Introduction
Lung adenocarcinoma is a highly malignant disease
with aggressive clinical behavior. In 2013, lung cancer was
predicted to account for 26% of all female cancer deaths and 28% of
all male cancer deaths (1).
Patients diagnosed with lung adenocarcinoma usually survive only a
few months, even with the assistance of individualized treatment
strategies, including surgery combined with radiation and
chemotherapy. The high mortality is mainly accounted for by drug
resistance during chemotherapy treatment. Thus, to achieve better
treatment outcome in lung adenocarcinoma patients, it is of the
utmost importance to explore the mechanisms of drug resistance and
to identify novel methods to overcome drug resistance.
Paclitaxel and cisplatin, as first-line treatment
for patients suffering from lung adenocarcinoma, improves survival
with manageable toxicity (2,3).
However, tumor cells acquire resistance to paclitaxel/cisplatin,
which causes eventual failure to prolong the survival of lung
adenocarcinoma patients. There is abundant evidence that
chemoresistance is associated with the acquisition of EMT
phenotypic of cancer cells (4).
The serial process of EMT involves epithelial cells switching to
mesenchymal phenotypic cells. The epithelial cell-cell junctions
become disassembled, and the expression of epithelial markers,
including E-cadherin and β-catenin, is reduced, while the motility
and invasion potential is increased, and mesen-chymal properties,
such as the high expression of molecular markers vimentin, Snail,
Slug, Twist, Zinc-finger E-boxbinding homeobox1 (ZEB1) and ZEB2,
become apparent (5–8). Additionally, EMT is often associated
with the loss of expression of the phosphatase and tensin homolog
deleted on chromosome ten (PTEN), an inhibitor of the
phosphatidylinositol 3-kinase/Akt pathway (9). Mutation or reduced expression of PTEN
is frequently observed in lung cancer, though the mechanism of PTEN
loss is not well understood (10,11).
MicroRNAs (miRNAs) have been see as key
post-transcriptional level regulators of gene expression (12). Involvement of miRNAs has been
demonstrated in tumorigenesis, metastasis, embryonic development,
metabolism and other pathological and normal physiological
(13,14). PTEN has been shown to be a tumor
suppressor gene in human hepatocellular carcinoma, and miRNA-21 has
been shown to regulate the expression of PTEN (14). MicroRNA-492 expression promotes the
progression of hepatic cancer by targeting PTEN (15). The PTEN gene is targeted by
miRNA-221 and miRNA-222 to regulate gastric carcinoma cell
proliferation and radioresistance by targeting (16). High-frequency miRNA dysfunction is
also associated with lung adenocarcinoma development and
progression (17). Therefore, it
is of value to determine whether dysregulation of the
miRNA-regulatory network is responsible chemoresistance in lung
adenocarcinoma.
The dysregulation of miR-181a was identified in the
present study on lung adenocarcinoma. We determined that miR-181a
responds to chemotherapy by changing the levels of its target PTEN.
The identification of miR-181a/PTEN as a novel regulatory circuit
that mediates EMT and chemoresistance in lung adenocarcinoma
provides a new molecular mechanism, which could be targeted as a
novel therapy for chemoresistance in lung adenocarcinoma.
Materials and methods
Cell culture, reagents and
antibodies
The cell line A549, which is a human lung
adenocarcinoma cell line, and its drug-resistant strain were
cultured for use in the present study. The culture conditions were
37°C in 5% CO2 in RPMI-1640 medium (Gibco, Gaithersburg,
MD, USA) supplemented with 10% fetal bovine serum (FBS; Gibco). The
cells were sub-cultured every 3–4 days. Cisplatin was purchased
from Qilu Pharmaceutical Co., Ltd. (Jinan, China). Paclitaxel was
purchased from Beijing SL Pharmaceutical Co., Ltd. (Beijing,
China). MTT [3-(4,5-dimethythiazol-2-yl)-2,5-diphenyl tetrazolium
bromide] was from Sigma (St. Louis, MO, USA). Primary antibodies
against E-cadherin (cat. AF0131, dilution, 1:500–1:3,000),
β-catenin (cat. AF0122, dilution, 1:500–1:3,000), MMP-2 (cat.
AF0577, dilution, 1:500–1:2,000), MMP-9 (cat. AF0577, dilution,
1:500–1:2,000), Snail (cat. AF6032, dilution, 1:500–1:2,000), Slug
(cat. AF4002, dilution, 1:500–1:2,000), vimentin (cat. AF7013,
dilution, 1:500–1:2000), ZEB1 (cat. DF7414, dilution,
1:500–1:2,000), PTEN (cat. AF6351, dilution, 1:500–1:2,000) and
β-actin (cat. AF0115, dilution, 1:500–1:3,000) were from Affinity
Biosciences, Inc. (Cincinnati, OH, USA). Secondary antibodies, goat
anti-rabbit IgG (H+L)-HRP (cat. S0001, dilution, 1:5,000–1:10,000),
were also obtained from Affinity Biosciences.
Cell proliferation assays
Cells (A549, A549/PTX and A549/DDP) were seeded in
96-well plate at 7×103 cells/well and cultured
overnight. The next day, cells were treated with different
concentrations of PTX/DDP for up to 72 h. MTT assays were conducted
as previously described (18). The
IC50 (drug concentration causing 50% inhibition of
viability) was calculated for each cell line, and the resistance
index (RI) was obtained by dividing the IC50 value of
the resistant cell lines by the IC50 value of the
non-resistant cell lines.
Woundhealingassays
Cells (A549, A549/PTX and A549/DDP) were seeded in
6-well plates and cultured to 90–95% confluency. Scratch wounds
were then generated on the surface of the plates using a pipette
tip. Photographic images were taken immediately after the scratch
wound and also 24 h later.
Transwell migration and invasion
assays
A 24-well Transwell chamber (Corning Inc., Corning,
NY, USA) with gelatin-coated polycarbonate membrane filters was
used to assess the migration ability of A549, A549/PTX and A549/DDP
cells. The invasive capacity of A549, A549/PTX and A549/DDP cells
was assessed using Transwell inserts with Matrigel (BD
Biosciences). After incubation for 24 h, the cells in the upper
surfaces of the Transwell chambers were removed with cotton swabs,
and the migrated and invaded cells were fixed with 4%
paraformaldehyde, and then stained with Giemsa solution. The
stained cells were photographed and counted under a light
microscope in four randomly selected fields.
RNA extraction and mRNA expression
analyzed by reverse transcription-PCR analysis
TRIzol (Invitrogen, Grand Island, NY, USA) was used
to isolate the total RNA from A549, A549/PTX and A549/DDP cells.
The obtained RNA was purified with RNeasy Mini kit and
RNase-free DNase Set (Qiagen) according to the protocols suggested
by the manufacturer. Table I shows
the primers used in the PCR reactions. GAPDH expression was used as
an internal control. The kit manufacturer's protocol was used for
RT-PCR amplifications.
 | Table IPrimer sequences and amplification
lengths of qRT-PCR products. |
Table I
Primer sequences and amplification
lengths of qRT-PCR products.
| Gene | Primer sequence
(5′-3′) | Size of product
(bp) |
|---|
| E-cadherin | F:
CATTTCCCAACTCCTCTCCTGGC
R: ATGGGCCTTTTTCATTTTCTGGG | 90 |
| β-catenin | F:
CACAAGCAGAGTGCTGAAGGTG
R: GATTCCTGAGAGTCCAAAGACAG | 146 |
| Vimentin | F:
AGATGGCCCTTGACATTGAG
R: TGGAAGAGGCAGAGAAATTC | 80 |
| Fibronectin | F:
CCCACCGTCTCAACATGCTTAG
R: CTCGGCTTCCTCCATAACAAGTAC | 264 |
| MMP-2 | F:
GATAACCTGGATGCCGTCGTG
R: CTTCACGCTCTTCAGACTTTGGTTC | 105 |
| MMP-9 | F:
CGGAGTGAGTTGAACCAG
R: GTCCCAGTGGGGATTTAC | 118 |
| Snail | F:
CCAGCTCTCTGAGGCCAAGGATC
R: TGGCTTCGGATGTGCATCTTGAG | 108 |
| Slug | F:
CCCTGAAGATGCATATTCGGAC
R: CTTCTCCCCCGTTGTAGTTCTA | 116 |
| Twist | F:
TGCGGAAGATCATCCCCA
R: TCCATCCTCCAGACCGAGAA | 187 |
| ZEB1 | F:
GCACAACCAAGTGCAGAAGA
R: GCCTGGTTCAGGAGAAGATG | 141 |
| GAPDH | F:
AAGGTGAAGGTCGGAGTCAAC
R: CTTGATTTTGGAGGGATCTCG | 252 |
miRNA microarray and quantitative miRNA
analysis
Cells (A549/PTX or A549/DDP) were cultured for one
week without treatment before the experiments. Total RNA from
drug-resistant and drug-sensitive cells was isolated with miRNeasy
Mini kit (Qiagen; cat. 217004) following the manufacturer's
instructions and RNA concentration was determined by NanoVue™ Plus
(Thermo Fisher Scientific, Inc., Waltham, MA, USA). For miRNA
analysis, the Human Cancer PathwayFinder miRNA PCR Array (Qiagen;
cat. MIHS-102Z) allows the simultaneous detection of 84 miRNAs
previously identified in human cancers. The fold change for each
miRNA was calculated by plugging the Crossing point (Cp) values
into the manufacturer's web-based software, and so microarray
images were acquired. For each array, a multiple controls were
used; i.e. RT negative and positive controls, and genomic
DNA contamination controls and endogenous controls. miScript PCR
primer (Qiagen) was used for validation of the miRNA samples. The
expression of hsa-miR181a (Qiagen; cat. MS00008827) in
drug-resistant and drug-sensitive cells was performed with a
similar approach. The cells were transfected as described above. U6
was used as endogenous control to normalize Ct values obtained for
each gene. The changes in the expression were calculated using the
2−ΔΔCt method.
Protein extraction and western
blotting
Cells were plated at a density of 4×105
cells/well in 6-well culture plates (Corning Life Sciences). RIPA
buffer (1X PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS
and protease inhibitor cocktail) was used to harvested and lyse
cells. A bicinchoninic acid (BCA) assay (Beyotime Institute of
Biotechnology, Beijing, China) was used to measure protein
concentrations. The resulting proteins (40 μg) were separated with
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
(SDS-PAGE). Proteins were then transferred onto polyvinylidene
difluoride (PVDF) membranes. The membranes were blocked with 5%
skim milk in TPBS, and probed with primary antibodies overnight at
4°C. HRP-conjugated secondary antibodies were incubated with the
membranes after they were washed. Chemiluminescent ECL reagent
(Millipore, Millipore, MA, USA) was used for visualization.
Finally, gel imaging equipment (Bio-Rad Laboratories, Hercules, CA,
USA) was used to image the membranes. β-actin was the loading
control. In fact, antibodies were dilution as follows: E-cadherin
(1:800), β-catenin (1:800), MMP-2 (1:1,000), MMP-9 (1:1,000), Snail
(1:1,000), Slug (1:1,000), vimentin (1:1,000), ZEB1 (1:1,000), PTEN
(1:1,000), β-actin (1:3,000) and goat anti-rabbit IgG (H+L)-HRP
(1:5,000).
Cell transfection
Six-well plates were used for cell seeding and cells
were transfected with miR-181a mimic, miR-181a inhibitor or their
negative controls. These miRNAs were synthesized by Shanghai
GenePharma Co. (Shanghai, China) (Table II) and cells were transfected with
the Lipofectamine 2000 (Invitrogen) using the manufacturer's
protocol. After the indicated incubation period, the cells were
subjected to further analysis by functional assays as
indicated.
 | Table IIThe sequence of the miR-181a mimic
and inhibitor and their negative controls. |
Table II
The sequence of the miR-181a mimic
and inhibitor and their negative controls.
| miRNA | | Sequence |
|---|
| hsa-miR-181a
mimic | 5′-3′ Sense |
AACAUUCAACGCUGUCGGUGAGU |
| Antisense |
UCACCGACAGCGUUGAAUGUUUU |
| hsa-miR negative
control | 5′-3′ Sense |
UUCUCCGAACGUGUCACGUTT |
| Antisense |
ACGUGACACGUUCGGAGAATT |
| hsa-miR-181a
inhibitor | 5′-3′ Sense |
ACUCACCGACAGCGUUGAAUGUU |
| Antisense |
AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR inhibitor
negative control | 5′-3′ Sense |
CAGUACUUUUGUGUAGUACAA |
| Antisense |
UUGUACUACACAAAAGUACUG |
Luciferase reporter assay
PCR was used to amplify the full-length 3′-UTR
segments of PTEN mRNA that contained the miR-181a binding site.
These segments were then inserted into the Xba1-site of the
pGL3 vector (Promega, Madison, WI, USA) and the vector pGL3-PTEN
was produced. A site-directed mutagenesis kit (Stratagene, La
Jolla, CA, USA) was used to construct the pGL3-PTEN-mut reporter
construct with point mutations in the seed sequence. A total of
1×106 cells were cotransfected with 50 pmol of miR-181a
inhibitor (or control inhibitor), 1 μg of pGL3-PTEN (or
pGL3-PTEN-mut) plasmid, and 1 μg of a Renilla luciferase
expression construct pRL-TK (Promega) to assess the endogenous
inhibitory activity of miR-181a, using Lipofectamine 2000. Cells
were cultured for 36 h after the transfection, and then luciferase
activity was assessed using a dual luciferase assay system
(Promega). Results were normalized to Renilla luciferase
activity.
Statistical analysis
Results were analyzed using GraphPad Prism 4.0
(Graphpad Software, La Jolla, CA, USA) and the data are shown as
means ± SEM. A Student's t-test was used for comparisons between
different groups. P<0.05 was considered to be statistically
significant.
Results
Establishment of paclitaxel- or
cisplatin-resistant A549 cells
To develop A549 lung carcinoma cells that are
resistant to paclitaxel or cisplatin, A549 cells were exposed to
increasing concentrations of paclitaxel or cisplatin for more than
12 months. After each round, the surviving cells that reached
>70% confluency were passaged by trypsinization, and the
concentration of paclitaxel or cisplatin was increased. The
procedure was performed repeatedly until the cells showed
resistance to the inhibitory activities of 200 μg/ml paclitaxel and
1,000 μg/ml cisplatin. The resulting cells (A549/PTX and A549/DDP
cells) were cultured for an additional 3 months in RPMI-1640 medium
containing 200 μg/ml paclitaxel or 1,000 μg/ml cisplatin.
To assess the resistance properties of A549 cells
and their derivatives, we exposed the cells to increasing amounts
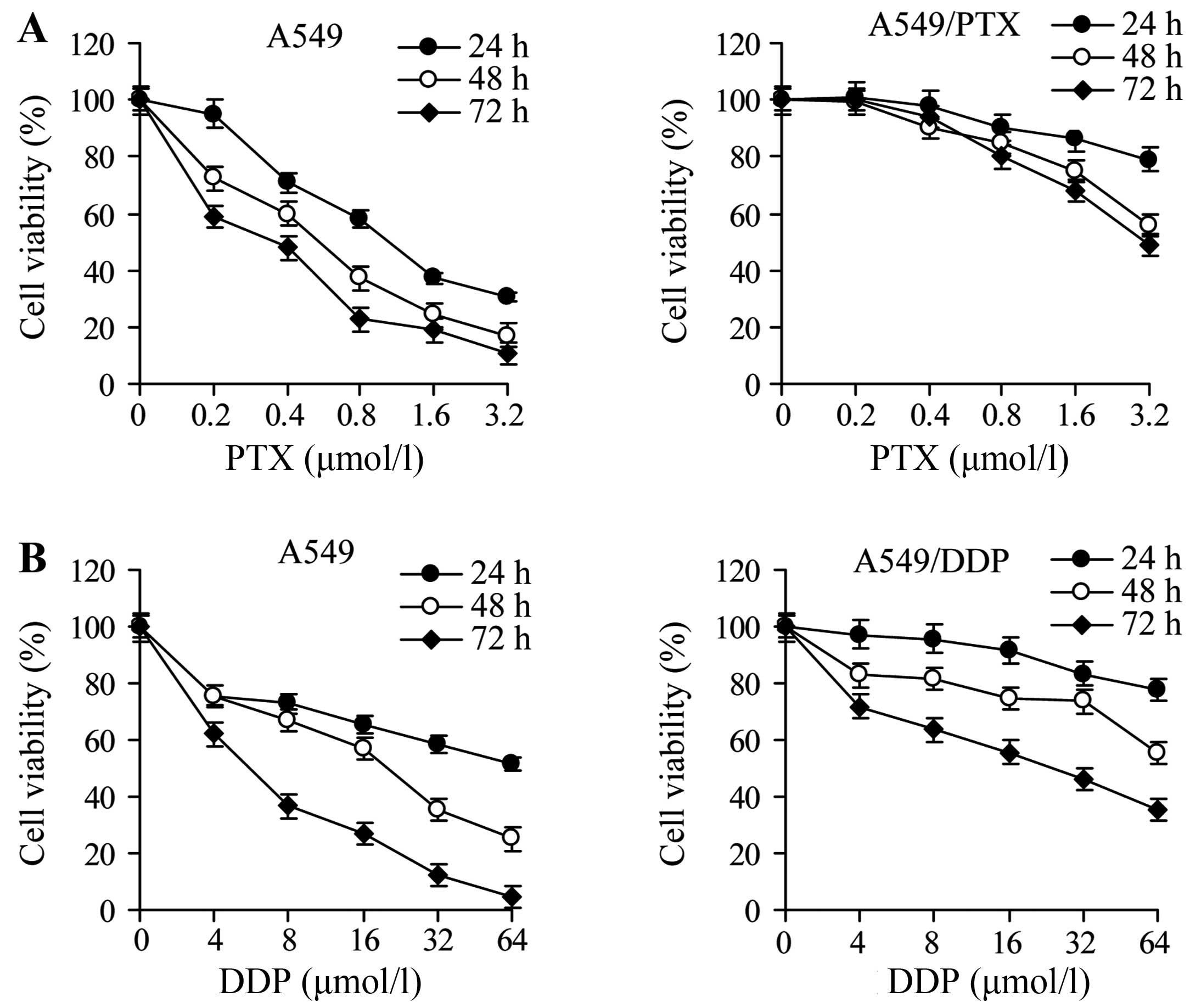
of paclitaxel or cisplatin for 24, 48 or 72 h. The viability was
decreased by drug exposure for all cell lines in a dose- and
time-dependent manner; however, A549/PTX and A549/DDP cells were
significantly more resistant than A549 cells (Fig. 1). The IC50 was greater
for A540/PTX and A549/DDP cells than for the parental A549 cells at
all times of drug treatment, and the RI ranged from 6 to 10.2 for
A549/PTX cells and from 3.3 to 9.4 for A549/DDP cells (Tables III and IV). These observations demonstrate that
A549/PTX and A549/DDP cells acquired chemoresistance.
 | Table IIIThe inhibitory 50% concentration
(IC50) and resistance index (RI) of A549 and A549/PTX
cells treated with paclitaxel for 24, 48 and 72 h. |
Table III
The inhibitory 50% concentration
(IC50) and resistance index (RI) of A549 and A549/PTX
cells treated with paclitaxel for 24, 48 and 72 h.
| IC50
(μmol·l−1) | |
|---|
|
| |
|---|
| Time (h) | A549 | A549/PTX | RI |
|---|
| 24 | 1.22±0.46 | 7.3±0.44 | 6 |
| 48 | 0.52±0.08 | 3.22±0.26 | 6.2 |
| 72 | 0.25±0.17 | 2.55±0.31 | 10.2 |
 | Table IVThe resistance index (RI) and
inhibitory 50% concentration (IC50) of A549 and A549/DDP
cells treated with cisplatin for 24 h, 48 h and 72 h. |
Table IV
The resistance index (RI) and
inhibitory 50% concentration (IC50) of A549 and A549/DDP
cells treated with cisplatin for 24 h, 48 h and 72 h.
| IC50
(μmol·l−1) | |
|---|
|
| |
|---|
| Time (h) | A549 | A549/DDP | RI |
|---|
| 24 | 75.83±0.53 | 250.24±0.94 | 3.3 |
| 48 | 17.62±0.12 | 66.96±0.59 | 3.8 |
| 72 | 5.84±0.32 | 54.90±0.63 | 9.4 |
A549/PTX and A549/DDP cells have
increased motility and invasion activity
The acquisition of metastatic properties by cancer
cells is characterized by increased migration and invasion
abilities. Wound healing, migration and invasion assays were
performed to assess the migratory potential of A549/PTX and
A549/DDP cells in relation to A549 cells. The results show that
significantly increased numbers of cells migrated across the
‘wound’ in a scratch assay, suggesting that A549/PTX and A549/DDP
cells acquired enhanced migration capacity (Fig. 2A). Transwell chamber assays were
performed to further compare the migration and invasion capacity of
A549/DDP and A549/PTX cells to A549 cells (Fig. 2B). A549/PTX had ~1.8-fold increased
migration and 2.9-fold increased invasion levels relative to A549
cells, and A549/DDP cells had ~1.5-fold increased migration and
2.2-fold increased invasion levels. Thus, A549/PTX and A549/DDP
cells have increased metastatic properties as compared with the
parental A549 cells.
A549/DDP and A549/PTX cells showed
molecular and morphological alterations that were consistent with
EMT
To determine whether EMT progression is detected in
A549/PTX and A549/DDP cells, we assessed the morphological changes
of the cells. Our results show that each of the resistant A549 cell
lines had marked morphologic changes compared with the parental
cell lines (Fig. 3A). Whereas A549
cells displayed a rounded shape and little formation of
pseudopodia, A549/PTX and A549/DDP cells displayed phenotypic
changes, including a loss of cell polarity and increased formation
of pseudopodia, leading to elongated, irregular fibroblastoid
appearance.
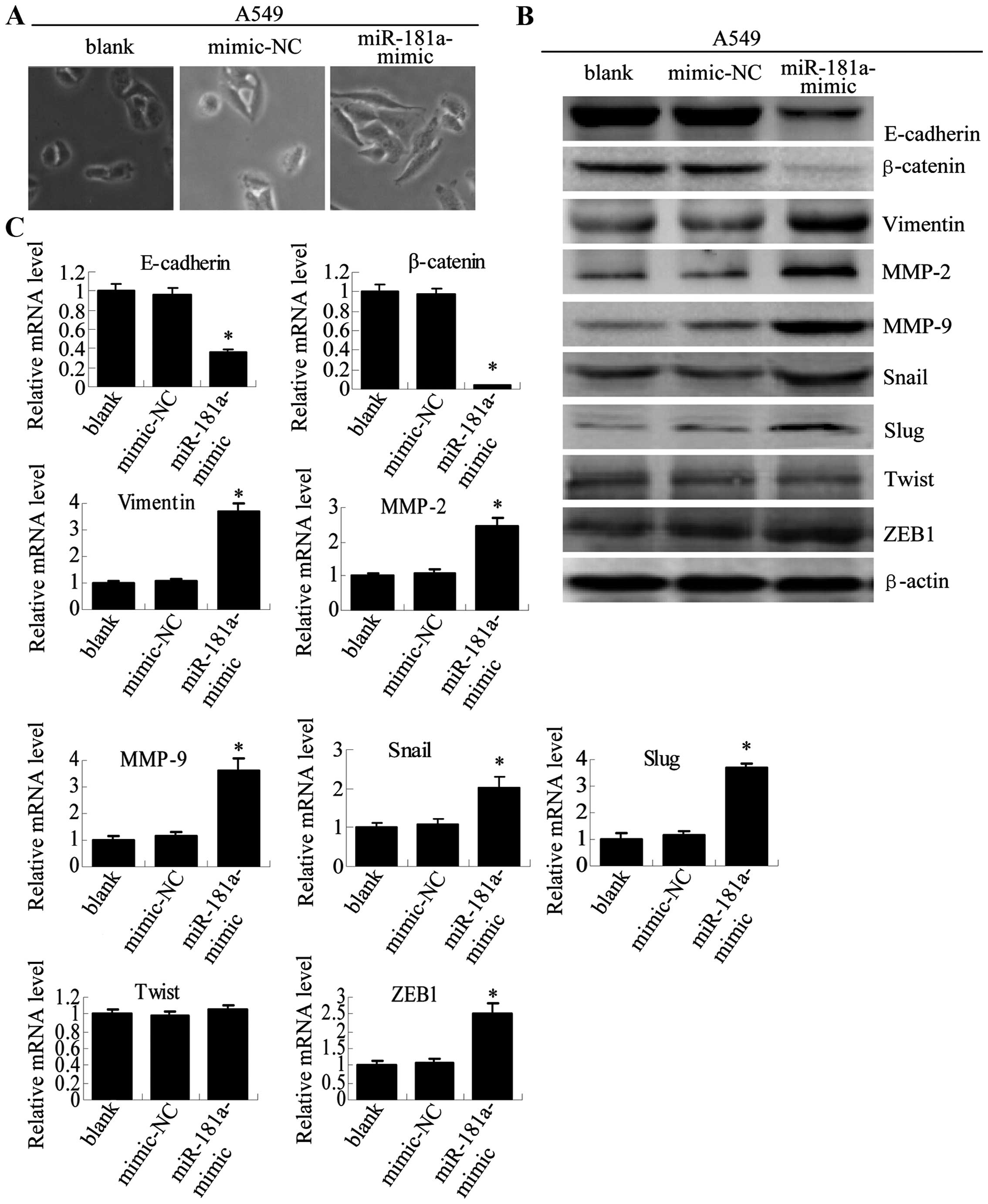
 | Figure 3A549/DDP and A549/PTX cells showed
molecular and morphological changes that were consistent with EMT.
(A) Microscopy at ×200 magnification was used to assess cell
morphology. The A549 cells (parental cells) had an epithelioid,
rounded cobblestone appearance and there was limited formation of
pseudopodia. A549/PTX and A549/DDP cells exhibited a spindle-shaped
morphology and an increased formation of pseudopodia, indicating a
loss of cell polarity. (B) E-cadherin, β-catenin, vimentin, MMP-2
and MMP-9 which are EMT-related proteins, were assessed in terms of
expression levels. EMT-related transcription factors (Snail, Slug,
Twist and ZEB1) were measured in A549/PTX and A549/DDP cells using
western blot analysis. (C) The expression changes were confirmed at
the mRNA level by qRT-PCR. Expression was standardized to the
expression of GAPDH and normalized to 1.0 in the parental cells
(compared with the parental A549 cells, means ± SEM, n=3,
*P<0.05). |
To further determine whether A549/PTX and A549/DDP
cells have specific molecular changes consistent with EMT, we
measured the expression of epithelial and mesenchymal phenotype
markers with western blotting (Fig.
3B) and RT-PCR analysis (Fig.
3C). Consistent with EMT progression in A549/PTX and A549/DDP
cells, the expression of epithelial adhesion molecules E-cadherin
and β-catenin were significantly reduced in A549/PTX and A549/DDP
cells. The expression of the mesenchymal markers (vimentin, MMP-2,
MMP-9, Snail, Slug and ZEB1) was elevated in A549/PTX and A549/DDP
cells, although the expression of Twist showed no significant
change. These results showed the expression levels of genes that
are known to play a critical role in EMT are modulated in lung
adenocarcinoma cells upon acquisition of either paclitaxel- or
cisplatin-resistance.
miR-181a is differentially expressed in
A549/PTX, A549/DDP, and A549 parental A549 cells
To identify miRNAs that are potentially involved in
the underlying mechanisms of drug-resistant cells and induction of
EMT-like properties, we used microarray analysis to assess the
differential expression of miRNA in A549/PTX and parental A549
cells. Ten miRNAs were significantly upregulated and 5 were
significantly down-regulated in A549/PTX cells relative to A549
cells (absolute log fold-change |logFC|>4; Table V). Among the differentially
expressed miRNAs, the miR-181 family (miR-181a, b and c) was
significantly upregulated, and among the miR-181 family members,
miR-181a was the most significantly dysregulated. qRT-PCR analysis
verified that miR-181a was upregulated 16-fold in A549/PTX compared
with A549 cells. Furthermore, miR-181a was also upregulated 12-fold
in A549/DDP compared to parental A549 cells (Fig. 4A). Therefore, we hypothesized that
miR-181a may represent a primary regulator in lung adenocarcinoma
cells.
 | Table VDifferentially expressed miRNAs in
A549/PTX compared with A549 cells (|LogFC|>4). |
Table V
Differentially expressed miRNAs in
A549/PTX compared with A549 cells (|LogFC|>4).
| miRNA | Up/down | Fold change
(log2) |
|---|
|
hsa-miR-135b-5p | Up | 16.2407 |
| hsa-miR-155-5p | Up | 15.7135 |
|
hsa-miR-181a-5p | Up | 8.0039 |
|
hsa-miR-301a-3p | Up | 7.9692 |
| hsa-miR-19a-3p | Up | 7.9104 |
| hsa-miR-205-5p | Up | 5.0039 |
|
hsa-miR-181c-5p | Up | 4.9968 |
|
hsa-miR-181b-5p | Up | 4.0564 |
| hsa-miR-23b-3p | Up | 4.0254 |
| hsa-miR-96-5p | Up | 4.006 |
| hsa-let-7g-5p | Down | −4.0231 |
| hsa-miR-128-5p | Down | −4.0284 |
| hsa-miR-191-5p | Down | −4.1711 |
|
hsa-miR-200c-3p | Down | −5.0792 |
| hsa-miR-25-3p | Down | −6.0224 |
miR-181a regulates metastatic properties
and EMT in human lung adenocarcinoma cells
To explore the potential role of miR-181a in
regulating metastasis and EMT in lung adenocarcinoma cells, we used
miR-181a mimic and inhibitor to modulate miR-181 expression
(Fig. 4B). Our results demonstrate
that miR-181a mimic increases the ability of A549 cells to migrate
and invade (Fig. 5A). Conversely,
miR-181a inhibitor decreases the migration and invasion abilities
in both A549/PTX and A549/DDP cells (Fig. 5B). These results suggest that
miR-181a is an upstream regulator of migration and invasion in lung
adenocarcinoma cells.
To further assess the role of miR-181a in EMT, we
examined the morphological and molecular characteristics of cells
that were transfected with miR-181a inhibitor or miR-181a mimic.
Morphological study showed that the A549 cells transfected with
miR-181a mimic had a more mesenchymal appearance (Fig. 6A). Furthermore, at both the protein
(Fig. 6B) and mRNA (Fig. 6C) levels, miR-181a mimic caused a
reduction in the expression of the epithelial adhesion molecules
(E-cadherin and β-cadherin) and an increase in the expression of
the mesen-chymal markers (vimentin, MMP-2, MMP-9, Snail, ZEB1 and
Slug), which was similar to the pattern observed for A549/PTX and
A549/DDP cells (Fig. 3).
Conversely, transfection of A549/PTX cells with miR-181a inhibitor
may reverse drug resistance, as indicated by MTT. These cells also
showed a modest but clearly visible change in morphological
characteristics, changing from mesenchymal-like spindle-cell shape
to epithelial-like shapes (Fig. 7A and
B), with opposite effects on the expression of epithelial and
mesenchymal markers (Fig. 7C and
D). Similar results were observed for A549/DDP cells (Fig. 8). These results directly
demonstrate that overexpression of miR-181a promotes the
acquisition of EMT phenotype in parental A549 cells and that
inhibition of miR-181a expression reverses the EMT phenotype in
A549/PTX or A549/DDP cells. Taken together, miR-181a plays an
important role in the regulation of the EMT in human lung
adenocarcinoma cells.
 | Figure 6Overexpression of miR-181a in A549
cells induces morphological and molecular changes characteristic of
EMT. (A) Twenty-four hours after transfection, cell morphology was
observed by microscopy at ×200 magnification for non-transfected
A549 cells (blank), A549 cells transfected with mimic-NC or
miR-181a-mimic. (B) E-cadherin, β-catenin, vimentin, MMP-9, MMP-2,
Snail, Slug, Twist and ZEB1 expression levels after transfection
were determined by western blot analysis. (C) The mRNA expression
levels were analyzed in A549 cells using qRT-PCR (compared with
blank or mimics-NC, means ± SEM, n=3, *P<0.05). |
 | Figure 7miR-181a downregulation in A549/PTX
cells reverses both drug resistance and morphological and molecular
changes. (A) Twenty-four hours after transfection, A549/PTX cells
transfected with miR-181a-inhibitor and control inhibitor were
treated with paclitaxel at increasing concentrations (0.2, 0.4,
0.8, 1.6 and 3.2 μmol/l) for 48 h, and viability was assessed by
MTT assay. Results are expressed as the relative number of control
levels at each point in time. Values represent the means ± SEM of 3
wells. (B) Twenty-four hours after transfection, microscopy at ×200
magnification was used to observed cell morphology in
non-transfected A549/PTX cells (blank), A549/PTX cells transfected
with negative control inhibitor (mimic-NC) or A549/PTX cells
transfected with miR-181 inhibitor (miR-181a-inhibitor). (C)
Western blot analysis was used to detect the expression of
E-cadherin, β-catenin, vimentin, MMP-9, MMP-2, Snail, Slug, Twist
and ZEB1 after transfection. (D) qRT-PCR was used to analyze mRNA
levels (means ± SEM, n=3, *P<0.05, compared with
blank or inhibitor-NC). |
 | Figure 8miR-181a downregulation in A549/DDP
cells reverses both drug resistance and morphological and molecular
changes. (A) Twenty-four hours after transfection, A549/DDP cells
transfected with miR-181a-inhibitor and control inhibitor were
treated with paclitaxel at increasing concentrations (4, 8, 16, 32
and 64 μmol/l) for 48 h, and viability was assessed by MTT assay.
Results are expressed as the relative number of control levels at
each point in time. Values represent the means ± SEM of 3 wells.
(B) Microscopy at ×200 magnification was used to observe cell
morphology 24 h after transfection for non-transfected A549/DDP
cells (blank), A549/DDP cells transfected with negative control
inhibitor (mimic-NC) or A549/DDP cells that were transfected with
miR-181a-inhibitor. (C) Expression of E-cadherin, β-catenin,
vimentin, MMP-2, MMP-9, Snail, Slug, Twist and ZEB1 in A549/DDP
cells was measured with western blotting. (D) The mRNA expression
levels were detected by qRT-PCR (compared with blank or
inhibitor-NC, means ± SEM, n=3, *P<0.05). |
miR-181a regulates the protein expression
of PTEN by directly targeting 3′UTR of PTEN
To further explore the molecular mechanism of
miR-181a in promoting EMT progression in lung adenocarcinoma cells,
miRNA target prediction databases (miRNA.org and TargetScan)
available online were used to identify potential targets of
miR-181a in humans. This analysis identified PTEN, an
inhibitor of the PI3 kinase/Akt pathway that is reported to play an
important role in the pathogenesis and drug resistance, as a
potential target of miR-181a (Fig.
9A). Consistent with a potential role for PTEN inhibition in
the development of drug resistance, the protein levels of
PTEN were reduced in the paclitaxel- or cisplatin-resistant
A549 cells compared with the parental A549 cells (Fig. 9B). We cloned the wild-type 3′UTR of
PTEN and a mutated version of the 3′UTR into the pGL3-report
plasmid to investigate whether the predicted binding site of
miR-181a to the 3′UTR of the PTEN gene was responsible for
this regulation. The activity of the reporter containing the
wild-type sequence, but not the mutant sequence, was decreased in
A549 cells by co-transfection with miR-181a mimic as demonstrated
by luciferase assays. Conversely, miR-181a inhibitor increased the
activity of wild-type, but mutant reporter activity did not change
in A549/PTX and A549/DDP cells (Fig.
9C). These results suggest that miR-181a may target the
PTEN promoter to inhibit its expression.
 | Figure 9miR-181a directly targets PTEN by
binding to its 3′UTR. (A) The predicted miR-181a binding site
within PTEN 3′UTR and its mutant version resulting from site
mutagenesis are presented. Red indicates the nucleotides that were
mutated to create a mismatch. (B) PTEN expression in lung
adenocar-cinoma cell lines was assessed by western blotting. (C)
Luciferase assays were performed for the indicated cell lines
transfected with a luciferase reported containing either the
wild-type or mutant PTEN 3′UTR sequence; together with a blank
control, a negative control, or miR-181 mimic/inhibitor. Results
were determined relative to a co-transfected Renilla
construct and were standardized to 1.0 in the blank control cells
(compared to blank or mimic/inhibitor-NC, mean ± SEM, n=3,
*P<0.05). (D) Western blots of PTEN are shown for
A549, A549/PTX or A549/DDP cells that were non-transfected (blank)
or that were transfected with negative control or miR-181a
mimic/inhibitor. β-actin was also tested as a loading control.
Shown are results that are representative of three independent
experiments. (E) A proposed model for miR-181a/PTEN signaling
pathway in A549/PTX and A549/DDP EMT-type cells, prolonged exposure
of A549 cells to either paclitaxel or cisplatin caused elevated
expression of miR-181a. miR-181a targets the promoter of PTEN to
inhibit its expression. Because PTEN is an inhibitor of the PI3
kinase pathway, which regulates metastatic processes, inhibition of
PTEN expression leads to the enhancement of migration, invasion,
and EMT progression. |
To verify these findings with endogenous
PTEN, we investigated whether miR-181a modulates the
expression of PTEN protein in A549 cells and its
drug-resistant derivatives. Western blotting results demonstrated
that PTEN protein levels are reduced in A549 cells
transfected with miR-181a mimic, but that conversely, the protein
expression of PTEN is increased when miR-181a inhibitor is
transfected into A549/PTX and A549/DDP cells (Fig. 9D). Overall, these results suggest
that miR-181a inhibits the protein expression of PTEN by directly
targeting 3′UTR of PTEN.
Discussion
Drug resistance is achieved by the sequential
acquisition of genetic alterations that re-route crucial pathways
promoting tumor cell invasive/aggressive phenotypes. Although the
causes of drug resistance have been explored for many years, there
remains no remedy to overcome it to improve clinical outcomes in
resistant cancers. There is growing evidence suggesting that
drug-resistance in cancer cells is associated with the EMT process
in human cancers, including lung adenocarcinoma (19). For example, gefitinib resistance of
cancer cells is correlated with TM4SF5-mediated EMT (20). Similarly, activation of the PI3
kinase/Akt/HIF-1α pathway contributes to hypoxia-induced EMT
and chemoresistance in hepatocellular carcinoma (21). Moreover, paclitaxel-resistant
ovarian cancer cells, cisplatin resistant colorectal cancer cells,
gemcitabine-resistant pancreatic cancer cells and
tamoxifen-resistant breast cancer cells have an EMT
phenotype which includes downregulation of E-cadherin and
upregulation of vimentin (22).
Similarly, we have observed that paclitaxel or cisplatin-resistant
lung adenocarcinoma cells acquire EMT features. Consistently, in
the present study, we found that multi-resistant lung
adenocarcinoma cells demonstrated altered morphological
characteristics of cells similar to EMT with decreased E-cadherin
and increased vimentin, Snail and Slug, suggesting that there is a
link between chemoresistance and EMT in lung adenocarcinoma.
Recently, miRNAs have emerged as crucial mediators
in regulating the cellular responses of cancer cells to therapy
(23). Patient response to
chemotherapy has been shown to be closely correlated to the
functional status of miRNAs. Although the mechanisms of
miRNA-regulated drug resistance are still largely unknown, current
evidence suggest several roles for miRNAs, including influencing
therapy-induced cell death, altering drug targets, regulating
multiple drug resistance (MDR)-related proteins, modifying
bioavailable drug concentrations and promoting angiogenesis
(24–28). It is also becoming increasingly
evident that miRNAs are key modulators of EMT, which is an
important process that drives cancer metastasis. Recent studies
have shown that miRNAs can play a role as important modulators of
EMT through the regulation of E-cadherin and other molecules such
as vimentin and ZEB (29). For
example, the members of the miR-429 family inhibit cells growth and
invasion and regulate EMT-related marker genes which happens
throngh the targeting of Onecut2 in colorectal carcinoma (30). Moreover, miR-205 is downregulated
during EMT with the associated downregulation of ZEB1 and ZEB2
expression in a panel of epithelial breast cancer cells (31). In the present study, we identified
10 miRNAs that are upregulated and 5 miRNAs are downregulated in
A549/PTX cells after compared with A549 cells. To determine a
potential relation with drug resistance in lung cancer cells, we
selected miR-181a, the most highly differentially expressed miRNA
among the miR-181a, miR-181b, and miR-181c genes. This miRNA was
also upregulated in A549/DDP cells, which supports its general
association with the multi-drug resistant phenotype.
Our results indicated that overexpression of
miR-181a promotes invasion and migration and the acquisition of EMT
in lung adenocarcinoma cells, but that inhibition of miR-181a
expression partly reverses EMT. Target prediction tools identified
PTEN as a putative target gene of miR-181a. The PTEN signaling
pathway has been reported to play a role in the control of various
cellular processes, such as cell proliferation, apoptosis,
invasion, metastases and EMT in human cancer (32,33);
therefore, a role for miR-181 in targeting PTEN is consistent with
its known biological effect. In fact, some other miRNAs are capable
of conferring drug resistance by targeting PTEN in other types of
cancer. For example, miR-21 induces cell survival and
chemoresistance, by binding the 3′UTR of PTEN mRNA (34). Loss of PTEN is a very frequent
genetic aberration in malignant tumors such as breast cancer,
gastric cancer and glioblastoma (34–36),
and PTEN loss is significantly associated with cytotoxic drug
resistance (37). In this study,
we found that miR-181a negatively regulates PTEN expression in lung
adenocarcinoma cell lines. As a result, these cells became more
aggressive and invasive after transfection with an miR-181a
expression construct. This finding is consistent with clinical
observations, which have revealed that more advanced stage patients
expressed higher levels of miR-181a (38). There is growing evidence suggesting
that dysfunction of PTEN has prognostic importance in several
malignancies, including lung adenocarcinoma. Our findings reveal
that targeting PTEN at the post-transcriptional level by miRNAs
such as miR-181a mediates PTEN inactivation (Fig. 9E). These results suggest a
mechanism whereby increased miR-181a expression may be associated
with reduced survival in lung adenocarcinoma patients, and also
support the development of miR-181a inhibitor as a potential target
for reversing the effects of drug resistance.
In conclusion, we demonstrated that miR-181a
responds to chemotherapy in lung adenocarcinoma cells by changing
the levels of both itself and its target PTEN. Downregulation of
miR-181a could successfully sensitize cancer cells to chemotherapy.
In the future, therapeutic strategies could be developed based on
the predictive levels of miR-181a. Additionally, miR-181a
repression could potentially be combined with chemotherapy to
prolong drug sensitivity in lung adenocarcinoma, and potentially
other types of cancer.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (nos. 81372899 and
81000992), the Twelfth Five-year Science and Technology Research
Program of Anhui Provincial Scientific Committee (no. 1301042200),
the Key Project of Natural Science Research of the Educational
Department of Anhui Province, China (no. 1208085MH130), and the
Natural Science Foundation of Anhui province (no.
1508085MH166).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Brown T, Pilkington G, Bagust A, Boland A,
Oyee J, Tudur-Smith C, Blundell M, Lai M, Martin Saborido C,
Greenhalgh J, et al: Clinical effectiveness and cost-effectiveness
of first-line chemotherapy for adult patients with locally advanced
or metastatic non-small cell lung cancer: A systematic review and
economic evaluation. Health Technol Assess. 17:1–278. 2013.
|
|
3
|
Macedo-Pérez EO, Morales-Oyarvide V,
Mendoza-García VO, Dorantes-Gallareta Y, Flores-Estrada D and
Arrieta O: Long progression-free survival with first-line
paclitaxel plus platinum is associated with improved response and
progression-free survival with second-line docetaxel in advanced
non-small-cell lung cancer. Cancer Chemother Pharmacol. 74:681–690.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hugo H, Ackland ML, Blick T, Lawrence MG,
Clements JA, Williams ED and Thompson EW: Epithelial-mesenchymal
and mesenchymal-epithelial transitions in carcinoma progression. J
Cell Physiol. 213:374–383. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Comijn J, Berx G, Vermassen P, Verschueren
K, van Grunsven L, Bruyneel E, Mareel M, Huylebroeck D and van Roy
F: The two-handed E box binding zinc finger protein SIP1
downregulates E-cadherin and induces invasion. Mol Cell.
7:1267–1278. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang X, Ling MT, Guan XY, Tsao SW, Cheung
HW, Lee DT and Wong YC: Identification of a novel function of
TWIST, a bHLH protein, in the development of acquired taxol
resistance in human cancer cells. Oncogene. 23:474–482. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Haslehurst AM, Koti M, Dharsee M, Nuin P,
Evans K, Geraci J, Childs T, Chen J, Li J, Weberpals J, et al: EMT
transcription factors snail and slug directly contribute to
cisplatin resistance in ovarian cancer. BMC Cancer. 12:912012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao W, Zhou Y, Xu H, Cheng Y and Kong B:
Snail family proteins in cervical squamous carcinoma: Expression
and significance. Clin Invest Med. 36:E223–E233. 2013.PubMed/NCBI
|
|
9
|
Bowen KA, Doan HQ, Zhou BP, Wang Q, Zhou
Y, Rychahou PG and Evers BM: PTEN loss induces epithelial -
mesenchymal transition in human colon cancer cells. Anticancer Res.
29:4439–4449. 2009.PubMed/NCBI
|
|
10
|
Soria JC, Lee HY, Lee JI, Wang L, Issa JP,
Kemp BL, Liu DD, Kurie JM, Mao L and Khuri FR: Lack of PTEN
expression in non-small cell lung cancer could be related to
promoter methylation. Clin Cancer Res. 8:1178–1184. 2002.PubMed/NCBI
|
|
11
|
Jin G, Kim MJ, Jeon HS, Choi JE, Kim DS,
Lee EB, Cha SI, Yoon GS, Kim CH and Jung TH: PTEN mutations and
relationship to EGFR, ERBB2, KRAS, and TP53 mutations in non-small
cell lung cancers. Lung Cancer. 69:279–283. 2010. View Article : Google Scholar
|
|
12
|
De Craene B and Berx G: Regulatory
networks defining EMT during cancer initiation and progression. Nat
Rev Cancer. 13:97–110. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Moustakas A and Heldin CH: Signaling
networks guiding epithelial-mesenchymal transitions during
embryogenesis and cancer progression. Cancer Sci. 98:1512–1520.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang J, Zhang Y, Yu C, Li Z, Pan Y and
Sun C: MicroRNA-492 expression promotes the progression of hepatic
cancer by targeting PTEN. Cancer Cell Int. 14:952014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gibbons DL, Lin W, Creighton CJ, Rizvi ZH,
Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y,
Pertsemlidis A, et al: Contextual extracellular cues promote tumor
cell EMT and metastasis by regulating miR-200 family expression.
Genes Dev. 23:2140–2151. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang P, Liu H, Xia F, Zhang QW, Zhang YY,
Zhao Q, Chao ZH, Jiang ZW and Jiang CC: Epithelial-mesenchymal
transition is necessary for acquired resistance to cisplatin and
increases the metastatic potential of nasopharyngeal carcinoma
cells. Int J Mol Med. 33:151–159. 2014.
|
|
19
|
Reka AK, Chen G, Jones RC, Amunugama R,
Kim S, Karnovsky A, Standiford TJ, Beer DG, Omenn GS and Keshamouni
VG: Epithelial-mesenchymal transition-associated secretory
phenotype predicts survival in lung cancer patients.
Carcinogenesis. 35:1292–1300. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee MS, Kim HP, Kim TY and Lee JW:
Gefitinib resistance of cancer cells correlated with
TM4SF5-mediated epithelial-mesenchymal transition. Biochim Biophys
Acta. 1823:514–523. 2012. View Article : Google Scholar
|
|
21
|
Jiao M and Nan KJ: Activation of PI3
kinase/Akt/HIF-1α pathway contributes to hypoxia-induced
epithelial-mesenchymal transition and chemoresistance in
hepatocellular carcinoma. Int J Oncol. 40:461–468. 2012.
|
|
22
|
Li Y, VandenBoom TG II, Kong D, Wang Z,
Ali S, Philip PA and Sarkar FH: Up-regulation of miR-200 and let-7
by natural agents leads to the reversal of
epithelial-to-mesenchymal transition in gemcitabine-resistant
pancreatic cancer cells. Cancer Res. 69:6704–6712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar
|
|
25
|
Cimmino A, Calin GA, Fabbri M, Iorio MV,
Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, et
al: miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dews M, Homayouni A, Yu D, Murphy D,
Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, et
al: Augmentation of tumor angiogenesis by a Myc-activated microRNA
cluster. Nat Genet. 38:1060–1065. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jiang K: Biotech comes to its ‘antisenses’
after hard-won drug approval. Nat Med. 19:2522013. View Article : Google Scholar
|
|
28
|
Bockhorn J, Dalton R, Nwachukwu C, Huang
S, Prat A, Yee K, Chang YF, Huo D, Wen Y, Swanson KE, et al:
MicroRNA-30c inhibits human breast tumour chemotherapy resistance
by regulating TWF1 and IL-11. Nat Commun. 4:13932013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ren J, Chen Y, Song H, Chen L and Wang R:
Inhibition of ZEB1 reverses EMT and chemoresistance in
docetaxel-resistant human lung adenocarcinoma cell line. J Cell
Biochem. 114:1395–1403. 2013. View Article : Google Scholar
|
|
30
|
Sun Y, Shen S, Liu X, Tang H, Wang Z, Yu
Z, Li X and Wu M: MiR-429 inhibits cells growth and invasion and
regulates EMT-related marker genes by targeting Onecut2 in
colorectal carcinoma. Mol Cell Biochem. 390:19–30. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lee JY, Park MK, Park JH, Lee HJ, Shin DH,
Kang Y, Lee CH and Kong G: Loss of the polycomb protein Mel-18
enhances the epithelial-mesenchymal transition by ZEB1 and ZEB2
expression through the downregulation of miR-205 in breast cancer.
Oncogene. 33:1325–1335. 2014. View Article : Google Scholar
|
|
32
|
Alimonti A, Carracedo A, Clohessy JG,
Trotman LC, Nardella C, Egia A, Salmena L, Sampieri K, Haveman WJ,
Brogi E, et al: Subtle variations in Pten dose determine cancer
susceptibility. Nat Genet. 42:454–458. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Aguissa-Touré AH and Li G: Genetic
alterations of PTEN in human melanoma. Cell Mol Life Sci.
69:1475–1491. 2012. View Article : Google Scholar
|
|
34
|
Liu ZL, Wang H, Liu J and Wang ZX:
MicroRNA-21 (miR-21) expression promotes growth, metastasis, and
chemo- or radioresistance in non-small cell lung cancer cells by
targeting PTEN. Mol Cell Biochem. 372:35–45. 2013. View Article : Google Scholar
|
|
35
|
Wu Z, He B, He J and Mao X: Upregulation
of miR-153 promotes cell proliferation via downregulation of the
PTEN tumor suppressor gene in human prostate cancer. Prostate.
73:596–604. 2013. View Article : Google Scholar
|
|
36
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Q, Li SH, Wang H, Xiao Y, Sahin O,
Brady SW, Li P, Ge H, Jaffee EM, Muller WJ, et al: Concomitant
targeting of tumor cells and induction of T-cell response
synergizes to effectively inhibit trastuzumab-resistant breast
cancer. Cancer Res. 72:4417–4428. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schwind S, Maharry K, Radmacher MD, Mrózek
K, Holland KB, Margeson D, Whitman SP, Hickey C, Becker H, Metzeler
KH, et al: Prognostic significance of expression of a single
microRNA, miR-181a, in cytogenetically normal acute myeloid
leukemia: A Cancer and Leukemia Group B study. J Clin Oncol.
28:5257–5264. 2010. View Article : Google Scholar : PubMed/NCBI
|