Introduction
G-protein coupled receptors (GPCRs) are
membrane-spanning protein receptors characterized by a variable
number of ligands that include endogenous neurotransmitters and
hormones, but also exogenous natural and artificial compounds
(1,2). More than 600 GPCRs have been
identified in humans, and their activation results in diverse
physiological changes. Upon binding to receptors, agonists trigger
the activation of signaling pathways; whereas, antagonists impede
the agonist-mediated activation of the receptors. Inverse agonists,
besides interfering with agonists as do antagonists, suppress the
constitutive activity of receptors (3,4).
In many cases, the dysregulation of GPCRs has been
linked to the development of cancer, and hence, these proteins are
of great interest to academia and the pharmaceutical industry. The
overexpression of GPCRs has been revealed in a variety of cancer
types. For example, an elevated GPER protein expression was
revealed in embryonal carcinomas as well as in testicular stromal
neoplasms (5,6). CXCR4, one of the best established
chemokine receptors, is highly expressed in breast cancer cells
(7). The HER2/Neu oncogene, which
occurs in ~30% of breast cancers, limits the degradation of CXCR4,
leading to its increased expression. In certain malignancies, some
GPCRs and G proteins may play tumor-suppressive roles, and
mutations may reflect the inactivation of the respective genes. For
example, inactivating mutations in the melanocortin 1 receptor
(MC1R), which is important for pigment production, increase the
risk of melanoma development (8).
Additionally, SIP2 receptor signaling through Gα13 in
diffuse large B cell lymphoma (DLBCL) may exert tumor-suppressive
functions (9).
Secretin receptor (SCTR) is a GPCR to which secretin
binds, and it has long been known to mediate the effect of the
gastrointestinal hormone on digestion and water homeostasis
(10,11). In physiological conditions, SCTR is
highly expressed in secretin target organs (e.g., pancreas, kidney
and small intestine), and it is expressed in the distal lung
regions and liver, with trace levels in the brain, heart and
ovaries (12). The dysregulation
of SCTR and consequent diseases have been documented in limited
cases. High SCTR expression has been observed in pancreatic ductal
adenocarcinomas, cholangiocellular carcinomas, gastrinomas and
bronchopulmonary carcinoid tumors (13–15).
In contrast, SCTR has been found to be downregulated in colorectal
cancer (16) and prostate cancer
(17).
Many tumor-related genes are known to be regulated
by epigenetic mechanisms, including DNA methylation. Genes that are
downregulated and hypermethylated in breast cancer include BRCA1,
PTEN and RASSF1 (18–20). Regarding SCTR, its CpG island
methylation has not yet been extensively reported in cancer. Only a
large chromosomal region at 2q14.2 that spans EN1, SCTR and INHBB
is known to be silenced in colorectal cancer (16).
In the present study, using an in silico
approach, we found that SCTR was downregulated in all stages of
breast cancer cell lines. After confirming the downregulation in
cancer tissue, we tried to elucidate the epigenetic mechanism of
the dysregulation by examining the promoter methylation.
Furthermore, the gene was induced to be downregulated using siRNA,
and a gene interaction network was constructed from the affected
gene pool. Finally, the cell proliferation and migration activities
of SCTR were monitored based on the information from the
siRNA-knockdown experiment.
Materials and methods
In silico database analysis
The Infinium Methylation Chip data for a normal
breast cell line, MCF-10A, and 10 cancer cell lines ranging from
tumor stage I–IV were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The adopted
accession number for normal breast cell is GSM443815. In cancer
cells, the data used in stage I is GSM443818, stage II is
GSM443811, GSM443817, GSM443812, GSM443819 and GSM664892, stage III
is GSM443813 and GSM443814 and stage IV is GSM443816, GSM664906 and
GSM443821. The Illumina Infinium Methylation Assay detects
genome-wide methylation covering 27,578 CpG sites in 14,495 genes.
Observations with adjusted P-values ≥0.05 were removed and thus
excluded from further analysis. Following adjustment, the genes of
the cancer cell lines were defined as differentially methylated if
they displayed an increased or decreased methylation level of at
least 2-fold compared to that of the normal cell line. The
Student's t-test was used to detect differences in mean levels of
methylation and the expression level between the normal and cancer
tissues using SPSS for Windows, version 17.0 (SPSS Inc., Chicago,
IL, USA). P-values <0.05 were considered statistically
significant.
Cell culture and transfection
The breast cancer cell line MCF-7 and the normal
breast cell line MCF-10A, were purchased from the American Type
Culture Collection (ATCC; Manassas, VA, USA). MCF-10A was grown in
MEBM (Lonza, Basel, Switzerland) supplemented with MEGM SingleQuots
(Lonza) and cholera toxin (List Biological Laboratories, Inc.,
Campbell, CA, USA). MCF-7 was grown in Dulbecco's modified Eagle's
medium supplemented with 10% fetal bovine serum (FBS). All cells
were cultured on the surface of a 75-cm2 culture
flask.
siRNA-based downregulation of SCTR was carried out
by transiently transfecting the siRNA against SCTR into the MCF-10A
cell cultures in a 60-mm culture dish. siRNA of the gene and a
control siRNA were purchased from Sigma-Aldrich (St. Louis, MO,
USA) and added to the culture media up to 200 nM in final
concentration using a Lipofectamine RNAiMAX reagent (Invitrogen,
Carlsbad, CA, USA). The cells were harvested 48 h after
transfection for RNA isolation.
To establish stable cell lines expressing SCTR, 2 μg
of SCTR-expressing vector (GeneCopoeia, Germantown, MA, USA) was
transfected into 1×106 of MCF-7 cells in a
75-cm2 culture flask using Lipofectamine 2000
(Invitrogen) according to the manufacturer's instructions. A vector
without the gene was used as a control. Two days after
transfection, neomycin (400 μg/ml) (Gibco-BRL, Carlsbad, CA, USA)
was added to select stable transfectants. The expression of SCTR
was monitored by reverse transcription-polymerase chain reaction
(RT-PCR).
Study subjects
Twenty-one pairs of breast cancer (BrCa) specimens
and their corresponding adjacent normal tissue specimens were
obtained from patients who had undergone surgery between 2011 and
2012 at the National Cancer Center (NCC) in Korea. All patients
provided written informed consent to donate removed tissue to the
NCC, and samples were obtained according to protocols approved by
the Research Ethics Board of the NCC. BrCa specimens were subjected
to histological examination by an expert pathologist for
independent confirmation of tumor grade.
Real-time RT-PCR
Total RNA from ~100 mg of tissue was isolated using
TRIzol reagent according to the manufacturer's protocol (Gibco-BRL)
with a final suspension in 30 μl of distilled water. Total RNA from
a 75-cm2 culture flask was isolated using the RNeasy
Plus Mini kit (Qiagen) with a final elution in 30 μl of distilled
water. Reverse transcription was conducted using 1–5 μg of total
RNA with a reverse transcription kit (Toyobo, Osaka, Japan). Gene
expression was measured by real-time quantitative RT-PCR analysis
using a Kapa SYBR FAST qPCR kit (Kapa Biosystems, Inc., Woburn, MA,
USA) on an ABI 7300 instrument (Applied Biosystems, Foster City,
CA, USA). One microliter of cDNA was used for PCR, which was
performed in duplicate. The primers used for SCTR are forward,
5′-GATGTCACCTACTGCGATGC-3′ and reverse, 5′-ACAAAAATGGCTGGAGAACC-3′.
RNA quantity was normalized to GAPDH content, and gene expression
was quantified according to the 2−ΔCt method. Primer
sequences for GAPDH are forward, 5′-CAGGAGGCATTGCTGATGAT-3′ and
reverse, 5′-GAAGGCTGGGGCTCATTT-3′.
Methylation-specific PCR
Chromosomal DNA from ~100 mg of tissue samples was
isolated using a genomic DNA purification kit (Promega, Madison,
WI, USA) according to the manufacturer's protocol with a 60-μl
elution volume. Sodium bisulfite modification of genomic DNA and
PCR were carried out as previously described (18). Briefly, 0.1 mg of DNA was treated
with sodium bisulfite, and then PCR was carried out using primers
and a Kapa SYBR Fast qPCR kit (Kapa Biosystems). The primer
sequences of methylated SCTR are forward,
5′-TTTGGAGTTTACGGATAGAAAGC-3′ and reverse,
5′-CCGAAAATAAATATTATCAAACGTA-3′. Unmethylated SCTR are forward,
5′-TTTGGAGTTTATGG ATAGAAAGTGT-3′ and reverse,
5′-TCCAAAAATAAATATTATCAAACATA-3′. A methylation index was
calculated for each sample using the following formula: Methylation
index = 1/[1+2−(CTu-CTme)] × 100%, where CTu and CTme
are the cycle threshold obtained using the unmethylated and the
methylated primer pair, respectively.
Expression microarrays and pathway
analysis
An Illumina HumanHT-12 v4 Expression BeadChip
(Illumina, San Diego, CA, USA) analysis covering 31,000 human genes
and containing 47,231 probes was performed by Macrogen (Seoul,
Korea). Briefly, biotinylated cRNA was prepared from 550 ng of
total RNA using the Illumina TotalPrep RNA amplification kit
(Ambion, Austin, TX, USA). Fluorescent signals were obtained by
scanning with iScan System, and data were extracted with Gene
Expression Module 1.9.0 in GenomeStudio v2011.1 (Illumina). Data
were processed by excluding genes with the detection P-values
≥0.05, and differentially expressed genes displaying at least a
2-fold difference between the control cells and the siRNA treated
cells were obtained. The array data were uploaded to the Gene
Expression Omnibus (GEO) database, and they can be accessed via
their website (http://www.ncbi.nlm.nih.gov/geo/; accession number
GSE60750).
Functional categorization and pathway construction
for the gene pool obtained from the affected genes by siRNA or
over-expressed SCTR were performed using the Ingenuity Pathway
Analysis (IPA) software tool (Ingenuity Systems, Redwood City, CA,
USA) (16). P-values for
individual networks were obtained by comparing the likelihood of
obtaining the same number of transcripts or greater in a random
gene set as were actually present in the input file (i.e., the set
of genes differentially methylated in normal and tumor tissue)
using Fisher's exact test, based on the hypergeometric
distribution. The highest confidence functional network was
designated as the top network.
Cell proliferation and migration
analysis
Cells cultured in a 60-mm dish at ~80% confluence
were harvested using 0.05% Trypsin-EDTA (Gibco-BRL), washed with
PBS, and then fixed in ice-cold 70% ethanol overnight. The cells
were treated with 50 μg/ml RNaseA (Sigma-Aldrich) for 1 h and then
stained with 50 μg/ml propidium iodide (Sigma-Aldrich) in the dark
for 30 min at room temperature. Samples were analyzed using a
FACSCanto II flow cytometer (Becton-Dickinson, Franklin Lakes, NJ,
USA) reading with a 488-nm laser. The cell proliferation index was
calculated using the following formula: Proliferation index =
(S+G2+M)/(G0/G1+S+G2+M) × 100%, where each letter represents the
number of cells at each stage.
For cell migration assay, cells were plated onto 8.0
μm Transwell Inserts (Corning Life Sciences, Tewksbury, MA, USA)
and cultured following the supplier's protocol. The migrated cells
were stained with crystal violet and counted under a microscope.
The migration ratio was calculated by dividing the number of
migrated cells by the number of cells seeded.
Results
Various GPCRs are aberrantly methylated
in breast cancer
To identify novel GPCRs that are deregulated by
aberrant promoter methylation in breast cancer, a series of in
silico analysis was performed on genome-wide methylation
databases. The Illumina methylation assay data from a normal breast
cell line and 10 breast cancer cell lines of stage I–IV were
extracted from the GEO database. After filtrating out statistically
insignificant CpG sites (P>0.05), the β-values of remaining
CpGs, indicative of the methylation level, were compared between
the normal cell line and each cancer cell line. Screening of CpG
sites fitting our criteria with 2-fold higher or lower methylation
in cancer than in normal cells presented 808–1,517 CpGs which
included 114 GPCRs. The GPCRs that showed the highest change of
hypermethylation and hypomethylation were LPAR2 (33.3-fold) and
GPR133 (−18.8-fold), GALR3 (19.9-fold) and GPR135 (−20.3-fold),
QRFPR (20.9-fold) and CELSR1 (−21.3-fold), and GPR56 (16.7-fold)
and BDKRB2 (18.1-fold) at stages I–IV, respectively.
SCTR is hypermethylated and downregulated
in breast cancer
Among the aberrantly methylated GPCRs, the ones that
frequently appear throughout the four cancer stages were selected
for further examination. Twenty-four GPCRs (e.g., GPR135, TAS1R2
and SCTR) hit more than three stages (Table I). Three GPCRs, TAS1R2, F2RL2 and
SCTR, whose methylation status is not yet known in any cancer
tissue, were selected for further examination in breast cancer
tissue. Twenty-one pairs of tumor and nearby normal tissue were
examined by real-time methylation-specific PCR (MSP) and RT-PCR to
monitor methylation and expression, respectively. The results
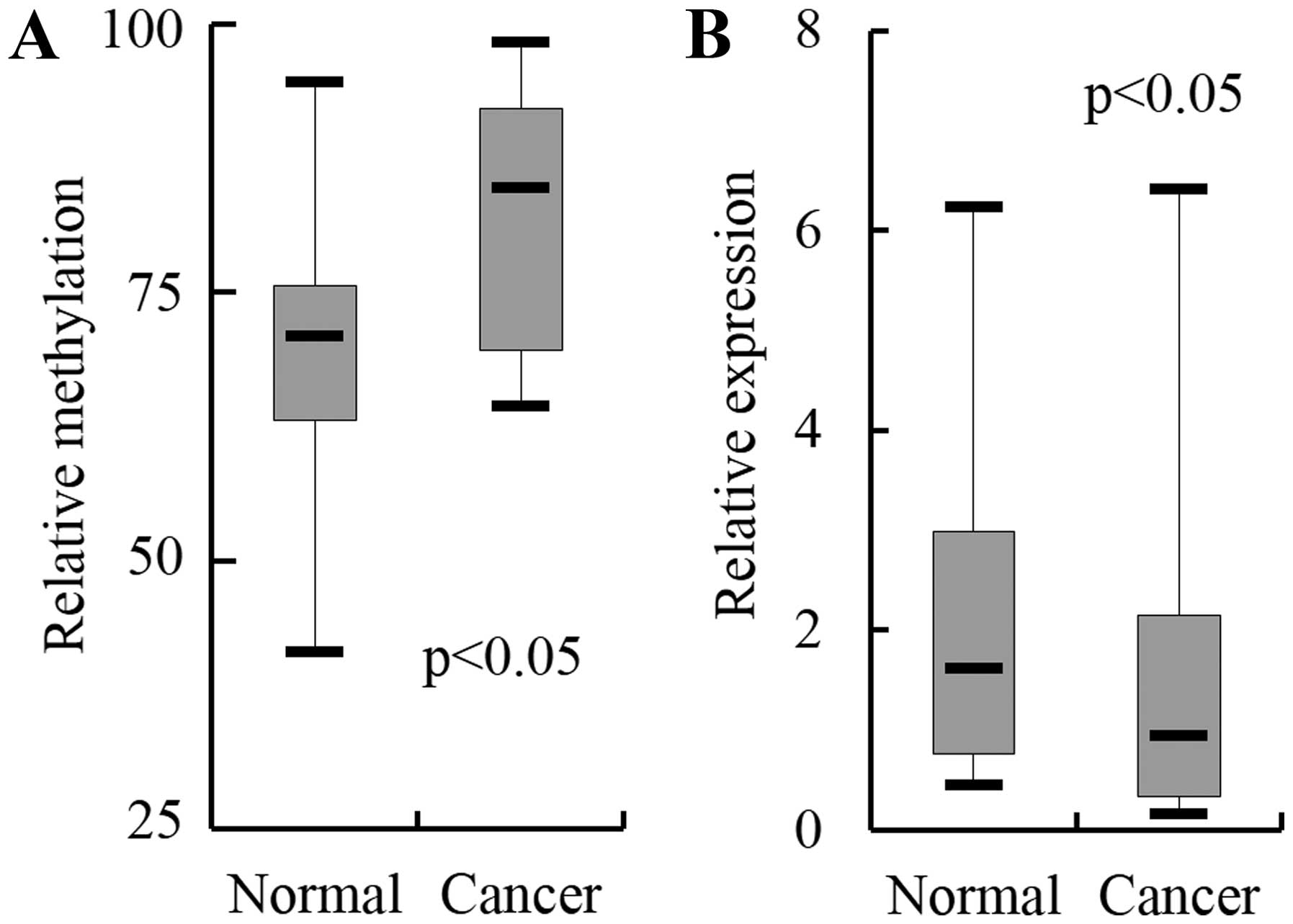
indicated that only SCTR showed higher methylation levels
(P<0.05) and downregulation (P<0.05) in cancer tissues than
in normal tissues with a statistical significance (Fig. 1). SCTR, therefore, was selected to
further address its role in breast cancer.
 | Table IGPCRs displaying differential
methylation at breast tumor stage I–IV. |
Table I
GPCRs displaying differential
methylation at breast tumor stage I–IV.
| | Fold changea |
|---|
| |
|
|---|
| | Stage I | Stage II | Stage III | Stage IV |
|---|
| |
|
|
|
|
|---|
| Gene | Accession no. | HCC-1395 | HCC-38 | HCC-1008 | HCC-1143 | HCC-2218 | HCC-1599 | MDA-MB-231 | BT-474 |
|---|
| GALR3 | NM_003614.1 | 19.9 | 19.4 | 18.1 | 19.7 | 19.2 | 15.1 | 12.4 | 19.7 |
| MC4R | NM_005912.1 | 17.9 | 19.7 | 1.3 | 16.2 | 19.8 | 19.5 | 16.0 | 19.0 |
| QRFPR | NM_198179.1 | 13.2 | 20.9 | −1.1 | 11.5 | 20.3 | 22.6 | 22.5 | 22.2 |
| GIPR | NM_000164.2 | 12.2 | 11.4 | 3.2 | 4.5 | 12.0 | 11.1 | 9.8 | 13.2 |
| F2RL2 | NM_004101.2 | 9.8 | 11.5 | 7.5 | 10.2 | 7.9 | 10.9 | −1.0 | 10.7 |
| FFAR2 | NM_005306.1 | 9.4 | 9.1 | −1.9 | 5.5 | 7.8 | −2.0 | 4.6 | −2.6 |
| HTR2A | NM_000621.2 | 8.8 | 4.3 | 1.2 | 7.5 | 5.2 | 7.7 | 2.7 | 4.2 |
| GHSR | NM_198407.1 | 6.4 | −3.1 | −1.2 | 6.3 | 6.3 | 6.3 | 5.7 | 6.3 |
| TAS1R2 | NM_152232.1 | 6.2 | 6.2 | 4.8 | 4.6 | 6.0 | 6.1 | 3.9 | 6.3 |
| CRHR2 | NM_001883.2 | 5.7 | 4.5 | −2.9 | 5.3 | 2.1 | 2.1 | 7.5 | 3.7 |
| SCTR | NM_002980.1 | 5.4 | 5.8 | −2.6 | 5.5 | 5.5 | 5.7 | 1.3 | 5.7 |
| GPR78 | NM_080819.2 | 1.4 | −3.1 | −3.2 | −4.8 | −1.1 | 2.1 | −1.4 | 2.1 |
| GPR135 | NM_022571.4 | 1.0 | −20.3 | −10.0 | −1.1 | −17.3 | −18.9 | −13.5 | −18.7 |
| GPR63 | NM_030784.1 | −1.1 | −18.5 | −21.1 | −1.5 | −8.1 | −1.9 | −1.1 | −1.1 |
| CELSR1 | NM_014246.1 | −1.5 | −21.3 | −12.9 | −1.1 | −17.1 | −20.6 | −2.2 | −22.8 |
| GPR160 | NM_014373.1 | −1.8 | −2.0 | −12.6 | −1.2 | −1.8 | −49.4 | −2.0 | −17.5 |
| HTR1E | NM_000865.1 | −1.9 | −20.0 | −23.8 | −11.3 | −13.5 | −3.6 | −1.9 | −1.8 |
| GALR1 | NT_025004.13 | −2.5 | −2.6 | −3.1 | −1.5 | −1.0 | 1.0 | 1.0 | −5.0 |
| NTSR1 | NM_002531.1 | −4.7 | −18.8 | −18.9 | −2.6 | −1.2 | 1.0 | −1.0 | −2.8 |
| FZD10 | NM_007197.2 | −6.6 | −10.4 | −51.8 | −9.0 | −1.2 | −4.7 | 1.0 | 1.0 |
| RXFP3 | NM_016568.1 | −7.2 | −31.1 | −27.9 | −1.9 | −24.2 | −1.8 | −1.1 | −1.6 |
| SSTR5 | NM_001053.1 | −7.4 | −7.7 | −56.2 | −3.8 | −2.1 | −1.8 | 1.0 | −17.6 |
| GPR133 | NM_198827.2 | −18.8 | −1.5 | −74.1 | −1.0 | −10.9 | −1.1 | −3.8 | −1.0 |
| GPR37 | NM_005302.2 | −23.2 | −57.7 | −48.1 | −80.1 | −64.8 | −44.9 | −9.6 | −87.1 |
SCTR regulates cell cycle-related
genes
To address the role of SCTR in breast cancer,
knockdown was induced by transiently transfecting siRNA, which
targeted the message in the MCF-10A cells, and then genome-wide
expression change was monitored through microarray analysis. In
total, 1,587 genes fitting our criteria of expression change
>2-fold with 993 upregulated and 594 downregulated genes were
submitted to the IPA. The results indicated the ‘cellular movement,
cancer, connective tissue disorders’ as the top network and ‘cell
cycle: G2/M DNA damage checkpoint regulation’ as the top pathway,
implying its role in tumorigenesis (Fig. 2 and Table II). Notably, the majority of the
cell cycle-related genes in the network were upregulated or
downregulated in a way increasing cell proliferation after SCTR was
downregulated by siRNA, suggesting SCTR has anti-proliferative
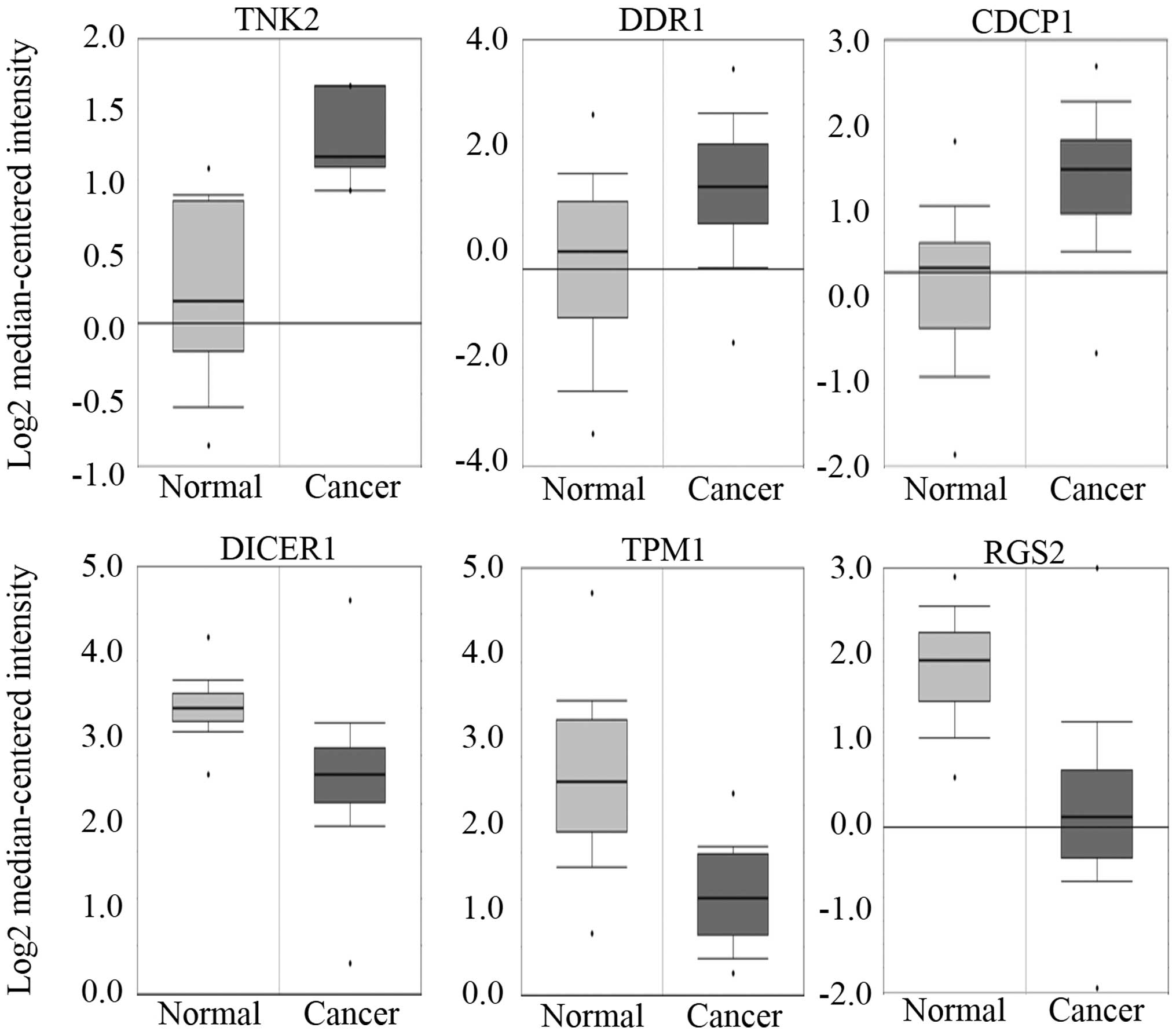
activity in normal cells. In the network, three kinases, TNK2
(ACK1), DDR1 and CDCP1, all of which are known to be oncogenic
proteins, were upregulated (21–23).
Whereas, DICER1, TPM1 and RGS2, all of which are known to be
related with tumor-suppressing activity, were downregulated in the
network (24–26). Therefore, it is expected that the
genes would show upregulation or downregulation in cancer if they
are oncogenic or tumor-suppressive, respectively. The in
silico expression analysis based on the Oncomine database
platform confirmed the upregulation of TNK2 (ACK1), DDR1, and CDCP1
and the downregulation of DICER1, TPM1 and RGS2 in the breast
cancer cells (Fig. 3).
 | Table IIGenes on top network displaying
differential expression by knockdown of SCTR in MCF-10A cells. |
Table II
Genes on top network displaying
differential expression by knockdown of SCTR in MCF-10A cells.
| Gene | Accession no. | Description | Fold change |
|---|
| IL8 | NM_000584.2 | Interleukin 8 | 21.17 |
| LAMC2 | NM_005562.1 | Laminin, γ 2 | 7.17 |
| SDC1 | NM_001006946 | Syndecan 1 | 6.62 |
| EPHA2 | NM_004431.2 | EPH receptor
A2 | 4.69 |
| LAMB3 | NM_000228.2 | Laminin, β 3 | 4.64 |
| ABCB6 | NM_005689.1 | ATP-binding
cassette, sub-family B | 4.62 |
| ZMAT3 | NM_152240.1 | Zinc finger,
matrin-type 3 | 3.83 |
| ANGPTL4 | NM_139314.1 | Angiopoietin-like
4 | 3.38 |
| LAMA3 | NM_198129.1 | Laminin, α 3 | 3.31 |
| DDR1 | NM_013993.2 | Discoidin domain
receptor tyrosine kinase 1 | 3.30 |
| GALNT3 | NM_004482.2 | GalNAc-T3 | 3.24 |
| SORL1 | NM_003105.3 | Sortilin-related
receptor | 3.18 |
| TNK2 | NM_005781.4 | Tyrosine kinase,
non-receptor, 2 | 3.16 |
| CDCP1 | NM_022842.3 | CUB domain
containing protein 1 | 2.97 |
| PALLD | NM_016081.3 | Palladin,
cytoskeletal associated protein | 2.93 |
| ADM | NM_001124.1 | Adrenomedullin | 2.81 |
| TNC | NM_002160.1 | Tenascin C | 2.76 |
| ABCG1 | NM_016818.2 | ATP-binding
cassette, sub-family G (WHITE), member 1 | 2.63 |
| IQGAP1 | NM_003870.3 | IQ motif containing
GTPase activating protein 1 | 2.63 |
| LAMA5 | NM_005560.3 | Laminin, α 5 | 2.63 |
| TNFRSF12A | NM_016639.1 | Tumor necrosis
factor receptor superfamily, member 12A | 2.54 |
| LILRB1 | NM_001081637.1 | Leukocyte
immunoglobulin-like receptor, subfamily B | 2.51 |
| DICER1 | NM_030621.2 | Dicer 1,
ribonuclease type III | −2.77 |
| PRUNE | NM_021222.1 | Prune homolog
(Drosophila) | −3.16 |
| TPM1 | NM_001018020.1 | Tropomyosin 1 | −3.52 |
| RGCC | NM_014059.2 | Regulator of cell
cycle | −3.54 |
| RGS2 | NM_002923.1 | Regulator of
G-protein signaling 2 | −7.41 |
To examine the cell cycle-related activity of SCTR,
the MCF-10A normal breast cells wherein the SCTR was downregulated
by siRNA were monitored for cell proliferation index by FACS
analysis. The result indicated, unexpectedly, that no remarkable
change of the cell proliferation index was observed (Fig. 4). To determine whether the effect
of SCTR on the cell cycle differs between normal and cancer cells,
the gene was overexpressed in the MCF-7 cancer cells by stable
transfection (Fig. 5A), and the
cells were monitored for cell-cycle status (Fig. 5B). The result indicated a 35%
increase of the cell proliferation index with a 9% decrease of G1
phase cells but a 39% increase of S and G2/M phase cells. Next, the
effect of SCTR on the migration of the cancer cells was examined
for the SCTR-overexpressing MCF-7 cells, and the cells showed a
2.1-fold increase of migration compared to the MCF-7 cells
(Fig. 5C). These facts suggest
that SCTR stimulates cell migration as well as cell proliferation
of the MCF-7 cancer cells by regulating cell cycle-related
genes.
Discussion
GPCRs modulate diverse cellular responses to the
majority of neurotransmitters and hormones within the human body
(27). Various GPCRs have been
found to be overexpressed in primary and metastatic tumors
(28,29). In breast cancer, specific GPCR
systems are excessively activated in malignant breast cancer due to
the overexpression of receptors, which contributes to the
progression and spread of breast cancer (30,31).
For example, GPR30, a seven membranespanning estrogen receptor, is
linked to estrogen binding and heparin-bound epidermal growth
factor release, and it induces the proliferation and migration of
breast cancer cells through connective tissue growth factor
(32,33). For these reasons, GPCRs have long
been the most promising targets for developing therapeutic agents
(34). For instance, an adhesion
GPCR, CD97, was targeted by lysophosphatidic acid in the MDA-MB-231
breast cancer cells (35).
In the present study, many GPCRs showed aberrant
methylation, with some being hypermethylated and others being
hypomethylated in breast cancer. These differential methylation
profiles of the GPCRs should direct their expression in such a way
as to specifically contribute to carcinogenesis. In the case of
SCTR, a high frequency of methylation at the gene's CpG island was
revealed in colorectal cancer, although its relationship to
expression has yet to be determined (36). The roles of SCTR have been
elucidated only in a few cancer types. When transfected into Y1
adrenocortical carcinoma cells, overexpressing SCTR stimulated cell
proliferation via the PI3K/AKT signaling cascade (37).
Whether SCTR is an oncogene or a tumor suppressor is
still controversial. Several studies support the idea that it is an
oncogene, as high expression was observed in a few cancer types,
such as pancreatic ductal adenocarcinoma and gastrinoma (13,15),
whereas, other studies indicate the downregulation of the gene in
colorectal cancer, cholangiocarcinoma and prostate cancer (14,16,17).
Notably, the differential regulation occurs in a tissue-specific
manner, and this drove us to profile the expression in breast
cancer.
In the present study, the authors suggested unique
regulatory activities of SCTR in breast cells (i.e.,
tumor-suppressive activity in normal cells and proliferation- and
migration-stimulating activity in cancer cells). Several
experimental results support our hypothesis. First, the
downregulation of SCTR by siRNA in normal breast MCF-10A cells,
dysregulated oncogenes or tumor suppressors toward suppressing
tumor development; oncogenes were upregulated, and tumor
suppressors were downregulated by siRNA. Second, the overexpression
of SCTR in MCF-7 breast cancer cells increased the proliferation
rate of the cells, while little effect on normal cells was observed
when the gene was downregulated. Third, the overexpression of SCTR
in MCF-7 cancer cells increased cell migration. The so-called
double-edge sword activity of tumor-related genes (i.e., upholding
both oncogenic and tumor-suppressive activity) are known for genes
involved in key signaling pathways, such as TGF-β1 and JNK. TGF-β1
is a potent growth inhibitor, with tumor-suppressing activity.
Cancers are often refractile to this growth inhibition because of
the downstream perturbation of the signaling pathway (38). JNK has pro- or anti-apoptotic
functions depending on the cell type, nature of the death stimulus,
duration of its activation, and activity of other signaling
pathways (39). The molecular
mechanism of SCTR for negative and positive regulation in terms of
the cell proliferation that appeared in normal and cancer cells is
to be elucidated in further studies.
The signaling pathway via GPCR is a highly
complicated process. NF-κB, β-catenin and PI3Kγ are major signaling
genes aberrantly regulated in cancer (40). In the present study, IL-8 was
upregulated after inhibiting SCTR, implying that SCTR is able to
suppress IL-8 expression. IL-8 is reported to promote breast cancer
by increasing cell invasion, angiogenesis, and metastases (41,42).
The signaling pathway connecting SCTR and IL-8 should be elucidated
in future studies. Genes known to interact with IL-8 in the network
(Fig. 2) (e.g., DDR1, TNFRSF12A
and DICER1) could be informative. Taken together, it is speculated
that SCTR acts as a tumor suppressor in normal breast tissue.
However, the gene has a tendency to proliferate breast cancer cells
although it is downregulated in cancer by promoter methylation.
Acknowledgements
The present study was supported by the Basic Science
Research Program through the National Research Foundation of Korea
(NRF) funded by the Ministry of Education, Science, and Technology
(NRF-2012R1A1A2040830). Dr H.S. Kang was supported by a grant
provided by the National Cancer Center, Korea.
Abbreviations:
|
SCTR
|
secretin receptor
|
|
CpG
|
cytosine guanine dinucleotide
|
|
GPCR
|
G-protein coupled receptor
|
|
MSP
|
methylation-specific PCR
|
|
RT-PCR
|
reverse transcription-polymerase chain
reaction
|
References
|
1
|
Pierce KL, Premont RT and Lefkowitz RJ:
Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 3:639–650.
2002. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Reiter E, Ahn S, Shukla AK and Lefkowitz
RJ: Molecular mechanism of β-arrestin-biased agonism at
seven-transmembrane receptors. Annu Rev Pharmacol Toxicol.
52:179–197. 2012. View Article : Google Scholar
|
|
3
|
Rajagopal S, Ahn S, Rominger DH,
Gowen-MacDonald W, Lam CM, Dewire SM, Violin JD and Lefkowitz RJ:
Quantifying ligand bias at seven-transmembrane receptors. Mol
Pharmacol. 80:367–377. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Urban JD, Clarke WP, von Zastrow M,
Nichols DE, Kobilka B, Weinstein H, Javitch JA, Roth BL,
Christopoulos A, Sexton PM, et al: Functional selectivity and
classical concepts of quantitative pharmacology. J Pharmacol Exp
Ther. 320:1–13. 2007. View Article : Google Scholar
|
|
5
|
Chevalier N, Bouskine A and Fenichel P:
Role of GPER/GPR30 in tumoral testicular germ cells proliferation.
Cancer Biol Ther. 12:2–3. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rago V, Romeo F, Giordano F, Maggiolini M
and Carpino A: Identification of the estrogen receptor GPER in
neoplastic and non-neoplastic human testes. Reprod Biol Endocrinol.
9:1352011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Friedl P and Wolf K: Tumour-cell invasion
and migration: Diversity and escape mechanisms. Nat Rev Cancer.
3:362–374. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mitra D, Luo X, Morgan A, Wang J, Hoang
MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, et al: An
ultraviolet-radiation-independent pathway to melanoma
carcinogenesis in the red hair/ fair skin background. Nature.
491:449–453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Green JA, Suzuki K, Cho B, Willison LD,
Palmer D, Allen CD, Schmidt TH, Xu Y, Proia RL, Coughlin SR, et al:
The sphingosine 1-phosphate receptor S1P2 maintains the homeostasis
of germinal center B cells and promotes niche confinement. Nat
Immunol. 12:672–680. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chey WY and Chang TM: Secretin, 100 years
later. J Gastroenterol. 38:1025–1035. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chu JY, Lee LT, Lai CH, Vaudry H, Chan YS,
Yung WH and Chow BK: Secretin as a neurohypophysial factor
regulating body water homeostasis. Proc Natl Acad Sci USA.
106:15961–15966. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Davis RJ, Page KJ, Dos Santos Cruz GJ,
Harmer DW, Munday PW, Williams SJ, Picot J, Evans TJ, Sheldrick RL,
Coleman RA, et al: Expression and functions of the duodenal peptide
secretin and its receptor in human lung. Am J Respir Cell Mol Biol.
31:302–308. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Körner M, Hayes GM, Rehmann R, Zimmermann
A, Friess H, Miller LJ and Reubi JC: Secretin receptors in normal
and diseased human pancreas: Marked reduction of receptor binding
in ductal neoplasia. Am J Pathol. 167:959–968. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Onori P, Wise C, Gaudio E, Franchitto A,
Francis H, Carpino G, Lee V, Lam I, Miller T, Dostal DE, et al:
Secretin inhibits cholangiocarcinoma growth via dysregulation of
the cAMP-dependent signaling mechanisms of secretin receptor. Int J
Cancer. 127:43–54. 2010. View Article : Google Scholar
|
|
15
|
Ding WQ, Kuntz S, Böhmig M, Wiedenmann B
and Miller LJ: Dominant negative action of an abnormal secretin
receptor arising from mRNA missplicing in a gastrinoma.
Gastroenterology. 122:500–511. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mayor R, Casadomé L, Azuara D, Moreno V,
Clark SJ, Capellà G and Peinado MA: Long-range epigenetic silencing
at 2q14.2 affects most human colorectal cancers and may have
application as a non-invasive biomarker of disease. Br J Cancer.
100:1534–1539. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Devaney J, Stirzaker C, Qu W, Song JZ,
Statham AL, Patterson KI, Horvath LG, Tabor B, Coolen MW, Hulf T,
et al: Epigenetic deregulation across chromosome 2q14.2
differentiates normal from prostate cancer and provides a regional
panel of novel DNA methylation cancer biomarkers. Cancer Epidemiol
Biomarkers Prev. 20:148–159. 2011. View Article : Google Scholar
|
|
18
|
Stefansson OA, Jonasson JG, Olafsdottir K,
Hilmarsdottir H, Olafsdottir G, Esteller M, Johannsson OT and
Eyfjord JE: CpG island hypermethylation of BRCA1 and loss of pRb as
co-occurring events in basal/triple-negative breast cancer.
Epigenetics. 6:638–649. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hino R, Uozaki H, Murakami N, Ushiku T,
Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada
K, et al: Activation of DNA methyltransferase 1 by EBV latent
membrane protein 2A leads to promoter hypermethylation of PTEN gene
in gastric carcinoma. Cancer Res. 69:2766–2774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sebova K, Zmetakova I, Bella V, Kajo K,
Stankovicova I, Kajabova V, Krivulcik T, Lasabova Z, Tomka M,
Galbavy S, et al: RASSF1A and CDH1 hypermethylation as potential
epimarkers in breast cancer. Cancer Biomark. 10:13–26.
2012.PubMed/NCBI
|
|
21
|
Mahajan K, Coppola D, Chen YA, Zhu W,
Lawrence HR, Lawrence NJ and Mahajan NP: Ack1 tyrosine kinase
activation correlates with pancreatic cancer progression. Am J
Pathol. 180:1386–1393. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Quan J, Yahata T, Adachi S, Yoshihara K
and Tanaka K: Identification of receptor tyrosine kinase, discoidin
domain receptor 1 (DDR1), as a potential biomarker for serous
ovarian cancer. Int J Mol Sci. 12:971–982. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Miyazawa Y, Uekita T, Ito Y, Seiki M,
Yamaguchi H and Sakai R: CDCP1 regulates the function of MT1-MMP
and invadopodia-mediated invasion of cancer cells. Mol Cancer Res.
11:628–637. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Díaz-García CV, Agudo-López A, Pérez C,
López-Martín JA, Rodríguez-Peralto JL, de Castro J, Cortijo A,
Martínez-Villanueva M, Iglesias L, García-Carbonero R, et al:
DICER1, DROSHA and miRNAs in patients with non-small cell lung
cancer: Implications for outcomes and histologic classification.
Carcinogenesis. 34:1031–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bharadwaj S and Prasad GL: Tropomyosin-1,
a novel suppressor of cellular transformation is downregulated by
promoter methylation in cancer cells. Cancer Lett. 183:205–213.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wolff DW, Xie Y, Deng C, Gatalica Z, Yang
M, Wang B, Wang J, Lin MF, Abel PW and Tu Y: Epigenetic repression
of regulator of G-protein signaling 2 promotes androgen-independent
prostate cancer cell growth. Int J Cancer. 130:1521–1531. 2012.
View Article : Google Scholar
|
|
27
|
Rosenbaum DM, Rasmussen SG and Kobilka BK:
The structure and function of G-protein-coupled receptors. Nature.
459:356–363. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
O'Hayre M, Degese MS and Gutkind JS: Novel
insights into G protein and G protein-coupled receptor signaling in
cancer. Curr Opin Cell Biol. 27:126–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lappano R and Maggiolini M: G
protein-coupled receptors: Novel targets for drug discovery in
cancer. Nat Rev Drug Discov. 10:47–60. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wülfing P, Kersting C, Tio J, Fischer RJ,
Wülfing C, Poremba C, Diallo R, Böcker W and Kiesel L:
Endothelin-1-, endothelin-A-, and endothelin-B-receptor expression
is correlated with vascular endothelial growth factor expression
and angiogenesis in breast cancer. Clin Cancer Res. 10:2393–2400.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Filardo EJ, Graeber CT, Quinn JA, Resnick
MB, Giri D, DeLellis RA, Steinhoff MM and Sabo E: Distribution of
GPR30, a seven membrane-spanning estrogen receptor, in primary
breast cancer and its association with clinicopathologic
determinants of tumor progression. Clin Cancer Res. 12:6359–6366.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pandey DP, Lappano R, Albanito L, Madeo A,
Maggiolini M and Picard D: Estrogenic GPR30 signalling induces
proliferation and migration of breast cancer cells through CTGF.
EMBO J. 28:523–532. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lagerström MC and Schiöth HB: Structural
diversity of G protein-coupled receptors and significance for drug
discovery. Nat Rev Drug Discov. 7:339–357. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park SJ, Lee KP, Kang S, Chung HY, Bae YS,
Okajima F and Im DS: Lysophosphatidylethanolamine utilizes LPA(1)
and CD97 in MDA-MB-231 breast cancer cells. Cell Signal.
25:2147–2154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Karpinski P, Ramsey D, Grzebieniak Z,
Sasiadek MM and Blin N: The CpG island methylator phenotype
correlates with long-range epigenetic silencing in colorectal
cancer. Mol Cancer Res. 6:585–591. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lee M, Waser B, Reubi JC and Pellegata NS:
Secretin receptor promotes the proliferation of endocrine tumor
cells via the PI3K/ AKT pathway. Mol Endocrinol. 26:1394–1405.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oft M, Peli J, Rudaz C, Schwarz H, Beug H
and Reichmann E: TGF-beta1 and Ha-Ras collaborate in modulating the
phenotypic plasticity and invasiveness of epithelial tumor cells.
Genes Dev. 10:2462–2477. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu J and Lin A: Role of JNK activation in
apoptosis: A double-edged sword. Cell Res. 15:36–42. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu J, Xie N, Zhao X, Nice EC and Huang C:
Dissection of aberrant GPCR signaling in tumorigenesis - a systems
biology approach. Cancer Genomics Proteomics. 9:37–50.
2012.PubMed/NCBI
|
|
41
|
Chen Y, Chen L, Li JY, Mukaida N, Wang Q,
Yang C, Yin WJ, Zeng XH, Jin W and Shao ZM: ERβ and PEA3
co-activate IL-8 expression and promote the invasion of breast
cancer cells. Cancer Biol Ther. 11:497–511. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim H, Choi JA, Park GS and Kim JH: BLT2
up-regulates interleukin-8 production and promotes the invasiveness
of breast cancer cells. PLoS One. 7:e491862012. View Article : Google Scholar : PubMed/NCBI
|