Introduction
The process of parthenogenesis is the development an
egg into an embryo without the participation of sperm. This form of
reproduction exists in reptiles, fish and birds, but not in
mammals. A mammalian parthenogenetic embryo cannot develop into a
fetus and is usually arrested at a particular developmental stage
(1). This developmental arrest
probably correlates to a lack of paternally imprinted genes
(1,2). However, the eggs of mammals can be
activated in vitro and develop into blastocysts. The inner
cell mass may be used to create embryonic stem cells (ESCs), known
as parthenogenetic embryonic stem cells (pESCs). Previous studies
(3,4) have reported that pESCs have been
successfully established in rats and primates. Revazova et
al (5) reported the first
successful derivation of human pESCs (hpESCs) in 2007, following
this, numerous groups have also achieved hpESC derivation (6, 7).
A major concern ahead of the clinical utilization of
hpESCs is whether these cells hold the same differentiation ability
as normal hESCs; as a chimerical study showed that pESCs only exist
in limited tissues (8). Therefore
the differentiation potential of hpESCs into specific terminal
cells needs to be tested further. Numerous data have shown that
pESCs from rodents and non-human primates can differentiate into
mid-brain dihydroxyphenyl-ethylamine neurons (9), hepatic endoderms, hematopoietic cells
including CD45+ cells, lymphocytes, monocytes and akaryocyte-like
cells (10), cardiocytes,
lipocytes and epithelium (11).
pESCs are derived entirely from maternal genes. It
was previously determined that there is a correlation between the
expression of imprinted genes and the likelihood whether pESCs will
develop into functional tissues or not (12). Due to the lack of paternal genes,
pESCs do not express paternally imprinted genes, including
insulin-like growth factor 2 (IGF2), which is the main growth
factor involved in the promotion of mitosis (13). The expression of IGF2 is also
essential for the long-term proliferation of all cell types
(14). Previously, the endogenous
expression levels of IGF2 were shown to be varied between mouse
pESCs, androgenetic ESCs and normal ESCs (11). In addition, studies have shown that
the differentiation potential of pESCs from nuclear transplantation
in vivo and in vitro were significantly enhanced as
compared with primitive pESCs (~2–5 times) (15). Therefore, it needs to be confirmed
whether the differentiation potential of hpESCs differs from that
of hESCs.
Therefore, the present study aimed to confirm
whether hpESCs can be induced into islet-like clusters (ILCs) and
compare the difference between normal ESCs and hpESCs in this
differentiation progress.
Materials and methods
Culture and differentiation
The present study was approved by the Ethics
Committee of Central South University, Changsha, China.
Undifferentiated chHES8 (normal ESCs), chHES32 (hpESCs) and chHES69
(hpESCs) were established, according to previous methods (16), and were maintained on human
embryonic fibroblasts in Dulbecco’s Modified Eagle Medium/Nutrient
Mixture F12 (DMEM/F12) (Invitrogen Life Technologies, Carlsbad, CA,
USA) supplemented with 15% (vol/vol) KnockOut™ serum replacement, 1
mM non-essential amino acids, Glutamax™, 0.1 mM β-mercaptoethanol
(Invitrogen Life Technologies), and 4 ng/ml recombinant human
fibroblast growth factor (FGF2) (Invitrogen Life Technologies,
Minneapolis, MN, USA). Cultures were manually passaged at a 1:4
ratio at seven day intervals, and began to differentiate on the
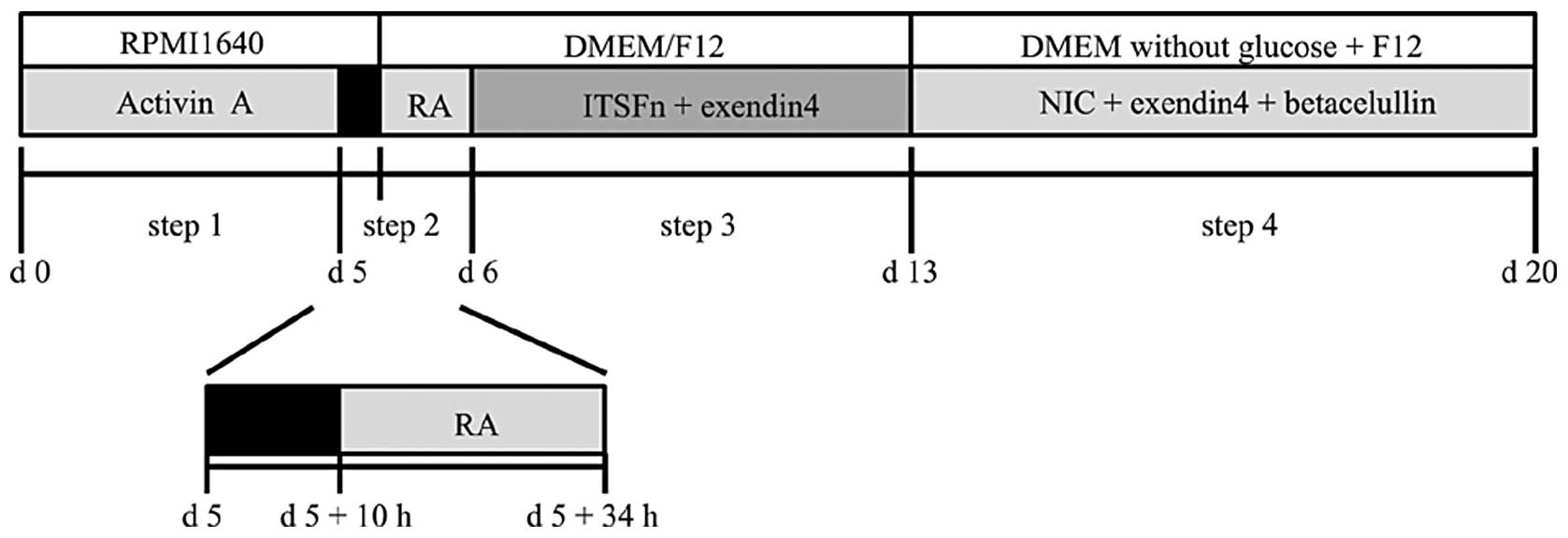
fifth day following the last passage. The induction protocol was
divided into four steps. (i) Activin A (100 ng/ml, R&D Systems,
Minneapolis, MN, USA) and low dosage Hyclone™ serum (GE Healthcare,
South Logan, UT, USA) were used to generate definitive endoderm
(DE) from hpESCs for five days in the first stage of the process
(17). (ii) Following withdrawal
of the Activin A and serum, the cells were cultured in RPMI-1640
medium for an interval of 10 hours, followed by the addition of
retinoic acid (RA, 10−5 M, Sigma-Aldrich, St Louis, MO,
USA) for 24 hours to initiate the pancreatic lineage specification.
(iii) A mixture of 1% ITS (100x), fibronectin (5 μg/ml) and
Exendin-4 (Ex-4, 50 ng/ml, Sigma-Aldrich) were added to the medium
for one week, in order to further differentiate the cells into
pancreatic precursor cells. (iv) The pancreatic precursor cells
were cultured in suspension with a medium containing 1%
N2 (100×), 1% B27 (50x, Gibco-BRL, Carlsbad, CA, USA),
nicotinamide (NIC, 10−2 M), Ex-4 (50 ng/ml) and
betacellulin (20ng/ml, R&D Systems) for an additional week to
obtain insulin producing cells. The induction method can be seen in
Fig. 1.
Semi quantitative polymerase chain
reaction (qPCR)
Total RNA was extracted using the TRIzol®
reagent (Invitrogen Life Technologies) and cDNA was synthesized
from 1 μg of total RNA using random primers and the Reverse
Transcriptase kit (Thermo Fisher Scientific, Rockford, IL, USA).
All PCR reactions were performed using Taq DNA polymerase with
various annealing temperatures and cycle numbers in a total
reaction volume of 10 μL. PCR products were separated using 2%
agarose gels and visualized with ethidium bromide staining. The
primer pairs and expected amplicon sizes are listed in Table 1.
 | Table IPrimer sequences used in the
quantitative polymerase chain reaction |
Table I
Primer sequences used in the
quantitative polymerase chain reaction
| Gene | Size (bp) | Sequence of primer
(5′-3′) | Temp (°C) | Cycles |
|---|
| Sox17 | 292 | F:
GGCGCAGCAGAATCCAGA | | |
| | R:
CCACGACTTGCCCAGCAT | 58.0 | 30 |
| Foxa2 | 588 | F:
GGGAGCGGTGAAGATGGA | | |
| | R:
TCATGTTGCTCACGGAGGAGTA | 57.5 | 30 |
| Pdx1 | 217 | F:
GGATGAAGTCTACCAAAGCTCACGC | | |
| | R:
CCAGATCTTGATGTGTCTCTCGGTC | 65.0 | 30 |
| Insulin | 244 | F:
CAGTGACCTGTCTTGGTTTTCCG | | |
| | R:
CAGCCGAGTAGTTTTCATCATTGC | 65.0 | 30 |
| Glucagon | 307 | F:
AGGCAGACCCACTCAGTGA | | |
| | R:
AACAATGGCGACCTCTTCTG | 55.0 | 30 |
| Somatostatin | 179 | F:
GTACTTCTTGGCAGAGCTGCTG | | |
| | R:
CAGAAGAAATTCTTGCAGCCAG | 55.0 | 30 |
| Amylase | 358 | F:
CTGACAACTTCAAAGCAAA | | |
| | R:
TACAGCATCCACATAAATACGA | 57.0 | 30 |
| GCK | 376 | F:
AGGGAATGCTTGCCGACTC | | |
| | R:
CACTGGCCTCTTCATGGGT | 57.1 | 30 |
| PC2 | 314 | F:
GCATCAAGCACAGACCTACACTCG | | |
| | R:
GAGACACAACCACCCTTCATCCTTC | 60.5 | 30 |
| PC1/3 | 456 | F:
TTGGCTGAAAGAGAACGGGATACATCT | | |
| | R:
ACTTCTTTGGTGATTGCTTTGGCGGTG | 65.4 | 30 |
| Kir6.2 | 499 | F:
CGCTGGTGGACCTCAAGTGGC | | |
| | R:
CCTCGGGCTGGTGGTCTTGCG | 65.0 | 30 |
| SUR1 | 429 | F:
GTGCACATCCACCACAGCACATGGCTTC | | |
| | R:
GTGTCTTGAAGAAGATGTATCTCCTCAC | 62.1 | 30 |
| Oct3/4 | 168 | F:
CTTGCTGCAGAAGTGGGTGGAGGAA | | |
| | R:
CTGCAGTGTGGGTTTCGGGCA | 64.0 | 28 |
| Nanog | 387 | F:
ACTGTCTCTCCTCTTCCCTCCTCC | | |
| | R:
GTAGAGGCTGGGGTAGGTAGGTG | 64.0 | 28 |
| Hb9 | 403 | F:
GCGCTCTCCTACTCGTACCC | | |
| | R:
CTTCTGTTTCTCCGCTTCCTG | 60.9 | 35 |
| Hnf6 | 457 | F:
AGTAATTCAGGGCAGATGGAAG | | |
| | R:
CGTTCATGAAGAAGTTGCTGAC | 56.0 | 35 |
| Cxcr4 | 80 | F:
CACCGCATCTGGAGAACCA | | |
| | R:
GCCCATTTCCTCGGTGTAGTT | 50.0 | 28 |
| Sox1 | 464 | F:
CAATGCGGGGAGGAGAAGTC | | |
| | R:
CTCTGGACCAAACTGTGGCG | 53.0 | 30 |
| Krt17 | 119 | F:
GGAGATTGCCACCTACCG | | |
| | R:
TGCCATCCTGGACCTCTT | 60.0 | 30 |
| Brachyury | 329 | F:
ACCCAGTTCATAGCGGTAGC | | |
| | R:
CAATTGTCATGGGATTCAG | 55.0 | 30 |
| FLK | 721 | F:
GAGGGCCACTCATGGTGATTGT | | |
| | R:
TGCCAGCAGTCCAGCATGGTCTG | 55.0 | 28 |
| Scl1 | 331 | F:
ATGGTGCAGCTGACTCCTCC | | |
| | R:
TCTCATTCTTGCTGAGCTTC | 55.0 | 35 |
| Runx1 | 516 | F:
CAGTGACCTGTCTTGGTTTTCCG | | |
| | R:
CAGCCGAGTAGTTTTCATCATTGC | 60.0 | 35 |
| IGF2 | 86 | F:
TCCCCTGATTGCTCTACCCA | | |
| | R:
GCAGTTTTGCTCACTTCCGATT | 58.0 | 28 |
| KCNQ10STSNP1 | 466 | F:
CAGCCACCTCTGTGGCGTGAATGTTCT | | |
| | R:
GCTCAAACCCGTCTCTGAAATGCACGG | 55.0 | 35 |
| SNRPN | 112 | F:
TGGCACCTTTAAGGCTTTTG | | |
| | R:
CCGCTTTTCTTCACGCTCT | 58.0 | 28 |
| IPW | 868 | F:
GGGAACTCTTCTGGGAGTGAATGTTATCA | | |
| | R:
GGGAGGTTCATTGCACAGAAATTTGG | 55.0 | 28 |
| H19 | 142 | F:
CCGGACACAAAACCCTCTAGCT | | |
| | R:
TGTTCCGATGGTGTCTTTGATG | 58.0 | 28 |
| CDKNIC | 146 | F:
TGGGACCGTTCATGTAGC | | |
| | R:
GGACCAGTGTACCTTCTCG | 50.5 | 28 |
| NES55 | 1141 | F:
TCGGAATCTGACCACGAGCA | | |
| | R:
CACGAAGATGATGGCAGTCAC | 55.0 | 35 |
| IGF1R | 540 | F:
GAATGGAGTGCTGTATGCCTCTGTGAACC | | |
| | R:
GTGAAATCTTCGGCTACCATGCAATTCCG | 55.0 | 28 |
| IGF2R | 284 | F:
GTTGTCTGCCCTCCAAAGAA | | |
| | R:
CCTTTGGAGTACGTGACAAG | 55.0 | 28 |
| β-actin | 200 | F:
CGCACCACTGGCATTGTCAT | | |
| | R:
TTCTCCTTGATGTCACGCAC | 60.0 | 28 |
Immunofluorescence staining
The cells were harvested on days 5, 13, and 20, and
were fixed in phosphate-buffered saline (PBS) containing 4%
paraformaldehyde for 15 min at room temperature, followed by three
washes with PBS containing 0.1% bovine serum albumin (BSA). The
cells were permeabilized using 0.1% Triton X-100 in PBS containing
0.1% BSA and 4% normal goat serum (Gibco-BRL), or 10% donkey serum
for Sox17. The cells were incubated with the primary antibodies
overnight at 4ºC, followed by a 1 h incubation with the secondary
antibodies at room temperature. The following antibodies and
dilutions were used: Goat anti-human sox17, 1:40 (R&D Systems);
guinea pig anti-human pdx1, 1:200 (Abcam, Cambridge, MA, USA);
mouse anti-human insulin l:200 (Sigma-Aldrich). Donkey anti-goat
antibody was used at 1:300 (Sigma-Aldrich); goat anti-guinea pig
antibody was used at 1:500 (abcam) and fluorescein isothiocyanate
anti-human insulin antibody was used at 1:400 (Chemicon, Temecula,
CA, USA). The cells were mounted in ten random fields of vision
using DAPI (BD Biosciences, Franklin Lakes, NJ, USA) dye, and
examined using a fluorescence microscope (Nikon Inc., Melville, NY,
USA). Each vision contained >200 cells and totaled >2000
cells per sample.
Insulin release assay
The ILCs were transferred into a four-well dish and
the number of clusters was recorded. The clusters were washed twice
with Hank’s Balanced Salt Solution (HBSS) containing 0.5% human
serum albumin (HSA), for 10 minutes each time. Following the wash
steps, the clusters were pre-incubated for 30 minutes in medium
containing 5.5 mM glucose. Subsequently, the clusters were
incubated in 5.5 and 25.0 mM glucose for one hour and stored at
−20ºC. The insulin released into the medium was detected using the
Insulin kit (12017547; Roche Diagnostics GmbH, Mannheim, Germany)
and Roche E170 equipment. According to the standard curve, the
detection range was between 2.6 and 24.9μU/ml.
Proliferation assay
The cells were harvested on differentiation days 5,
13 and 20, and fixed in PBS containing 4% paraformaldehyde for 15
min, followed by three washes with PBS containing 0.1% BSA. The
cells were incubated with anti-Ki67 antibody (mouse anti-human,
Sigma-Aldrich, dilution 1:10) for 30 min at room temperature.
Following the initial incubation with the primary antibody, the
cells were incubated with the appropriate secondary antibody, as
described previously. The cells were mounted in 10 random fields of
vision using DAPI. Each vision contained >200 cells and totalled
>2000 cells/sample.
Statistical analyses
All experiments were repeated three times and data
are expressed as the mean±standard deviation. Statistical analyses
were performed using the Student’s t-test. P<0.05 was considered
to indicate a statistically significant difference.
Results
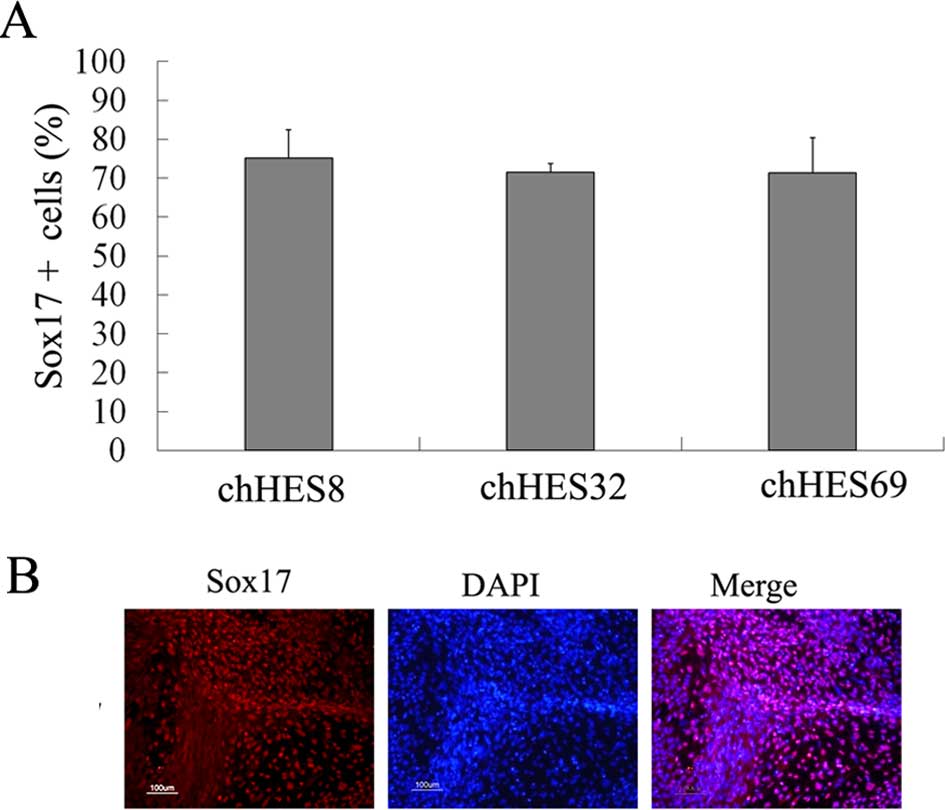
Generation of definitive endoderm (DE)
cells
HpESCs were treated with 100 ng/ml Activin A and a
low concentration of serum in order to induce DE. After 5 days,
endoderm-specific Sox17 and Foxa2 genes were shown to be expressed.
Expression of the mesoderm-related gene brachyury, and the
ectoderm-related gene Pax6 were not detected on day 5. Furthermore,
there were no pancreatic-related genes detected. The DE marker
Sox17 reached ~71.6±2.1% on day 5 (Fig. 2A and B)
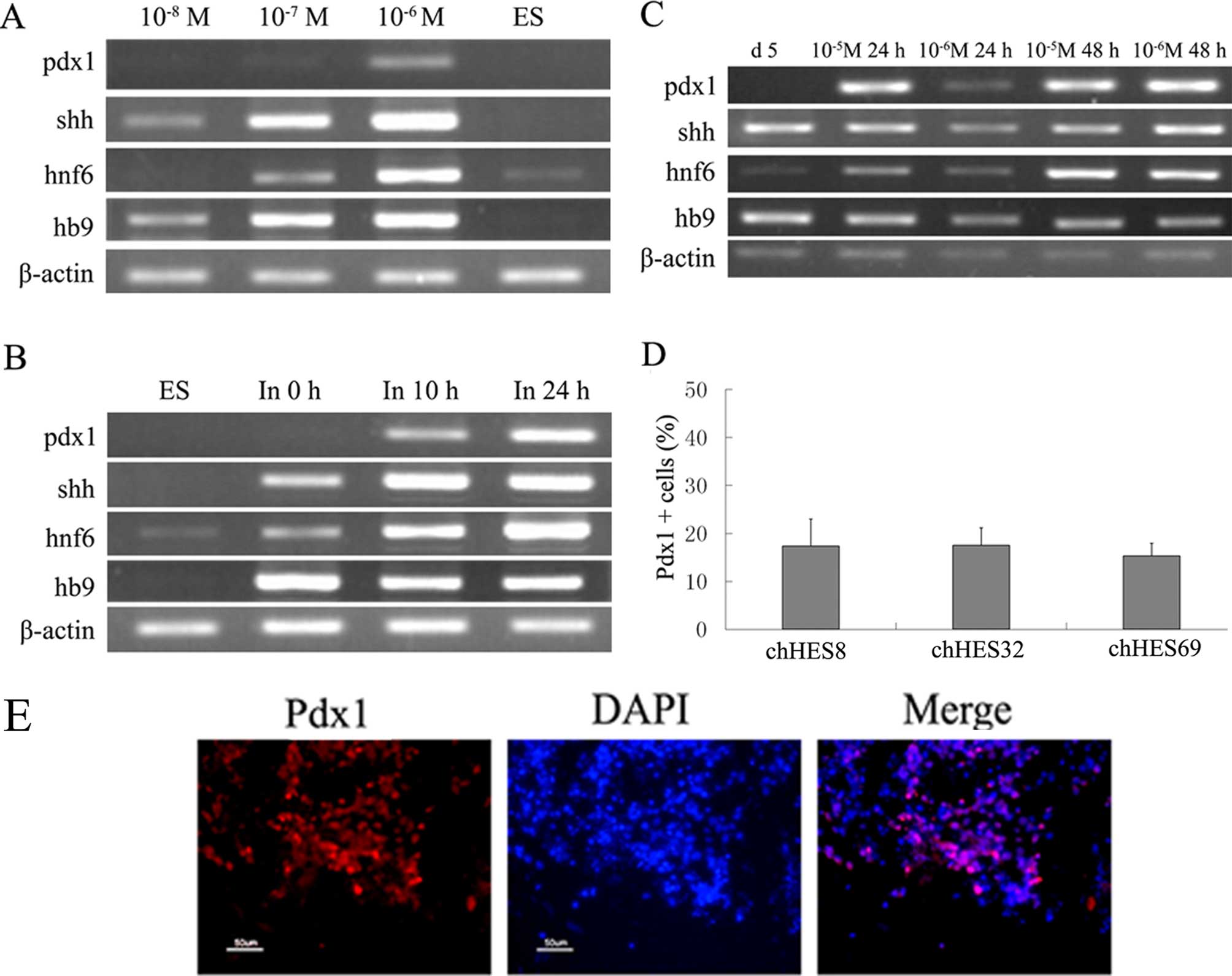
Differentiation of pancreatic precursor
cells
After treatment with Activin A for 5 days, cells
were transferred into media containing L-glutamine and RPMI-1640
for ~10 hours. Fresh media containing DMEM/F12 and RA was used to
initiate of pancreatic specialization. Retinoic acid (RA) was used
at several concentrations (10−4, 10−5,
10−6, 10−7 and 10−8 M). RA was
shown to rapidly upregulate the gene expression levels of Pdx1 when
the concentration of the RA was >10−6 M. However, at
concentrations approaching 10−4 M, cell death was
observed and the cultures could not continue (data not shown).
Suitable concentrations of RA were chosen (10−5 M and
10−6 M) to determine the gene expression levels of Pdx1
over varying durations (24 and 48 hours). Following a 24 hour
period of treatment, the gene expression levels of Pdx1, in the
cells treated with 10−5 M RA was stronger as compared
with the expression levels when the cells were treated with
10−6 M RA. When the treatment lasted 48 hours, there was
no significant difference in the expression levels, between the two
concentrations. The expression levels of Pdx1 were the same between
the two time periods when the cells were treated with
10−5 M RA (Fig. 3A–C),
however more dead cells appeared following 48 hours of treatment,
as compared with 24 hours of treatment (data not shown).
For further differentiation, 1% ITS and 5 μg/ml
fibronectin was added to the medium for 7 days for the
proliferation of pancreatic progenitors. It has previously been
reported that Ex-4 enhanced the expression levels of Pdx1 and Ngn3
during β cell regeneration in STZ-treated mice (18), and 50 ng/ml Ex-4 was used for
pancreatic hormone-expressing endocrine cell specification
(19). Therefore, the present
study selected Ex-4 as an accelerant for further differentiation of
the pancreatic precursor cells. On day 13 Pdx1, the marker of
pancreatic progenitors, was shown to be upregulated, and ~17.5±3.7%
Pdx1 positive cells were detected (Fig. 3D and E). Furthermore,
mesoderm-related genes including Flk1, were detected on day 13,
however ectoderm-related genes, such as Krt17, were not detected
(Fig. 4B). The imprinting gene,
including IGF12 and H19 were active (Fig. 4C).
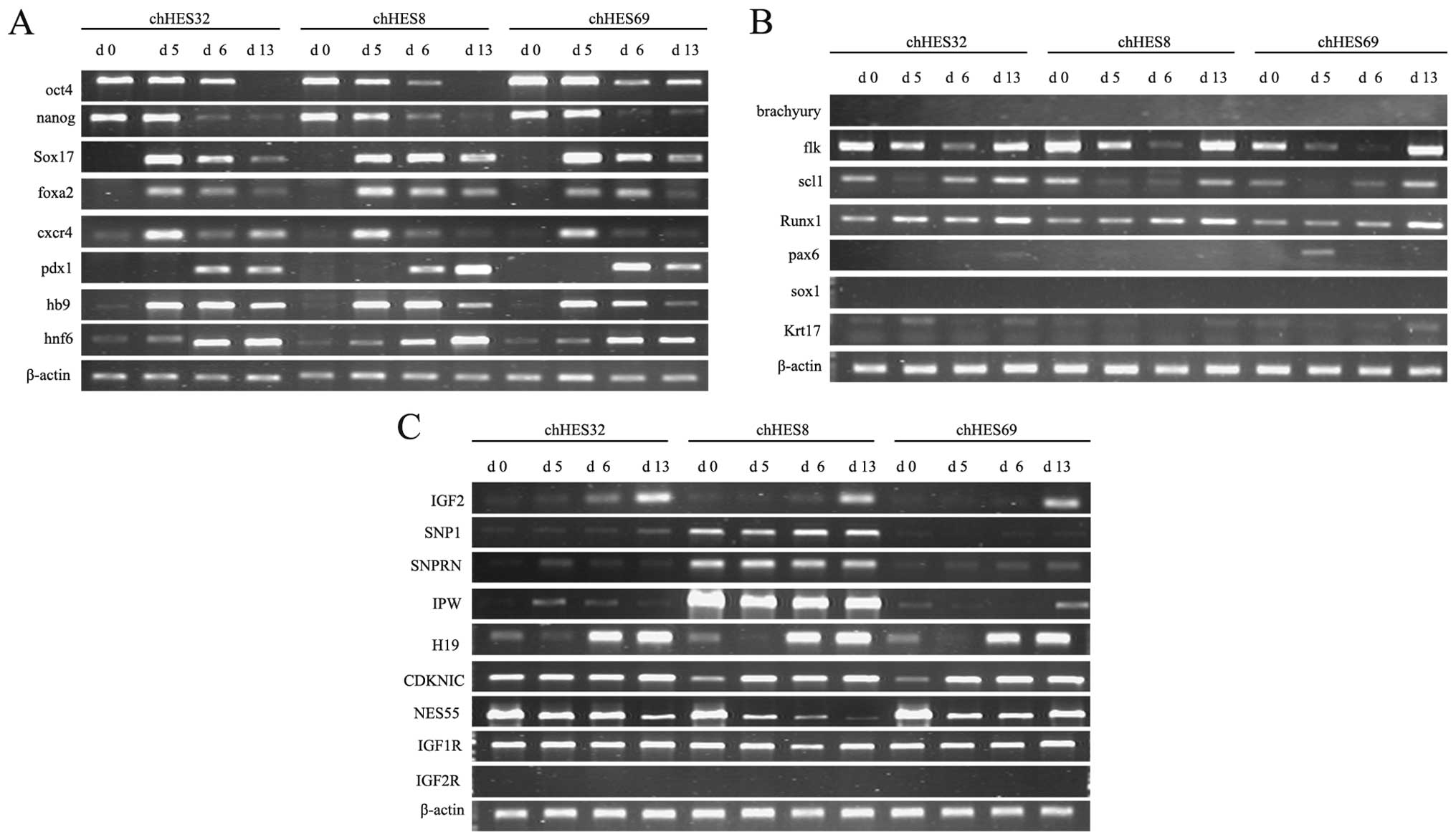 | Figure 4Expression of marker genes, mesoderm
and ectoderm-related genes, and imprinted genes on days 0, 5, 6 and
13, as determined by quantitative polymerase chain reaction. (A)
The expression levels of marker genes for embryonic stem cells
(ESCs) (Oct4, Nanog), definitive endoderm (Sox17, Foxa2, Cxcr4) and
pancreatic precursors (Pdx1, Hb9, Hnf6) in the three embryonic stem
cell lines. (B) The expression levels of marker genes of the
mesoderm (Brachyury, Flk, Scl1, Runx1) and ectoderm (Pax6, Sox1,
Krt17) in the three embryonic stem cell lines. (C) The expression
levels of paternally imprinted (IGF2, SNP1, SNPRN, IPW), maternally
imprinted (H19, CDKNIC, NES55), and receptor (IGF1R, IGF2R) genes
on days 5, 13 and 20, in the three embryonic stem cell lines. DE,
defined endoderm; chHES8, normal human ESCs; chHES32, human
parthenogenetic ESCs; chHES69, human parthenogenetic ESCs. |
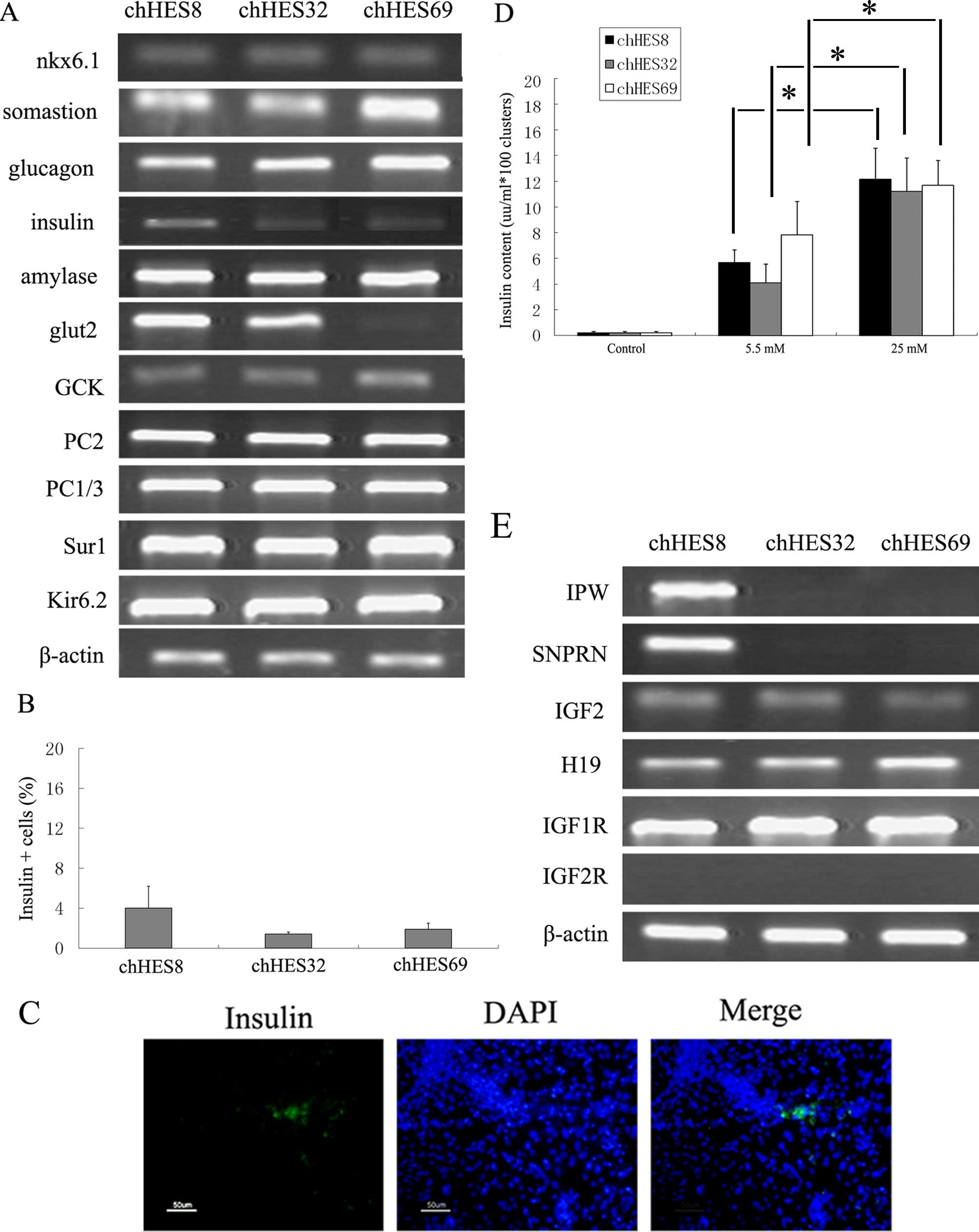
Formation of ILCs
On day 20, the pancreatic markers insulin, glucagon,
somatostatin and the insulin secretion-related genes, Kir6.2 and
PC1/3 were shown to be upregulated (Fig. 5A). The expression levels of these
marker genes were consistent with the progression of pancreatic
development. There were ~4.0±2.2% insulin-positive cells on day 20
(Fig. 5B and C). In the insulin
release test, the culture medium prior to stimulation with glucose
was used as the control sample. The insulin content in the control
group was 0.02 μU/ml. Upon treatment of the cells with 5.5 and 25
mM glucose, the contents of insulin increased to 8.23±2.3 μU/ml and
17.36±2.4 μU/ml, respectively. The insulin content of the media was
1.5–4 times higher in the high glucose concentration group, as
compared with the insulin content of the media obtained from the
low glucose concentration group. The results indicated that the
insulin release of the hpES-induced clusters corresponded to
variations in the glucose concentration (Fig. 5D).
Expression of imprinted genes and cell
proliferation assay
Various imprinted genes were detected in the three
ES cell lines. Paternally imprinted genes (SNP1, SNRPN, IPW) were
detected in chHESC8, but not in chHESC32 and chHESC69 cells.
Maternally imprinted genes (H19, CDKNIC, NES55) were expressed in
all three of the ES cell lines. The IGF2 gene has been reported not
to be expressed in hPESCs (8,20).
However, in the present study, IGF2 was expressed in all three of
the ES cell lines, with the expression being initiated on day 6 and
lasting until day 20. The expression levels of IGF1R and IGF2R were
also determined. Expression of IGF1R was observed in all of the ES
cell lines, whereas IGF2R was not detected in any of the cell lines
(Figs. 4C and 5E).
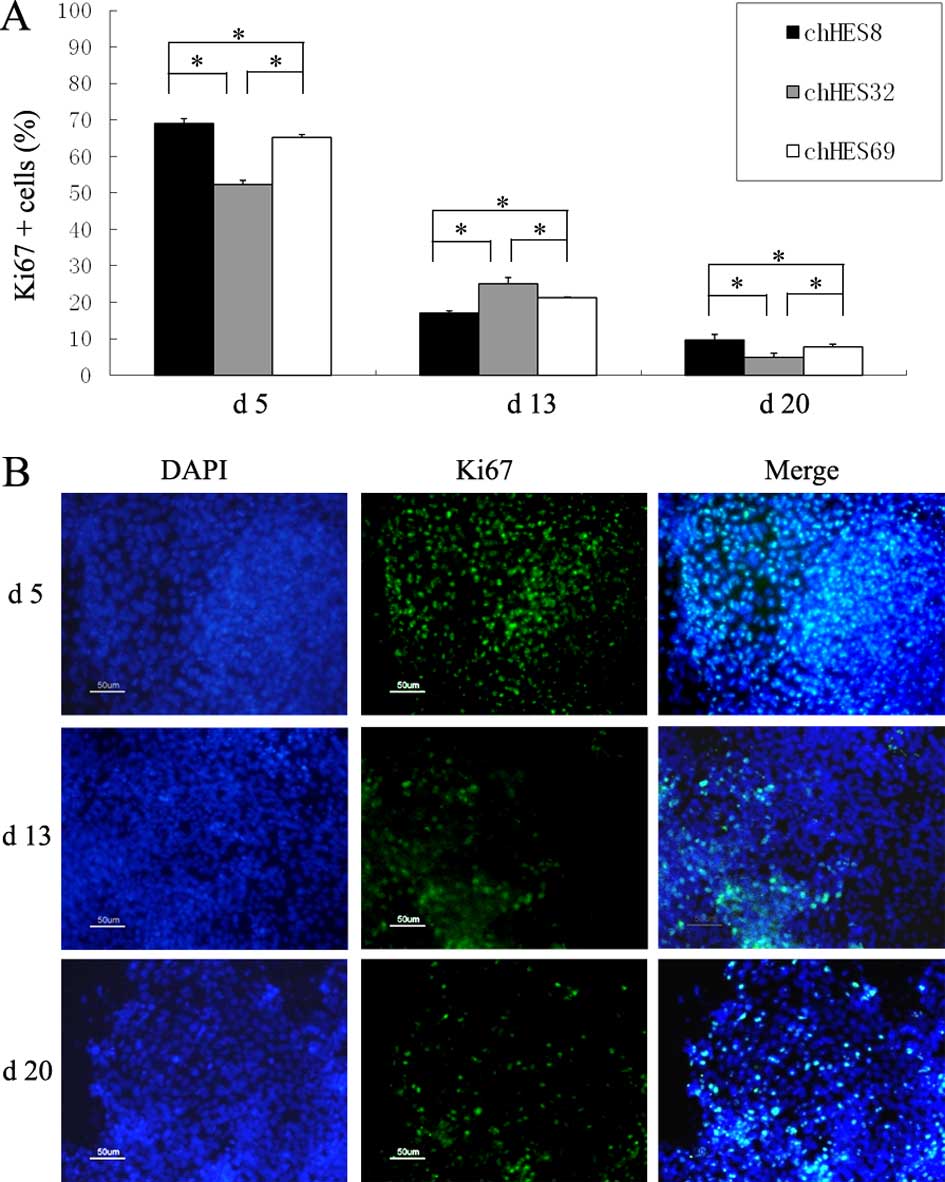
Ki67 was chosen as the marker for the evaluation of
the proliferative ability of hpESCs on days 5, 13, and 20. The
results of the present study demonstrated that the proliferative
ability of hpESCs reduced gradually, along with the extension of
induction time. There were statistically significant differences
between the three ES cell lines at the same stage of every
differentiation (Fig. 6A and
B).
Discussion
Mammals can not reproduce by parthenogenesis due to
the absence of paternal genetic material; the development of the
mammalian embryo is dependent on the expression of the paternal
genes. Embryological studies, using mouse parthenogenetic embryos,
have demonstrated that they can only develop to an early stage, and
that development arrests following 10 days gestation (21–24).
In humans, a lack of some paternally imprinted genes will lead to
developmental delay and mental retardation, manifesting in
conditions including Prader-Willi Syndrome. Genetic imprinting has
an important role in the progression of human development. However,
there is currently no clear way of assessing the extent to which
the expression and regulation of imprinted genes influences
development, or the role of imprinted genes in differentiation, due
to a lack of suitable in vitro research models. hpESCs offer
a promising model for study in vitro. pESCs have a similar
capacity for totipotency and proliferation as normal ESCs (25). The present study demonstrated that
hpESCs can be induced into ILCs. These results support further
study of pESCs and offer a feasible method for their introduction
into a clinical setting.
For differentiation, Activin A was found to be a key
factor in DE specification( 17),
which acts in the same manner as the DE differentiation from
hPESCs. A previous study reported that RA is important in the
anteroposterior patterning of neuroectoderm and mesoderm in
vertebrates (26). RA is also
involved in the regulation of early pancreas bud formation and it
is able to improve the expression of insulin in pancreatic β cells
and in the INS-1 cell line (27,28).
RA and FGF4 direct the differentiation of hESCs into foregut
endoderm in a time- and concentration-dependent manner (29). In the present study, the initiation
of pancreas progenitors was achieved by adding 10-6M RA for 48 h
after a 10 h intermission following 5 days DE formation. The
results demonstrated that the induced ILCs expressed islet-specific
hormones and associated functional markers. Additionally,
assessment of insulin release demonstrated that the degree of
insulin release corresponded with alterations in glucose
concentration. These results confirm that, following 20 days
differentiation, functional insulin-producing cells were obtained
from the hPESCs.
At a different differentiation stage, no
statistically significant differences were observed in the
expression level of the stage specific markers sox17, pdx1 and
insulin among the three ES cell lines. However, the expression
level of Ki67 was higher on day 5 and day 20 in the normal hESCs
compared with the hPESCs. It was previously reported that the
proliferative ability of partheno source fibroblasts was lower
compared with the proliferative ability of normal fibroblasts
(14). Whether hPESCs develop into
functional cells and organs or not is associated with
characteristics of gene imprinting (12). The paternally imprinted genes SNP1,
SNRPN and IPW were detected in the chHESC8 cells, but not in the
chHESC32 or chHESC69 cells. The maternally imprinted genes H19,
CDKNIC and NES55 were expressed in all three of the ES cell lines.
The expression of these imprinted genes was consistent with
previously observed characteristics of hPESCs (8,20).
The present study found that the paternal gene IGF2
recovered its expression levels in hpESCs during the
differentiation process, resulting in similar expression levels to
normal ESCs. It has been observed that mouse β cells overexpressing
IGF2 have higher levels of cell proliferation and islet
hypertrophia (30). It has been
suggested that IGF2 may be important in the determination of
endocrine cell fate, proliferative ability and amylon metabolism in
the perinatal period (31). A
previous study compared the gene expression levels of IGF2, H19,
IGF2R and SNRPN in androgenetic ESCs, pESCs and normal ESCs
(20). The expression levels of
these genes in pESCs and androgenetic ESCs was found to differ from
the normal ESCs. The markers of genomic imprinting were lost in the
somatic cells of the parthenogenetic embryonic chimera, but were
maintained in its stem cells. The loss of imprinting markers
correlates with the expression levels of imprinted genes (32). Other studies have also suggested
that the biallele methylation difference of IGF2 is established
gradually throughout development, including the postnatal period.
Previous data have revealed that the expression of IGF2 is
monallelic in the fetal stage of human and mouse development
(33, 34). In the human IGF2 gene, there are
four independent promoters switching on allele expression in the
paternal chromosome in an individual developmental period (33–35).
Following birth, the distal promoter of the human hepatic IGF2 was
shown to be active on both chromosomes, which leads to biallelic
expression in adults. It was presumed, in the present study, that
promoters of maternal IGF2 were activated during the
differentiation of hpESCs. IGF2 promoter activity may be influenced
by the external environment, resulting in the recovery of the
absent IGF2 expression and the ensuing effect of IGF2 in pancreatic
development.
In conclusion, parthenogenetic activation of human
oocytes is a novel method and may be used to produce
histocompatible cells for cell-based therapy. Furthermore, stem
cells derived from human parthenogenetic embryos could alleviate
some of the ethical concerns over embryonic and stem cell research,
and hpESCs provide a valuable in vitro model to explore the
influence of imprinted genes on cell differentiation. It will be
important to confirm the safety of this treatment at the epigenetic
level, prior to formal clinical application (36).
Acknowledgements
This work was supported by the National Basic
Research Program of China (no. 2011CB964900) and the National High
Technology Research and Development Program of China. (nos.
2011AA020113 and 2006AA02A102).
References
|
1
|
Brevini TA and Gandolfi F: Parthenotes as
a source of embryonic stem cells. Cell Prolif. 41:20–30. 2008.
|
|
2
|
Kono T: Genomic imprinting is a barrier to
parthenogenesis in mammals. Cytogenet Genome Res. 113:31–35. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shao H, Wei Z, Wang L, et al: Generation
and characterization of mouse parthenogenetic embryonic stem cells
containing genomes from non-growing and fully grown oocytes. Cell
Biol Int. 31:1336–1344. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vrana KE, Hipp JD, Goss AM, et al:
Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci
USA. 100:11911–11916. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Revazova ES, Turovets NA, Kochetkova OD,
et al: Patient-specific stem cell lines derived from human
parthenogenetic blastocysts. Cloning Stem Cells. 9:432–449. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin G, OuYang Q, Zhou X, et al: A highly
homozygous and parthenogenetic human embryonic stem cell line
derived from a one-pronuclear oocyte following in vitro
fertilization procedure. Cell Res. 17:999–1007. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mai Q, Yu Y, Li T, et al: Derivation of
human embryonic stem cell lines from parthenogenetic blastocysts.
Cell Res. 17:1008–1019. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Allen ND, Barton SC, Hilton K, Norris ML
and Surani MA: A functional analysis of imprinting in
parthenogenetic embryonic stem cells. Development. 120:1473–1482.
1994.PubMed/NCBI
|
|
9
|
Sánchez-Pernaute R, Studer L, Ferrari D,
et al: Long-term survival of dopamine neurons derived from
parthenogenetic primate embryonic stem cells (cyno-1) after
transplantation. Stem Cells. 23:914–922. 2005.PubMed/NCBI
|
|
10
|
Lin H, Lei J, Wininger D, et al:
Multilineage potential of homozygous stem cells derived from
metaphase II oocytes. Stem Cells. 21:152–161. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cibelli JB, Grant KA, Chapman KB, et al:
Parthenogenetic stem cells in nonhuman primates. Science.
295:8192002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kono T, Obata Y, Wu Q, et al: Birth of
parthenogenetic mice that can develop to adulthood. Nature.
428:860–864. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Morali OG, Jouneau A, McLaughlin KJ,
Thiery JP and Larue L: IGF-II promotes mesoderm formation. Dev
Biol. 227:133–145. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hernandez L, Kozlov S, Piras G and Stewart
CL: Paternal and maternal genomes confer opposite effects on
proliferation, cell-cycle length, senescence, and tumor formation.
Proc Natl Acad Sci USA. 100:13344–13349. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hikichi T, Wakayama S, Mizutani E, et al:
Differentiation potential of parthenogenetic embryonic stem cells
is improved by nuclear transfer. Stem Cells. 25:46–53. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lin G, Xie Y, Ouyang Q, et al:
HLA-matching potential of an established human embryonic stem cell
bank in China. Cell Stem Cell. 5:461–465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
D’Amour KA, Agulnick AD, Eliazer S, Kelly
OG, Kroon E and Baetge EE: Efficient differentiation of human
embryonic stem cells to definitive endoderm. Nat Biotechnol.
23:1534–1541. 2005.
|
|
18
|
Kodama S, Toyonaga T, Kondo T, et al:
Enhanced expression of PDX-1 and Ngn3 by exendin-4 during beta cell
regeneration in STZ-treated mice. Biochem Biophys Res Commun.
327:1170–1178. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
D’Amour KA, Bang AG, Eliazer S, et al:
Production of pancreatic hormone-expressing endocrine cells from
human embryonic stem cells. Nat Biotechnol. 24:1392–1401.
2006.PubMed/NCBI
|
|
20
|
Szabo P and Mann JR: Expression and
methylation of imprinted genes during in vitro differentiation of
mouse parthenogenetic and androgenetic embryonic stem cell lines.
Development. 120:1651–1660. 1994.PubMed/NCBI
|
|
21
|
Surani MA and Barton SC: Development of
gynogenetic eggs in the mouse: implications for parthenogenetic
embryos. Science. 222:1034–1036. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Surani MA, Barton SC and Norris ML:
Development of reconstituted mouse eggs suggests imprinting of the
genome during gametogenesis. Nature. 308:548–550. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Surani MA, Kothary R, Allen ND, et al:
Genome imprinting and development in the mouse. Dev Suppl. 89–98.
1990.
|
|
24
|
McGrath J and Solter D: Completion of
mouse embryogenesis requires both the maternal and paternal
genomes. Cell. 37:179–183. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L and Li H: Progress in the studies
of parthenogenetic embryonic stem cells. Zhonghua Nan Ke Xue.
10:55–58. 2004.(In Chinese).
|
|
26
|
Maden M: Role and distribution of retinoic
acid during CNS development. Int Rev Cytol. 209:1–77. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Stafford D and Prince VE: Retinoic acid
signaling is required for a critical early step in zebrafish
pancreatic development. Curr Biol. 12:1215–1220. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Blumentrath J, Neye H and Verspohl EJ:
Effects of retinoids and thiazolidinediones on proliferation,
insulin release, insulin mRNA, GLUT 2 transporter protein and mRNA
of INS-1 cells. Cell Biochem Funct. 19:159–169. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Johannesson M, Stahlberg A, Ameri J, Sand
FW, Norrman K and Semb H: FGF4 and retinoic acid direct
differentiation of hESCs into PDX1-expressing foregut endoderm in a
time- and concentration-dependent manner. PLoS One. 4:e47942009.
View Article : Google Scholar
|
|
30
|
Petrik J, Pell JM, Arany E, et al:
Overexpression of insulin-like growth factor-II in transgenic mice
is associated with pancreatic islet cell hyperplasia.
Endocrinology. 140:2353–2363. 1999.PubMed/NCBI
|
|
31
|
Lopez MF, Dikkes P, Zurakowski D,
Villa-Komaroff L and Majzoub JA: Regulation of hepatic glycogen in
the insulin-like growth factor II-deficient mouse. Endocrinology.
140:1442–1448. 1999.PubMed/NCBI
|
|
32
|
Horii T, Kimura M, Morita S, Nagao Y and
Hatada I: Loss of genomic imprinting in mouse parthenogenetic
embryonic stem cells. Stem Cells. 26:79–88. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vu TH and Hoffman A: Alterations in the
promoter-specific imprinting of the insulin-like growth factor-II
gene in Wilms’ tumor. J Biol Chem. 271:9014–9023. 1996.PubMed/NCBI
|
|
34
|
Christofori G, Naik P and Hanahan D:
Deregulation of both imprinted and expressed alleles of the
insulin-like growth factor 2 gene during beta-cell tumorigenesis.
Nat Genet. 10:196–201. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sussenbach JS, Steenbergh PH and
Holthuizen P: Structure and expression of the human insulin-like
growth factor genes. Growth Regu. 2:1–9. 1992.PubMed/NCBI
|
|
36
|
De Sousa PA and Wilmut I: Human
parthenogenetic embryo stem cells: appreciating what you have when
you have it. Cell Stem Cell. 1:243–244. 2007.PubMed/NCBI
|