Introduction
Cervical cancer is the second most common type of
cancer in females worldwide (1).
In China, there is an increasing trend in the incidence and
mortality of cervical cancer. Numerous risk factors have been
linked to this disease, including sexually transmitted diseases,
sexual debut time, menstruation and childbirth, smoking, immune
deficiency and viral infection (2).
Oral contraceptives (OCs) are one of the widely used
contraceptive methods for females. As the main components of OCs
are hormones (estrogen and progesterone), OC use may alter the gene
expression and thereby increase the incidence of cervical cancer
(3). Analysis of data from a total
of 28 studies including 12,531 females with cervical cancer
indicated that the risk of cervical cancer decreases following
termination of OC use (4). Another
pooled study including 16,573 females with and 35,509 with no
cervical cancer reported that the relative risk of cervical cancer
was increased in current users of OCs and declined after their use
was terminated (5).
The mechanism underlying the increased risk of
cervical cancer in OC users remains elusive. However, the most
prevalent mechanism is a correlation between OC and human papilloma
virus (HPV). HPV infection is thought to be the main but not the
sole cause of the majority of cases of cervical cancer. Steroid
contraception has been postulated to be one mechanism by which HPV
exerts its tumorigenic effect on cervical tissue. It is believed
that steroids bind to specific DNA sequences within transcriptional
regulatory regions on the HPV DNA, either to increase or suppress
the transcription of various genes (6). In females using combined OCs, the
cervical mucus remains scanty, thick and highly viscous (7). Therefore, Guven et al
(8) hypothesized that cervical
mucus changes may contribute to the pathogenesis of cervical
cancer, since scanty, thick and highly viscous cervical mucus may
modulate and prolong the effect of carcinogenic agents.
To assess changes in global gene expression
resulting from OC use, the transcriptome of cervical cancer
patients with and without OC use were compared and
differentially-expressed genes (DEGs) correlated to OC use were
identified. Further investigations were performed on their
biological functions and their relevance to clinical
manifestations, which may be beneficial for the development of
novel therapies.
Materials and methods
RNA-seq data and clinical
information
Level 3 RNA-seq data, which had been obtained using
a Illumina HiSeq (Illumina, San Diego, CA, USA), for cervical
squamous-cell carcinoma and endocervical adenocarcinoma as well as
the clinical information were downloaded from The Cancer Genome
Atlas (TCGA; National Human Genome Research Institute, National
Institute of Health, Bethesda, MD, USA; http://cancergenome.nih.gov/). The raw data provided
reads per kilobase of exon per million mapped reads (RPKM, a
measure of relative gene expression) for each gene. Data for 35
samples were obtained (17 samples from patients who never used OC,
16 samples from former users and 2 samples from current users).
Given the small sample size, samples from former and present users
were combined. Clinical data were also collected, including the
time of diagnosis, time of death and follow-up time, from which the
survival time could be calculated. The pathological response
status, histological grade, age and race were also contained in the
Biotab file.
Screening of DEGs
R software, version 3.0.1 (9) was used for statistical analysis. A
t-test was used to identify DEGs. In multiple testing corrections,
the false discovery rates (FDRs) were estimated using a
Beta-Uniform Mixture (BUM) model (10).
Functional enrichment analysis
Gene Ontology (GO) enrichment analysis was performed
with the Database for Annotation, Visualization and Integrated
Discovery tool (DAVID; National Institute of Health; http://david.abcc.ncifcrf.gov/) (11) with hypergeometric distribution. The
P-value was adjusted by the Benjamini & Hochberg method
(12). BioCarta (San Diego, CA,
USA; http://www.biocarta.com) was also used
to identify enriched regulatory networks in DEGs. All statistical
analyses were conducted using R software, version 3.0.1 (9)
Gene expression and clinical
manifestations
The relevance of gene expression to clinical
manifestations was investigated. The t-test was applied to examine
the correlation between genes and the pathological response.
Analysis of variance (ANOVA) was used to analyze the gene
expression among different histological grades. The Cox model was
selected to explore the significance of the correlation between
gene expression and the survival time.
Results
Differentially-expressed genes
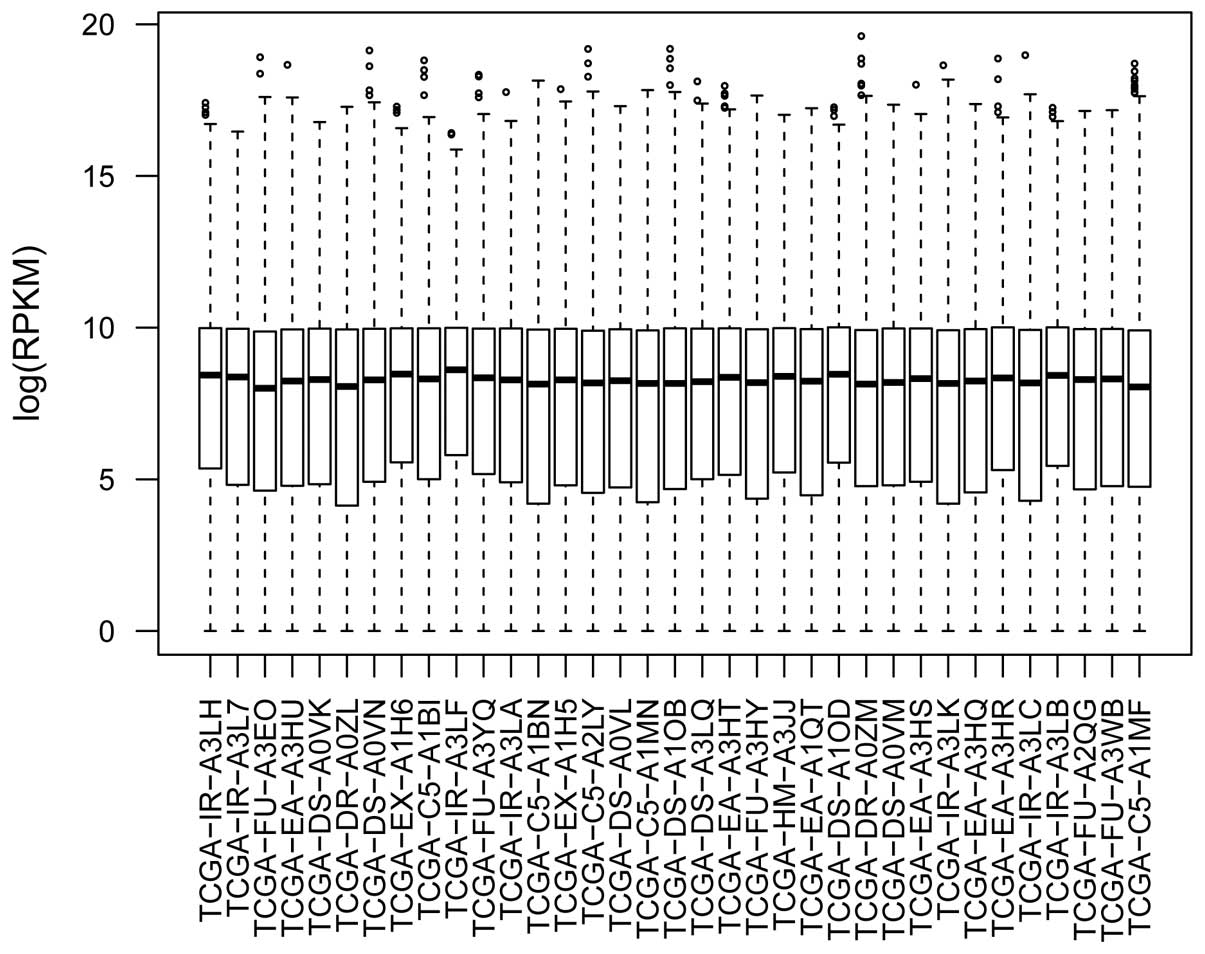
The distribution of RPKM was similar for each sample
following data pre-treatment (Fig.
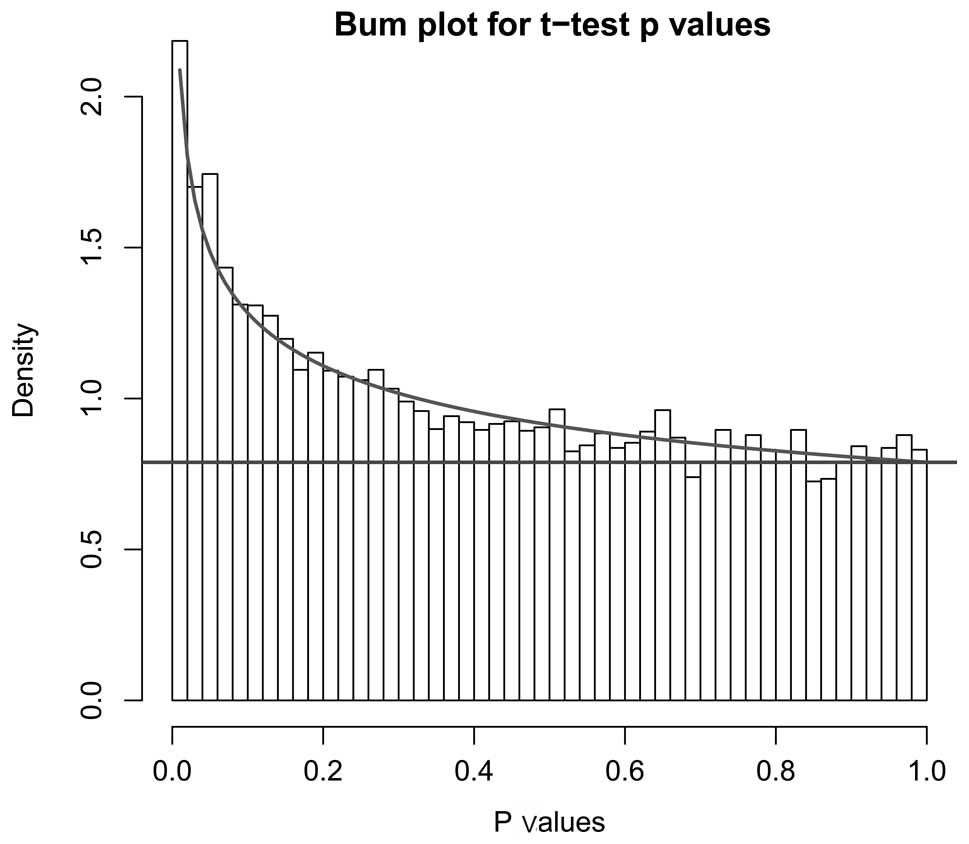
1). DEGs were screened using the t-test. The distribution of
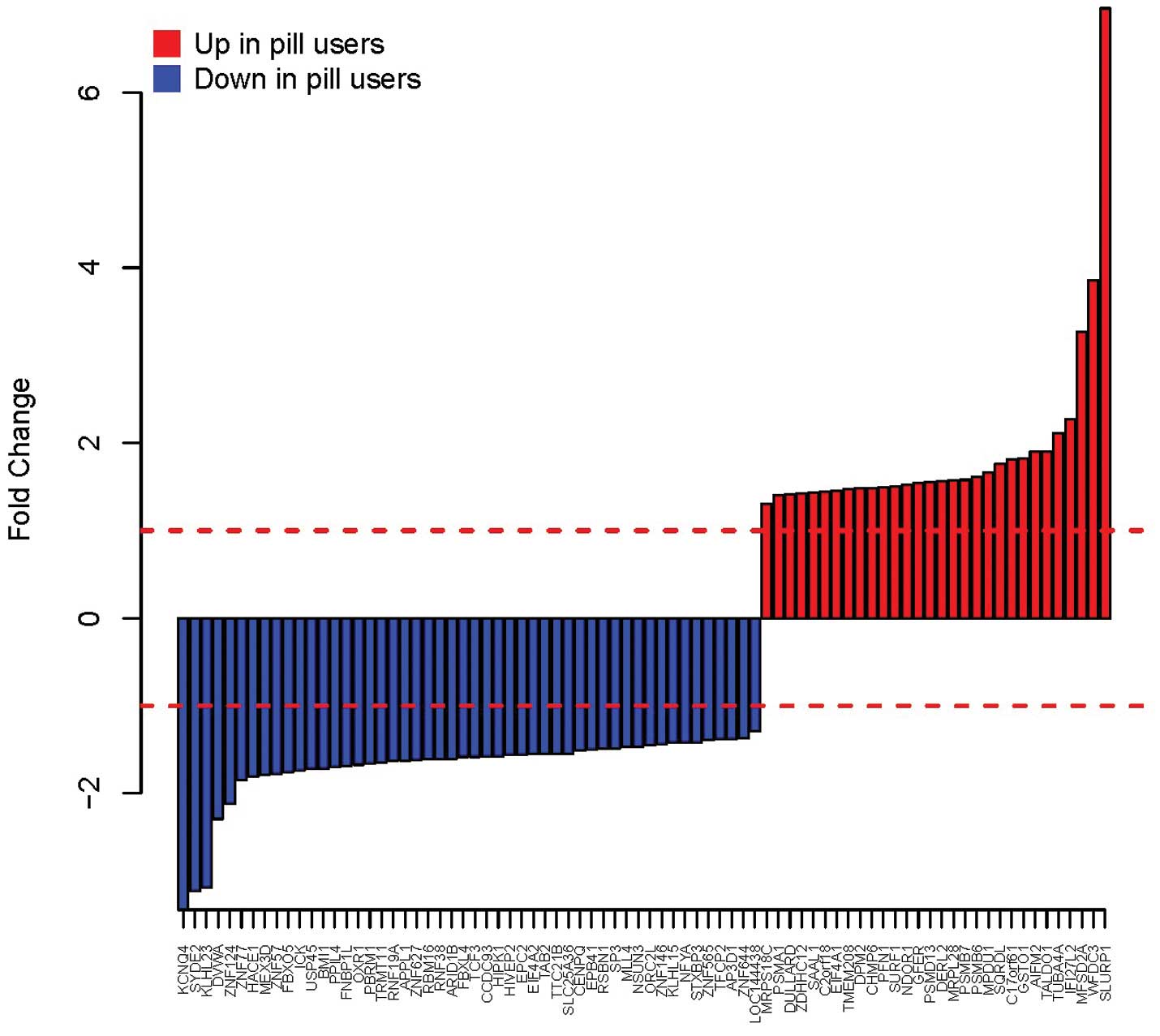
the P-value and the fitted BUM curve are shown in Fig. 2. A total of 80 DEGs were disclosed,
while FDR=0.3 was set as the threshold value. The fold changes of
these genes are shown in Fig.
3.
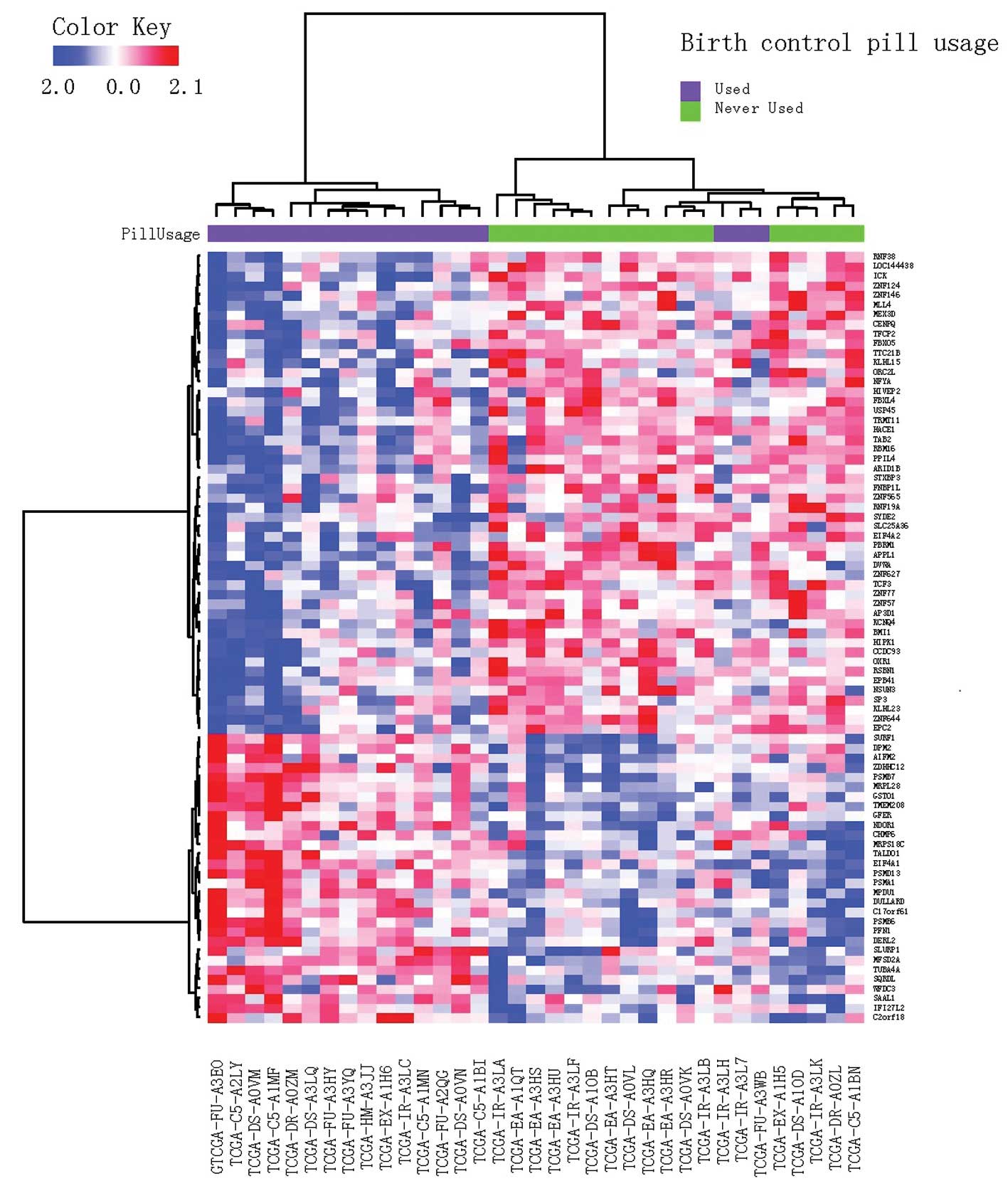
Cluster analysis results
A cluster analysis was performed using DEGs. The
distance was defined as 1-Pearson correlation coefficient and the
Ward’s method was applied in hierarchical clustering. The result is
shown in Fig. 4. A total of three
samples from former OC users were grouped with the control. It is
speculated that the disturbance of gene expression by OCs may
diminish with time.
Enriched biological functions
A GO enrichment analysis indicated that cellular
macromolecule metabolic processes were over-represented in DEGs
(Table I). BioCarta analysis
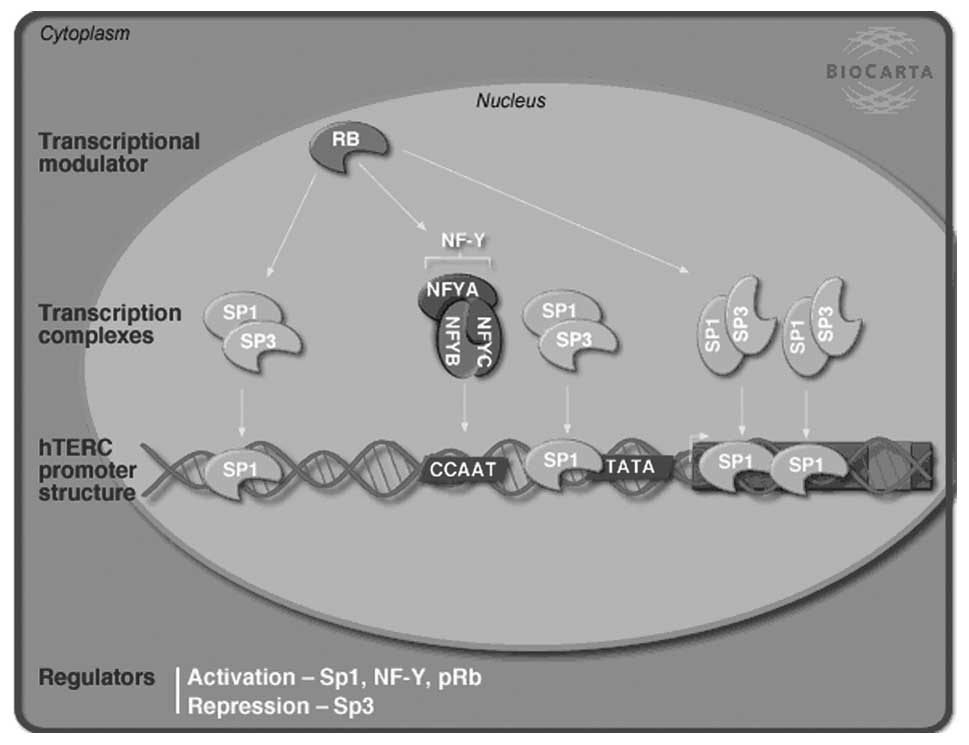
indicated that DEGs were involved in the transcriptional regulation
of the human telomerase RNA gene (hTERC) (Fig. 5; P=0.038, Fisher’s exact test).
 | Table IGene Ontology (GO) enrichment analysis
result for the differentially-expressed genes. |
Table I
Gene Ontology (GO) enrichment analysis
result for the differentially-expressed genes.
| Category | Term | Count (n) | P-value | Adjusted
P-valuea |
|---|
| GOTERM_BP_FAT | Protein catabolic
process | 11 |
5.50×10−4 |
7.20×10−2 |
| GOTERM_BP_FAT | Cellular protein
catabolic process | 11 |
4.30×10−4 |
7.50×10−2 |
| GOTERM_CC_ALL | Intracellular | 64 |
3.20×10−5 |
2.60×10−3 |
| GOTERM_CC_ALL | Intracellular
membrane-bounded organelle | 53 |
3.20×10−5 |
1.80×10−3 |
| GOTERM_CC_ALL | Membrane-bounded
organelle | 53 |
3.30×10−5 |
1.40×10−3 |
| GOTERM_CC_ALL | Intracellular
organelle | 55 |
2.40×10−5 |
8.00×10−3 |
| GOTERM_CC_ALL | Organelle | 55 |
2.50×10−5 |
7.00×10−3 |
| GOTERM_BP_ALL | Cellular
macromolecule metabolic process | 40 |
4.80×10−5 |
3.10×10−2 |
| GOTERM_BP_ALL | Macromolecule
metabolic process | 41 |
1.80×10−5 |
5.70×10−2 |
| GOTERM_BP_ALL | Metabolic
process | 49 |
2.50×10−5 |
5.30×10−2 |
Relevance of secreted LY6/PLAUR domain
containing 1 (SLURP1) and synapse defective 1 Rho GTPase homolog 2
(SYDE2) to clinical manifestations
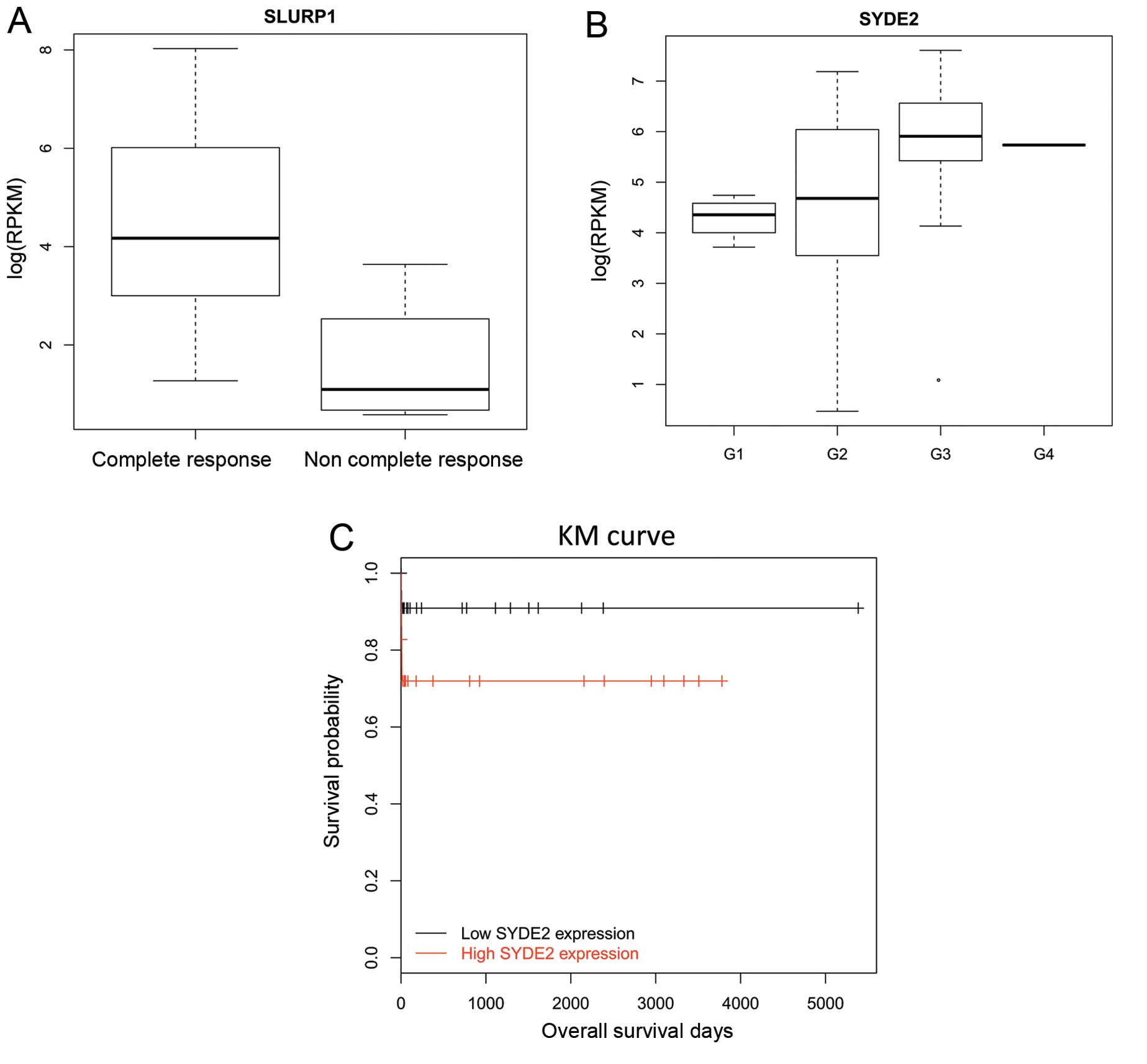
SLURP1 was the most upregulated gene, whereas SYDE2
was the second most downregulated gene. Their relevance to clinical
manifestations was explored. SLURP1 was correlated to the
pathological response (Fig. 6;
P=0.014, t-test), while SYDE2 was associated with the histological
grade (P=0.0028, ANOVA) and survival time (P=0.042, Log Rank test).
An upregulation of SLURP1 was observed in OC users and it was
linked to the reduced frequency of the complete response (Fig. 6A). Low expression of SYDE2 was
observed in patients with histological grade 1 tumors (Fig. 6B) and it was linked with a longer
survival time (Fig. 6C).
Therefore, it was believed that SLURP1 and SYDE2 were potential
therapeutic targets for cervical cancer. Additionally, the
inhibition of SYDE2 may improve the survival time.
Discussion
In the present study, the transcriptome of cervical
cancer patients with and without OC use were compared in order to
identify any relevant DEGs. A total of 80 genes were identified as
DEGs, while FDR=0.3 was set as the cut-off value. This finding
confirmed that OC use had a large impact on the gene expression in
the cervix and thereby stimulated the pathogenesis of cervical
cancer. GO enrichment analysis revealed that the metabolic process
was enriched in the DEGs. BioCarta analysis indicated that DEGs
were involved in the hTERC transcriptional regulation.
This finding was in accordance with previous
studies, which have indicated that OCs have an impact on the lipid
and carbohydrate metabolism (13).
Hormones present in OCs significantly affect the plasma lipoprotein
metabolism, which can raise the levels of plasma triglycerides,
low-density lipoprotein, and high-density lipoprotein 3. They also
affect the carbohydrate metabolism, primarily through the activity
of progestin, causing conditions including insulin resistance,
increases in plasma insulin levels and relative glucose intolerance
(14).
Further interesting DEGs were identified, including
adaptor-related protein complex 3 delta 1 (AP3D1). Downregulation
of genes encoding for subunits of adaptor complex-3 has been
reported in cervical cancer compared with normal controls (15). In the present study, AP3D1 was
identified to be further downregulated in the OC group compared
with non-OC group. AP3D1 is a subunit of the AP3 adaptor-like
complex, which is associated with the golgi region and more
peripheral structures. The AP-3 complex facilitates the budding of
vesicles from the golgi membrane, and may be directly involved in
trafficking to lysosomes (16).
BMI1 polycomb ring finger oncogene (BMI1) is a member of the
polycomb group, which participates in axial patterning,
hematopoiesis, cell cycle regulation and senescence. It may
contribute to malignant cell transformation and its overexpression
has been reported in cervical cancer (17,18).
However, according to the present study, BMI1 was downregulated in
OC users. Further studies on these genes may be helpful in
elucidating the molecular mechanism.
Telomerase is essential for the immortalization of
the majority of human cancer cells. A study by Guilleret et
al (19) indicated that hTERC
expression is regulated during carcinogenesis. This gene is often
amplified in certain types of solid human tumors, including the
cervix (20). Guo et al
(21) reported that hTERC
amplification testing is a promising evaluation method for cervical
cancer screening. This gene may be activated by the transcription
complex nuclear transcription factor (NF)-Y as well as the
transcription factors specificity protein (Sp)1 and retinoblastoma
protein (pRB) and could be repressed by Sp3 (22). In the present study, Sp3 was
downregulated in the OC group, which may contribute to the
increased incidence of cervical cancer through pathways including
hTERC. NF-Y alpha was also downregulated in OC users. Therefore,
further studies are required in order to elucidate the effect of OC
use on the hTERC transcriptional regulation.
SLURP1 and SYDE2 exhibited considerable fold changes
in OC users and thereby, their relevance to clinical manifestations
was investigated in the present study. The expression of SLURP1 was
identified to be negatively correlated with the pathological
response, whereas the expression of SYDE2 was negatively correlated
with the survival time. SLURP1 and SYDE2 may be potential
therapeutic targets for cervical cancer.
SLURP1 is a member of the Ly6/uPAR family that lacks
a GPI-anchoring signal sequence. It is thought that this secreted
protein contains antitumor activity. A study by Pettersson et
al (23) indicated that SLURP1
participates in the regulation of the gut immune functions and
motility, as well as possibly having a role in colon
carcinogenesis/cancer progression. Kalantari-Dehaghi et al
(24) also identified that
cancer-associated genes upregulation by nitrosamine
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone can be abolished by
SLURP1 (24). SYDE2, a GTPase
activator, has also been reported to be a tumor suppressor
(25). Methylated SYDE1 was
demonstrated to be significantly associated with the relapse-free
survival of patients with breast cancer (26) However, there are currently no
studies regarding the implications of SLURP1 and SYDE2 in cervical
cancer. Therefore, further studies should investigate their roles
in this cervical cancer.
In conclusion, the present study identified a range
of DEGs associated with OC use in patients with cervical cancer.
Further study of these genes may aid in elucidating the underlying
mechanism of the generation of cervical cancer associated with OC
use. Furthermore, SLURP1 and SYDE2 may be potential therapeutic
targets for cervical cancer.
References
|
1
|
Brake T and Lambert PF: Estrogen
contributes to the onset, persistence, and malignant progression of
cervical cancer in a human papillomavirus-transgenic mouse model.
Proc Natl Acad Sci USA. 102:2490–2495. 2005. View Article : Google Scholar
|
|
2
|
Kjellberg L, Hallmans G, Hren A, et al:
Smoking, diet, pregnancy and oral contraceptive use as risk factors
for cervical intra-epithelial neoplasia in relation to human
papillomavirus infection. Br J Cancer. 82:13322000.PubMed/NCBI
|
|
3
|
Ness RB, Grisso JA, Klapper J, et al: Risk
of ovarian cancer in relation to estrogen and progestin dose and
use characteristics of oral contraceptives. Am J Epidemiol.
152:233–241. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Smith JS, Green J, De Gonzalez AB, et al:
Cervical cancer and use of hormonal contraceptives: a systematic
review. The Lancet. 361:1159–1167. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sasieni P: Cervical cancer and hormonal
contraceptives: Collaborative reanalysis of individual data for 16
573 women with cervical cancer and 35 509 women without cervical
cancer from 24 epidemiological studies. Commentary Lancet.
370:2007.
|
|
6
|
Moodley M, Moodley J, Chetty R and
Herrington C: The role of steroid contraceptive hormones in the
pathogenesis of invasive cervical cancer: a review. Int J Gynecol
Cancer. 13:103–110. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rivera R, Yacobson I and Grimes D: The
mechanism of action of hormonal contraceptives and intrauterine
contraceptive devices. Am J Obstet Gynecol. 181:1263–1269. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guven S, Kart C, Guvendag Guven ES and
Serdar Gunalp G: The underlying cause of cervical cancer in oral
contraceptive users may be related to cervical mucus changes. Med
Hypotheses. 69:550–552. 2007. View Article : Google Scholar
|
|
9
|
The R Development Core Team. R: A language
and environment for statistical computing. http://www.R-project.org.
Accessed March 26, 2013
|
|
10
|
Pounds S and Morris SW: Estimating the
occurrence of false positives and false negatives in microarray
studies by approximating and partitioning the empirical
distribution of p-values. Bioinformatics. 19:1236–1242. 2003.
View Article : Google Scholar
|
|
11
|
Da Wei Huang BTS and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2008.PubMed/NCBI
|
|
12
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc Series B Stat Methodol. 289–300.
1995.
|
|
13
|
Godsland IF, Crook D, Simpson R, et al:
The effects of different formulations of oral contraceptive agents
on lipid and carbohydrate metabolism. N Engl J Med. 323:1375–1381.
1990. View Article : Google Scholar
|
|
14
|
Krauss RM and Burkman R Jr: The metabolic
impact of oral contraceptives. Am J Obstet Gynecol. 167:11771992.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Petrenko A, Pavlova L, Karseladze A,
Kisseljov F and Kisseljova N: Downregulation of genes encoding for
subunits of adaptor complex-3 in cervical carcinomas. Biochemistry
(Moscow). 71:1153–1160. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Odorizzi G, Cowles CR and Emr SD: The AP-3
complex: a coat of many colours. Trends Cell Biol. 8:282–288. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Honig A, Weidler C, Häusler S, et al:
Overexpression of polycomb protein BMI-1 in human specimens of
breast, ovarian, endometrial and cervical cancer. Anticancer Res.
30:1559–1564. 2010.PubMed/NCBI
|
|
18
|
Gavrilescu MM, Todosi AM, Anitei MG, Filip
B and Scripcariu V: Expression of bmi-1 protein in cervical, breast
and ovarian cancer. Rev Med Chir Soc Med Nat Iasi. 116:1112–1117.
2012.PubMed/NCBI
|
|
19
|
Guilleret I, Yan P, Guillou L,
Braunschweig R, Coindre J-M and Benhattar J: The human telomerase
RNA gene (hTERC) is regulated during carcinogenesis but is not
dependent on DNA methylation. Carcinogenesis. 23:2025–2030. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cairney C and Keith W: Telomerase
redefined: integrated regulation of hTR and hTERT for telomere
maintenance and telomerase activity. Biochimie. 90:13–23. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Guo Q, Sui L and Feng Y: Cervical cancer
screening: hTERC gene amplification detection by FISH in comparison
with conventional methods. Open Journal of Obstetrics and
Gynecology. 2:11–17. 2012. View Article : Google Scholar
|
|
22
|
Zhao JQ, Glasspool RM, Hoare SF, et al:
Activation of telomerase rna gene promoter activity by NF-Y, Sp1,
and the retinoblastoma protein and repression by Sp3. Neoplasia.
2:531–539. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Pettersson A, Nordlander S, Nylund G, et
al: Expression of the endogenous, nicotinic acetylcholine receptor
ligand, SLURP-1, in human colon cancer. Auton Autacoid Pharmacol.
28:109–116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kalantari-Dehaghi M, Bernard HU and Grando
SA: Reciprocal effects of NNK and SLURP-1 on oncogene expression in
target epithelial cells. Life Sci. 91:1122–1125. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iorns E, Ward TM, Dean S, et al: Whole
genome in vivo RNAi screening identifies the leukemia inhibitory
factor receptor as a novel breast tumor suppressor. Breast Cancer
Res Treat. 135:79–91. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hill VK, Ricketts C, Bieche I, et al:
Genome-wide DNA methylation profiling of CpG islands in breast
cancer identifies novel genes associated with tumorigenicity.
Cancer Res. 71:2988–2999. 2011. View Article : Google Scholar
|