of PCa.
Next, we treated DU145 cells with 5-Aza-CdR,
reducing the methylation levels of NDRG1 and observed changes in
cell biological behavior. The expression level and methylation rate
of NDRG1 was significantly increased. The survival rate of DU145
cells was significantly decreased. Thus, we conclude that DNA
methylation silenced NDRG1 expression.
In conclusion, the key finding of this study is that
the low expression of NDRG1 can increase the proliferation and
invasion of PCa cells. Methylation of CGIs in the NDRG1 promoter
may be one of the reasons which lead to the low expression of
NDRG1. This result indicates that the methylation of NDRG1 promoter
plays an essential role in regulating the proliferation and
invasion of PCa cells. NDRG1 may serve as a novel therapeutic
target for the treatment of PCa.
Acknowledgements
The present study was supported by grants from the
Key Project of Tianjin Municipal Science and Technology Commission
(no. 15JCZDJC35900), the Tianjin Municipal Education Commission
(no. 20140122) and the Key Project of Tianjin Health Bureau (no.
13KG141).
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Malvezzi M, Bertuccio P, Rosso T, Rota M,
Levi F, La Vecchia C and Negri E: European cancer mortality
predictions for the year 2015: Does lung cancer have the highest
death rate in EU women? Ann Oncol. 26:779–786. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Okuda T and Kondoh H: Identification of
new genes ndr2 and ndr3 which are related to Ndr1/RTP/Drg1 but show
distinct tissue specificity and response to N-myc. Biochem Biophys
Res Commun. 266:208–215. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Han LL, Hou L, Zhou MJ, Ma ZL, Lin DL, Wu
L and Ge YL: Aberrant NDRG1 methylation associated with its
decreased expression and clinicopathological significance in breast
cancer. J Biomed Sci. 20:522013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kovacevic Z, Fu D and Richardson DR: The
iron-regulated metastasis suppressor, Ndrg-1: Identification of
novel molecular targets. Biochim Biophys Acta. 1783:1981–1992.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Angst E, Dawson DW, Nguyen A, Park J, Go
VL, Reber HA, Hines OJ and Eibl G: Epigenetic regulation affects
N-myc downstream-regulated gene 1 expression indirectly in
pancreatic cancer cells. Pancreas. 39:675–679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim-Fuchs C, Winterhalder S, Winter A,
Malinka T, Born D, Schäfer S, Stroka D, Gloor B, Candinas D and
Angst E: The silencing of N-myc downstream-regulated gene-1 in an
orthotopic pancreatic cancer model leads to more aggressive tumor
growth and metastases. Dig Surg. 31:135–142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Youns M and Fathy GM: Upregulation of
extrinsic apoptotic pathway in curcumin-mediated antiproliferative
effect on human pancreatic carcinogenesis. J Cell Biochem.
114:2654–2665. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chang X, Zhang S, Ma J, Li Z, Zhi Y, Chen
J, Lu Y and Dai D: Association of NDRG1 gene promoter methylation
with reduced NDRG1 expression in gastric cancer cells and tissue
specimens. Cell Biochem Biophys. 66:93–101. 2013. View Article : Google Scholar
|
|
10
|
Li Q and Chen H: Transcriptional silencing
of N-Myc downstream-regulated gene 1 (NDRG1) in metastatic colon
cancer cell line SW620. Clin Exp Metastasis. 28:127–135. 2011.
View Article : Google Scholar
|
|
11
|
Geng XX, Quan LN, Ma R and Tang LP:
Effects of As2O3 and all-trans retinoic acid
on the growth of HeLa cell line and their relation with gene NDRG1.
Zhonghua Zhong Liu Za Zhi. 33:8–12. 2011.(In Chinese). PubMed/NCBI
|
|
12
|
Hosoya N, Sakumoto M, Nakamura Y, Narisawa
T, Bilim V, Motoyama T, Tomita Y and Kondo T: Proteomics identified
nuclear N-myc downstream-regulated gene 1 as a prognostic tissue
biomarker candidate in renal cell carcinoma. Biochim Biophys Acta.
1834:2630–2639. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Dixon KM, Lui GY, Kovacevic Z, Zhang D,
Yao M, Chen Z, Dong Q, Assinder SJ and Richardson DR: Dp44mT
targets the AKT, TGF-β and ERK pathways via the metastasis
suppressor NDRG1 in normal prostate epithelial cells and prostate
cancer cells. Br J Cancer. 108:409–419. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu W, Iiizumi-Gairani M, Okuda H,
Kobayashi A, Watabe M, Pai SK, Pandey PR, Xing F, Fukuda K, Modur
V, et al: KAI1 gene is engaged in NDRG1 gene-mediated metastasis
suppression through the ATF3-NFkappaB complex in human prostate
cancer. J Biol Chem. 286:18949–18959. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ghalayini MK, Dong Q, Richardson DR and
Assinder SJ: Proteolytic cleavage and truncation of NDRG1 in human
prostate cancer cells, but not normal prostate epithelial cells.
Biosci Rep. 33:332013. View Article : Google Scholar
|
|
16
|
Symes AJ, Eilertsen M, Millar M, Nariculam
J, Freeman A, Notara M, Feneley MR, Patel HR, Masters JR and Ahmed
A: Quantitative analysis of BTF3, HINT1, NDRG1 and ODC1 protein
over-expression in human prostate cancer tissue. PLoS One.
8:e842952013. View Article : Google Scholar
|
|
17
|
Lee JC, Chung LC, Chen YJ, Feng TH and
Juang HH: N-myc downstream-regulated gene 1 downregulates cell
proliferation, invasiveness, and tumorigenesis in human oral
squamous cell carcinoma. Cancer Lett. 355:242–252. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kovacevic Z, Sivagurunathan S, Mangs H,
Chikhani S, Zhang D and Richardson DR: The metastasis suppressor,
N-myc downstream regulated gene 1 (NDRG1), upregulates p21 via
p53-independent mechanisms. Carcinogenesis. 32:732–740. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hosoi F, Izumi H, Kawahara A, Murakami Y,
Kinoshita H, Kage M, Nishio K, Kohno K, Kuwano M and Ono M: N-myc
downstream regulated gene 1/Cap43 suppresses tumor growth and
angiogenesis of pancreatic cancer through attenuation of inhibitor
of kappaB kinase beta expression. Cancer Res. 69:4983–4991. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Holliday R: The inheritance of epigenetic
defects. Science. 238:163–170. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ushijima T: Detection and interpretation
of altered methylation patterns in cancer cells. Nat Rev Cancer.
5:223–231. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cottrell SE: Molecular diagnostic
applications of DNA methylation technology. Clin Biochem.
37:595–604. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hypermethylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
24
|
Yang B, Guo M, Herman JG and Clark DP:
Aberrant promoter methylation profiles of tumor suppressor genes in
hepatocellular carcinoma. Am J Pathol. 163:1101–1107. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Edamoto Y, Hara A, Biernat W, Terracciano
L, Cathomas G, Riehle HM, Matsuda M, Fujii H, Scoazec JY and Ohgaki
H: Alterations of RB1, p53 and Wnt pathways in hepatocellular
carcinomas associated with hepatitis C, hepatitis B and alcoholic
liver cirrhosis. Int J Cancer. 106:334–341. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kalaydjieva L, Gresham D, Gooding R,
Heather L, Baas F, de Jonge R, Blechschmidt K, Angelicheva D,
Chandler D, Worsley P, et al: N-myc downstream-regulated gene 1 is
mutated in hereditary motor and sensory neuropathy-Lom. Am J Hum
Genet. 67:47–58. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guan RJ, Ford HL, Fu Y, Li Y, Shaw LM and
Pardee AB: Drg-1 as a differentiation-related, putative metastatic
suppressor gene in human colon cancer. Cancer Res. 60:749–755.
2000.PubMed/NCBI
|
|
28
|
Gravina GL, Ranieri G, Muzi P, Marampon F,
Mancini A, Di Pasquale B, Di Clemente L, Dolo V, D'Alessandro AM
and Festuccia C: Increased levels of DNA methyltransferases are
associated with the tumorigenic capacity of prostate cancer cells.
Oncol Rep. 29:1189–1195. 2013.
|
|
29
|
Gillio-Tos A, Fiano V, Zugna D, Vizzini L,
Pearce N, Delsedime L, Merletti F and Richiardi L: DNA
methyltransferase 3b (DNMT3b), tumor tissue DNA methylation,
Gleason score, and prostate cancer mortality: Investigating causal
relationships. Cancer Causes Control. 23:1549–1555. 2012.
View Article : Google Scholar : PubMed/NCBI
|