Introduction
Regarding treatments for the full spectrum of skin
defects, autologous skin grafts are the best option. One major
limiting factor of autologous skin grafts is the availability of
autologous skin. In recent years, there have been significant
advancements in tissue-engineered skin replacements (1–3);
however, a good source of autogeneic cells remains to be
determined. Nuclear reprogramming offers a promising method,
whereby extracts from a specific type of cell are introduced into
donor cells and the donor cells subsequently acquire the
characteristics of the cells from which the extracts were derived
(4). This approach consists of
incubating reversibly permeabilized donor cells in nuclear and
cytoplasmic extracts derived from another type of somatic cell.
Following exposure to the extracts, the donor cells are resealed
and subsequently cultured. The extracts are considered to provide
the regulatory components required to initiate a transcriptional
program specific to the target cell type. Through reprogramming,
the epigenetic state of somatic cells can be changed to mirror the
epigenetic status of the cell types essential for the treatment of
clinical diseases. This approach has been demonstrated for a number
of specific types of cell. Håkeliien et al (5) reported that 293T cells permeabilized
with the cholesterol-binding toxin Streptolysin O (SLO) and exposed
to Jurkat T cell extracts were induced to adopt T cell-specific
properties. These T cell-specific properties were maintained for
~100 population doublings in vitro, suggesting that the
Jurkat extracts induced nuclear reprogramming to an extent.
Similarly, extracts derived from rat cardiomyocytes have been shown
to elicit expression of cardiomyocytic markers in human adipose
stem cells (6). These results
indicate that nuclear and cytoplasmic extracts from one target cell
type can induce some nuclear reprogramming in other types of
cells.
In previous reprogramming studies, the type of donor
cell used is often mesenchymal stem cells (MSCs) from adult tissues
or fibroblasts, which are considered to be able to differentiate
outside their predicted developmental lineage (7,8).
Human adipose tissue obtained by liposuction has previously been
identified as a convenient alternative source for MSCs. Adipose
tissue has been processed to obtain a fibroblast-like population of
cells or adipose tissue stem cells (ATSCs), which are easily
isolated by differential sedimentation from processed liposuction
material as previously reported by Zuk et al (9). ATSCs and bone marrow-derived
mesenchymal stem cells have the capacity for renewal and can be
cultured for several months in vitro with a regular doubling
time and low level of senescence. ATSCs are able to differentiate
into multiple cell lineages including adipocytes, osteoblasts,
chondrocytes, neurons, and multinucleated myocytes in response to
lineage-specific induction factors (9–11).
ATSCs migrate, engraft, and differentiate into functional cells
in vivo.
In the present study, the possibility of using
epidermal keratinocyte cell extracts to promote differentiation of
ATSCs was investigated.
Materials and methods
Isolation and culture of human ATSCs
ATSCs were isolated from liposuction aspirates from
subcutaneous adipose tissue sites obtained from male and female
subjects undergoing selective procedures in the Departments of
Plastic Surgery at Shanghai Ninth People’s Hospital, Shanghai Third
People’s Hospital and Shanghai Sixth People’s Hospital, Shanghai
Jiaotong University, Shanghai China). For all patients, written
informed consent was obtained from the patient or patient’s family
and the study was approved by the the Ethical Committee of Shanghai
Sixth People’s Hospital. The lipoaspirate was washed 3–4 times with
phosphate-buffered saline (PBS) to remove red blood cells and
tissue debris. Washed adipose tissue was suspended in an equal
volume of PBS supplemented with 1% bovine serum (Hyclone, Logan,
UT, USA) and 0.075% collagenase type I (Worthington, Lakewood, NJ,
USA), prewarmed to 37°C and then continuously agitated on a shaker
at 37°C for 1 h. Collagenase activity was neutralized by adding an
equal volume of Dulbecco’s modified Eagle’s medium (DMEM;
Gibco-BRL, Carlsbad, CA, USA) with 10% fetal bovine serum. The
tissue was centrifuged for 5 min at 197 × g at room temperature.
The supernatant, which contained the mature adipocytes, was
aspirated. Following centrifugation, cell pellets were resuspended
in culture medium, added to tissue culture dishes, and cultured for
24 h at 37°C in 5% CO2. Unattached cells and debris were
then removed, and fresh medium (DMEM/Ham’s F-12; Gibco-BRL)
containing 10% fetal bovine serum (Hyclone) was added to the
adherent cells. This initial passage of the primary cell culture
was referred to as passage 0. These cells were cultured to 75–90%
confluency, passaged using a 0.25%
trypsin-ethylenediaminetetracetic acid (EDTA) solution
(Sigma-Aldrich, St. Louis, MO, USA) and plated at a density of
5,000 cells/cm2 (passage 1). Cell viability and the
number of cells at the time of passage were determined using trypan
blue (Sigma-Aldrich) exclusion and hemacytometer cell counts
respectively. The culture medium was replaced twice a week.
Following passage 4, cells were passaged repeatedly once 75–90%
confluency was achieved.
Human epidermal keratinocyte
extracts
Human epidermal keratinocytes were obtained from
normal skin biopsied from the foreskin or other sites. The skin was
washed 3–4 times with PBS, the majority of the subcutaneous tissue
was removed with surgical scissors, and the remaining skin was
minced finely into pieces <1 mm in diameter. The skin fragments
were digested with 2.4 U/ml Dispase (Roche, Mannheim, Germany) at
37°C with occasional agitation. Following 2 h of digestion, the
dermis was removed from the epidermis. The epidermal keratinocytes
were dissociated from the epidermis via incubation with 0.05%
trypsin for 15 min and the cells were cultured in keratinocyte
serum-free medium (KSFM; Gibco-BRL). The epidermal keratinocytes
were frozen in liquid nitrogen and stored at −80°C for no longer
than four weeks. To prepare the epidermal keratinocyte extracts,
cells were thawed on ice and cold lysis buffer (50 mmol/l NaCl, 5
mmol/l MgCl2, 20 mmol/l
4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid, pH 8.2, and 1
mmol/l dithiothreitol) was added to the cells. Cells were pelleted
at 87 × g and resuspended in 1.5 volumes of cell lysis buffer
(Beyotime, Jiangsu, China) containing protease inhibitors
(Beyotime). Cells were homogenized by pulse-sonication until the
cells and their nuclei were completely lysed (monitored by phase
contrast microscopy; Nikon TS100, Tokyo, Japan) and the lysate was
sedimented at 21,890 × g for 15 min at 4°C. The supernatant was
collected and either used fresh or snap-frozen in liquid nitrogen
and stored at −80°C. The pH was measured with the aid of a micro-pH
meter (Sentron SI600, Roden, The Netherlands) and protein
concentration of the extract was measured using The microplate BCA
method using a BCA protein assay kit (Pierce, Rockford, IL)
according to the manufacturer’s instructions.
Cell permeabilization
ATSCs were washed in cold PBS and in cold
Ca2+- and Mg2+-free Hanks’ balanced salt
solution (HBSS; Gibco-BRL). The cells were resuspended in aliquots
of 20,000 cells per 15.5 μl HBSS, and 4.5 μl SLO (Sigma; 100 g/ml
stock diluted 1:100 in ice-cold HBSS) was added to yield a final
SLO concentration of 230 ng/ml. Samples were incubated for 50 min
at 37°C, and then sedimented at 22 × g for 15 min at 4°C. The cells
were resuspended in culture medium for use. In each experiment,
permeabilization efficiency was evaluated by monitoring uptake of a
fluorescein isothiocyanate (FITC)-protein (AnaTag™ 5-FITC Protein
Labeling Kit *Ultra Convenient*; AnaSpec
Inc., Freemont, CA, USA) in a separate sample. The protein was a
mixer that included all human epidermal keratinocyte extract
proteins. To reseal plasma membranes, cells were plated for 2 h in
RPMI-1640 medium containing 10% fetal calf serum and 2 mM
CaCl2. Cells that failed to reattach were removed during
medium replacement. Permeabilization of live cells was examined by
fluorescence microscopy (Olympus BX51, Tokyo, Japan).
Incubation of ATSCs in epidermal
keratinocyte extracts
The supernatant was removed when the ATSCs wre
cultured in passage 6, and ATSCs were suspended in 20 μl epidermal
keratinocyte extract containing an ATP regenerating system
(Sigma-Aldrich) and 1 mM of each NTP (ATP, CTP, GTP and UTP;
Sigma-Aldrich). The cells were incubated for 1 h at 37°C in a water
bath with occasional agitation, allowing for the keratinocyte
extract to permeate the ATSCs. To reseal plasma membranes, the
extract was diluted in DMEM medium containing 10% FCS, antibiotics
(Gibco-BRL), and 2 mmol/l CaCl2. Following
centrifugation at 197 × g for 5 min, the cells were transferred to
culture dishes, and fresh KSFM medium was added. Dead cells were
removed through decanting, and the remaining cells were cultured
until use. Control cells were permeabilized through exposure to
NaCl rather than epidermal keratinocyte extracts.
Cell viability analysis
The viability of cultured or reprogrammed ATSCs was
measured using the Cell Counting kit-8 (Dojindo, Kumamoto, Japan)
according to the manufacturer’s instructions. Each experiment was
performed in triplicate.
Laser scanning confocal microscopy
Cultured ATSCs were seeded on a glass slide and
incubated with FITC-labeled epidermal keratinocyte protein. At
different time points, the cells were fixed with acetone for 10 min
at room temperature. Following fixation, the cells were observed
using scanning confocal microscopy (TCS-SPE, Leica Microsystems
GmbH, Wetzlar, Germany).
Immunofluorescence
The cells were washed in PBS and fixed with 95%
acetone for 15 min. Following fixation, the cells were washed in
PBS, permeabilized with 0.25% Triton X-100 and 5% dimethylsufoxide
(DMSO)-PBS for 10 min at room temperature and washed in PBS.
Subsequently, 0.75/1.5% PBS-H2O2 was added to
the cells for 15 min at 37°C, the cells were washed in PBS, and
blocked in 10% sheep serum albumin (Abcam, Cambridge, UK). The
cells were incubated overnight with a 1:200 dilution of the primary
monoclonal antibodies (mAbs; mouse IgG, K19, involucrin and K1/10
Santa Cruz Biotechnology, Inc, Santa Cruz, CA, USA) and
subsequently incubated with the secondary antibody (FITC-conjugated
goat anti-mouse antibody; Santa Cruz Biotechnology, Inc.) at 37°C
for 30 min. Images were acquired using an Olympus BX51 microscope
(Olympus, Tokyo, Japan) and the AnalySIS software, version 1.8
(Soft Imaging Systems, Olympus) and processed with Adobe Photoshop
CS5 (Adobe, Mountain View, CA, USA).
Fluorescence-activated cell sorting
(FACS) analysis
Flow cytometric analysis was performed on ATSCs
incubated in epidermal keratinocyte extracts and the control cells
cultured from passages 0 through 4. Cells were trypsinized and
centrifuged for 3 min at 97 × g in an Eppendorf centrifuge. The
pellets were resuspended in PBS. The samples were fixed in a
solution of 70% ethanol and incubated overnight with 1% Tween-20.
The cells were washed in PBS containing 1% bovine serum albumin,
and then incubated for 45 min at room temperature with the primary
mAbs K19, involucrin and K1/10. Following washing, the secondary
antibody (phycoerythrin-conjugated goat anti-mouse antibody, Santa
Cruz Biotechnology, Inc.) was added to the cell suspension and
incubated for 30 min at room temperature. Cells were washed once
more and resuspended in PBS prior to analysis. Cells were analyzed
using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA,
USA). Gates were set based on stainings with combinations of
relevant and irrelevant mAbs so that no more than 1% of the cells
were positive using irrelevant antibodies. Flow cytometric
experiments were repeated three times.
RNA extraction and reverse
transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from cells using TRIzol
(Invitrogen, Gaithersburg, MD, USA), treated with DNase I (Qiagen,
Venlo, Netherlands), and processed according to the manufacturer’s
instructions. For cDNA synthesis, 1–5 μg total RNA was reverse
transcribed (1 h at 37°C) using oligo-dT primers. RT-PCR was
performed using the following primers: Forward,
5′-GACGGGAGGGCGAGAAATG-3′, and reverse, 5′-GCCATAGGACATCTGGGAAGC-3′
(246 bp) for K19; forward, 5′-CCAGCCTTCTACACCTCAC-3′, and reverse,
5′-ACCCATTCCTCCCACTCC-3′ (241 bp) for involucrin; forward,
5′-TCCAAGGAAATGGCAACTCA-3′, and reverse, 5′-AGGAACGGCAGGCGAGAT-3′
(288 bp) for K1/10; and forward, 5′-GCGGGAGGGCGAGAAATGA-3′, and
reverse, 5′-CGATAGGACATCTGGGAAGCC-3′ (280 bp) for β-actin. PCR
conditions were as follows: 94°C for 3 min, 28 cycles of 94°C for
30 sec, 55°C for 30 sec and 72°C for 45 sec, followed by 72°C for 5
min. Each sample (2.5 μl cDNA in a total reaction volume of 25 μl)
was run in triplicate. RT-PCR products were analyzed by
electrophoresis using a 1% agarose gel. Gels were stained with
ethidium bromide and photographed (Image Master VDS, Amersham
Pharmacia Biotech, USA) under a UV lamp.
Statistical analysis
The data are presented as the mean ± standard error
of the mean (SEM). One-way analysis of variance test was performed
by SAS 6.12 (Software, Inc., San Diego, CA, USA). Mortality was
evaluated by the log rank method. P<0.05 was considered to
indicate a statistically significant difference.
Results
Uptake of FITC-labeling protein by
ATSCs
Incubation of ATSCs with epidermal keratinocyte
extracts containing FITC-labeling protein resulted in complete
cytoplasmic localization of FITC-labeling protein after 12–24 h,
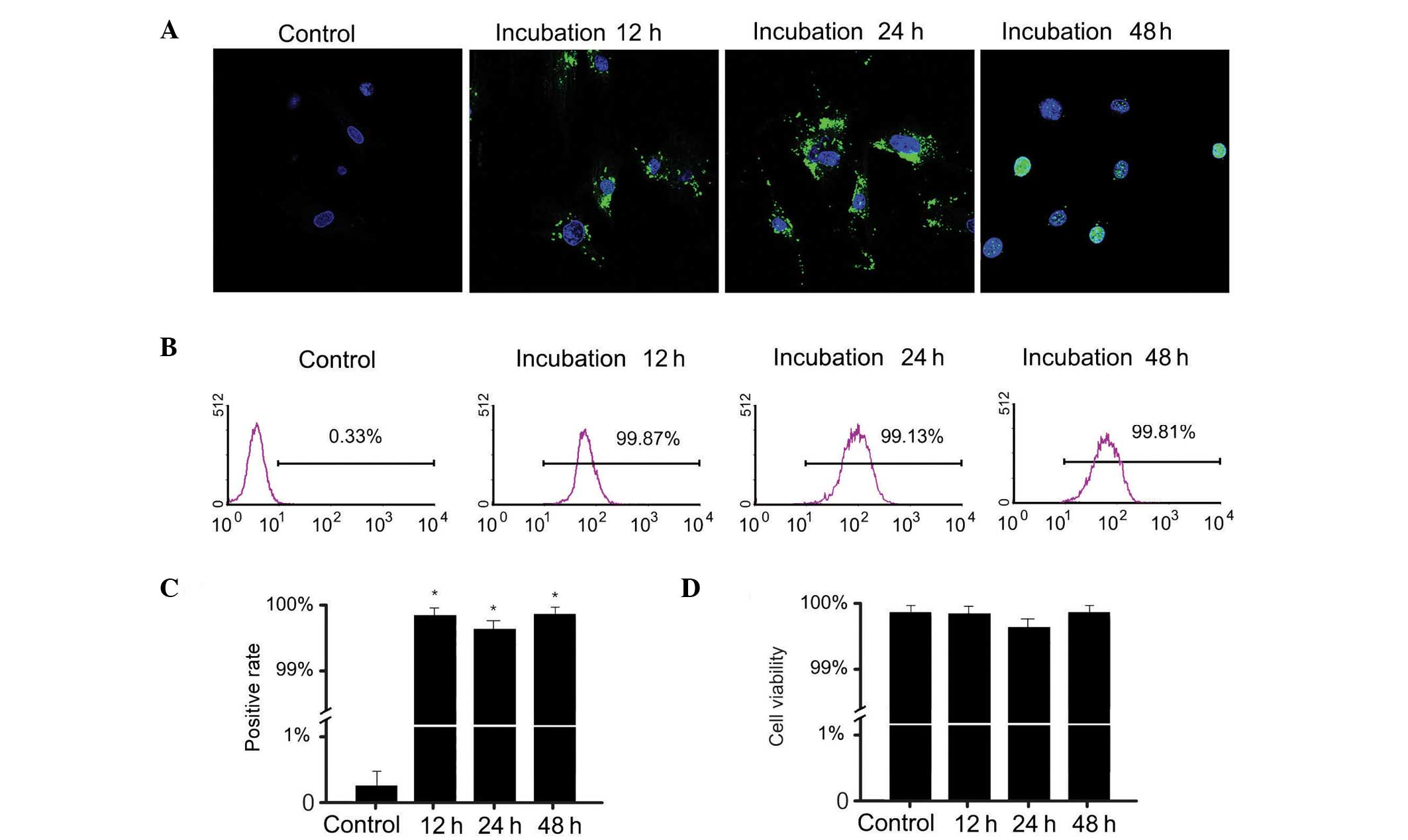
followed by nuclear localization after 48 h (Fig. 1A). After 12 h, the SLO-treated and
cultured ATSCs displayed bright intracellular FITC fluorescence
(Fig. 1A) indicating that a high
proportion of cells took up the FITC-labeling protein and resealed.
As determined by FACS analysis, the rate of FITC-labeling protein
uptake in ATSCs was ~99% (Fig. 1B and
C). Cell viability assays demonstrated that uptake of epidermal
keratinocyte extracts does not affect cell viability (Fig. 1D).
Alteration in cell morphology
Permeabilized ATSCs were exposed for 1 h to
epidermal keratinocyte extracts, resealed, and expanded in KSFM
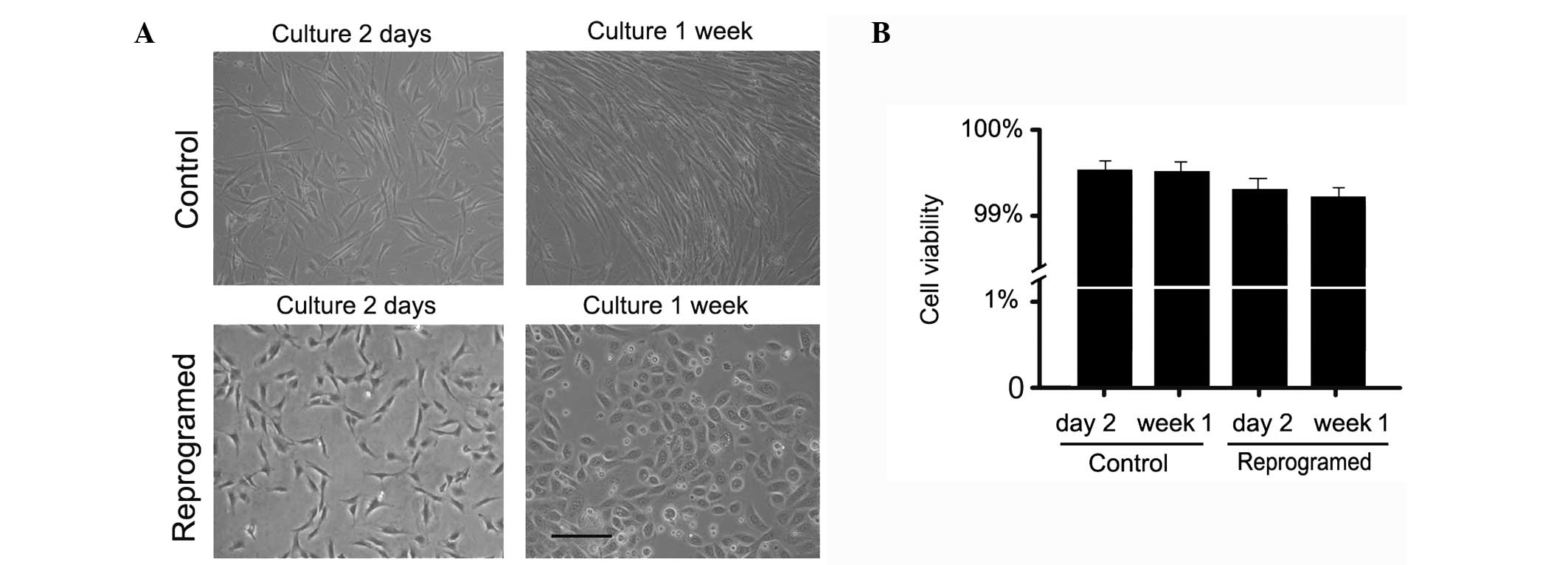
culture medium. After one day, cultured ATSCs changed shape and
acquired a rounder morphology associated with a marked reduction in
cell size. After one week of exposure to epidermal keratinocyte
extracts, ATSCs tended to resemble epidermal keratinocytes and
exhibited different cell sizes that were closely packed (Fig. 2A). This morphology was observed for
several weeks. No such changes were observed in the control
permeabilized ATSCs. Statistical analysis of cell viability showed
no significant difference between the reprogrammed and control
cells (Fig. 2B).
Expression of epidermal keratinocyte
markers
Permeabilized and epidermal keratinocyte
extract-treated ATSCs were cultured for seven days prior to protein
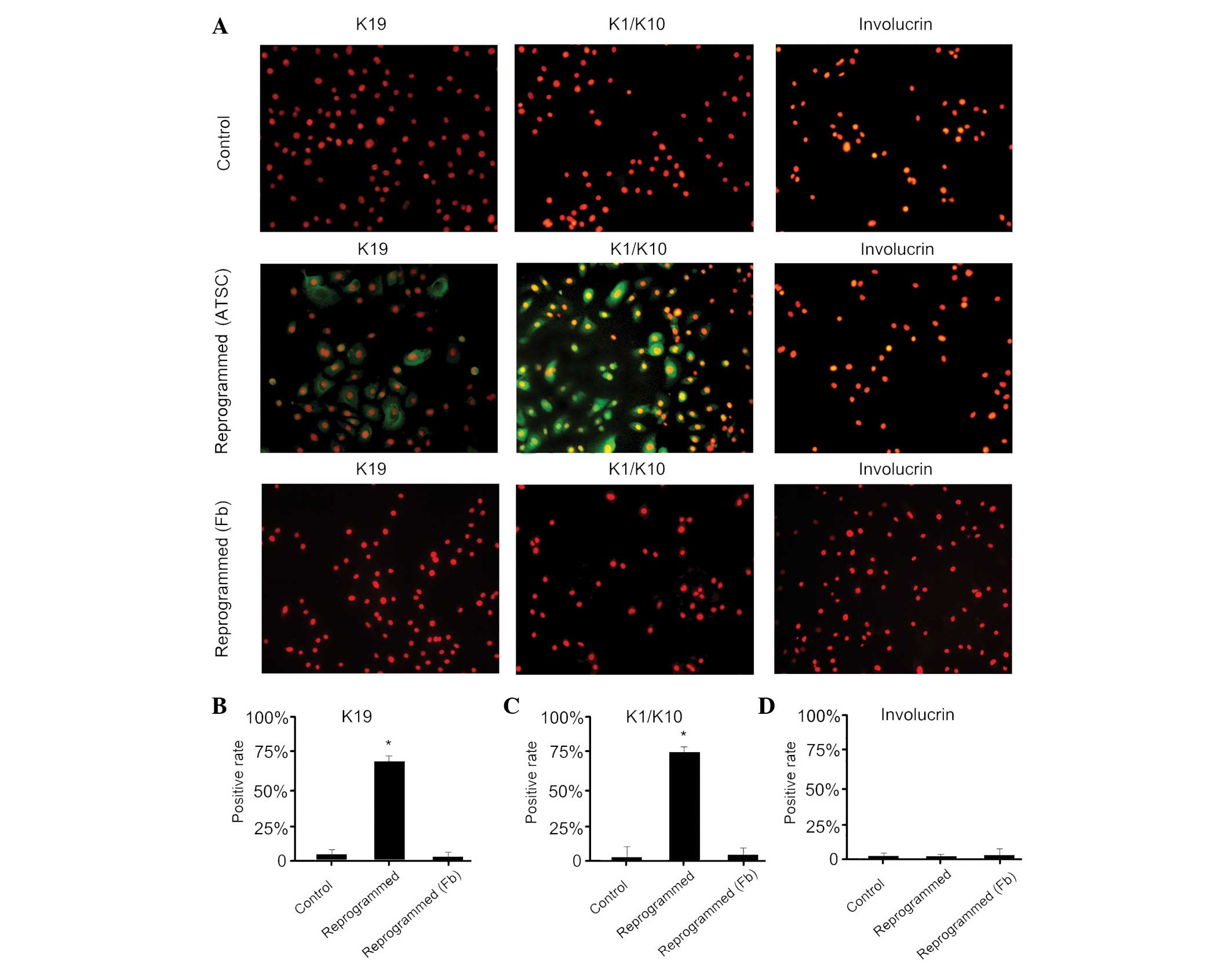
expression analysis. Immunofluorescence analysis revealed that
reprogrammed cells expressed K19, a marker of epidermal stem cells,
and K1/10, a characteristic of epidermal keratinocytes, but did not
express involucrin (Fig. 3). The
control cells did not express any of the markers analyzed. These
results indicate that human ATSCs can be induced to take on
properties of epidermal keratinocytes after a transient exposure to
nuclear and cytoplasmic extracts from epidermal keratinocytes.
mRNA expression in reprogrammed
ATSCs
The gene expression levels of K19, K1/10,
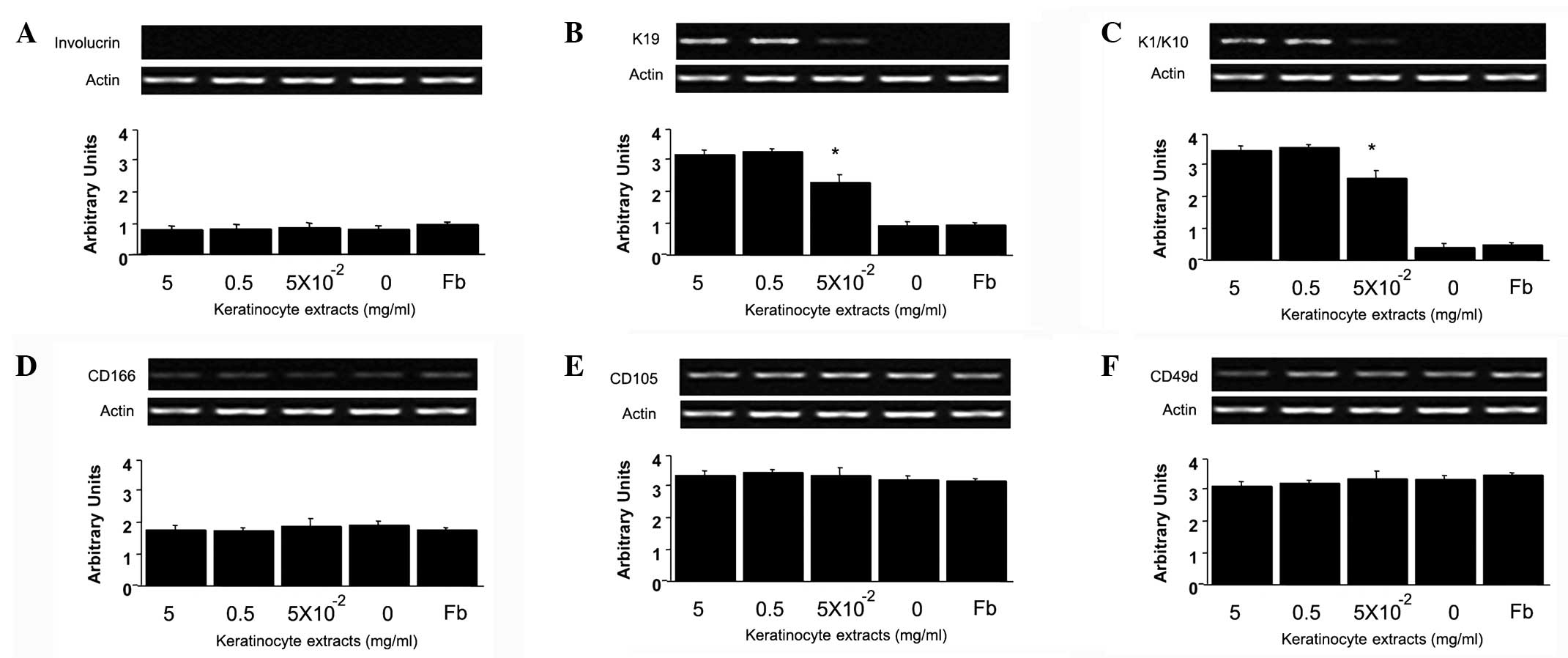
involucrin, CD166, CD105 and CD49d were analyzed by RT-PCR. The
results are shown and summarized in Fig. 4. Involucrin mRNA was not detected
in the ATSCs exposed to epidermal keratinocyte extracts (Fig. 4A). As expected, K19 and K1/10 mRNAs
were expressed after treatment with ≥5×10−2 mg/ml of
epidermal keratinocyte extracts, indicating that detection of K19
and K1/10 in ATSCs was dependent on extract protein concentration
(Fig. 4B and C). In all cell
groups, CD166, CD105 and CD49d mRNA were detected (Fig. 4D–F).
Discussion
ATSCs can be easily obtained in large quantities
from processed liposuction material by differential sedimentation
(9,10). The cells that are obtained from
lipoaspirates most likely represent a stem cell population. These
cells express surface markers which are characteristic of ATSCs and
are able to differentiate into bone, cartilage and fat, strongly
suggesting they are ATSCs (9). By
altering the epigenetic status of somatic cells, it is possible to
produce cells that are essential for the treatment of clinical
diseases. It has been shown that when nuclear and cytoplasmic
extracts from donor cells are introduced into target cells, the
target cells can express characteristics of the donor cells
(12).
During organ development, different cell types have
different epigenetic states that determine the on or off state of
specific genes. Changing the epigenetic status of mature cells
allows for the production of cells that are essential for the
treatment of clinical diseases. Nuclear reprogramming is a method
that can change the epigenetic state and function of cells and it
is of great medical interest as it has the potential to generate a
source of patient-specific cells. The ability to reprogram specific
somatic cells into other types of cells has previously been
demonstrated (5,6,11).
In the present study, reversibly permeabilized ATSCs were incubated
for 1 h in nuclear and cytoplasmic extracts from human epidermal
keratinocytes, resealed with CaCl2 and cultured.
Following ~2 h of incubation, the FITC-labeling protein of
epidermal keratinocyte extracts began to gradually permeate into
the ATSCs. After ~12–24 h, FITC fluorescence was found throughout
the cytoplasm and 48 h later the fluorescence was observed in the
nucleus. Using FACs analysis, the ATSC uptake rate of
FITC-conjugated protein was determined to reach 99%, thus
demonstrating the feasibility of this method. These results
demonstrate that ATSC reprogramming through exposure to nuclear and
cytoplasmic epidermal keratinocyte extracts is a promising method.
The results of multiple experiments support the hypothesis that
differentiation is induced in ATSCs. Changes in cell shape were
observed after one day, and cultured ATSCs acquired a rounder
morphology associated with a small reduction in cell size.
Following one week of exposure to epidermal keratinocyte extracts,
ATSCs tended to resemble epidermal keratinocytes and exhibited the
pavestone appearance. Immunofluorescence analysis demonstrates that
reprogrammed cells expressed K19, an epidermal stem cell marker,
and K1/10, an epidermal keratinocyte marker. The control cells did
not express any of the markers analyzed. K19 and K1/10 mRNA levels
in reprogrammed ATSCs increased with increasing concentrations of
protein from epidermal keratinocyte extracts. These results are
consistent with the results from the immunofluorescence and FACS
analysis. Reprogramming appears to be elicited by direct uptake of
factors from the extract, although how the extract affects ATSCs
and the nature of the cell extract molecules responsible for
induction of ATSC differentiation remain to be investigated.
Notably, K19 and K1/10 were detected in the
experiments, while involucrin was not. One possible explanation is
the heterogeneity of the ATSC population within a lipoaspirate,
similar to the heterogeneity within bone marrow-derived MSCs
(13). Only a fraction of ATSCs
may be responsive to the differentiation factors provided by the
extract.
In the present study, the nature of the cell extract
molecules responsible for the induction of ATSC differentiation in
the in vitro approach remains largely unknown. Transcription
factors, signaling pathways and chromatin remodeling complexes are
likely involved in differentiation. Previous studies indicate that
chromatin remodeling is facilitated by induction of histone H4
hyperacetylation at the IL2 locus in 293T nuclei incubated in
Jurkat extracts, which correlates with activation of the IL2 gene
(5,14). Furthermore, induced expression of
previously repressed genes has also been observed, indicating that
the extract is capable of reprogramming the epigenetic state of the
293T genome. Nevertheless, a role for nucleic acids, including
small noncoding nuclear RNAs, in promoting changes in gene
expression following the incubation of cells in extracts has not
been excluded in light of their function in transcriptional
regulation (15,16). Mikkelsen et al (17) suggested that certain cells may
become trapped in partially reprogrammed states due to incomplete
repression of transcription factors and that DNA demethylation is
an inefficient step in the transition to pluripotency. They
demonstrated that RNA inhibition of transcription factors
facilitates reprogramming and that treatment with DNA
methyltransferase inhibitors improves the overall efficiency of the
reprogramming process. In addition, Huangfu et al (18,19)
demonstrated that histone deacetylase and DNA methyltransferase
inhibitors greatly improve the efficiency of reprogramming mouse
embryonic fibroblasts by genetic factors. Biochemical fractionation
of the extracts is expected to help elucidate the nature of
molecules involved in altering cell fate.
In conclusion, the present study determined that
chromatin remodeling and transcription factors are key elements
involved in the induction of reprogramming. However, certain
factors are yet to be elucidated, including how nuclear
reprogramming is induced, the mechanism behind induction and how
long the developmental programming persists.
Acknowledgements
This study was supported by the National Scientific
Fund (grant nos.: 81071580 and 81000837).
References
|
1
|
Nayak S, Dey S and Kundu SC: Skin
equivalent tissue-engineered construct: co-cultured
fibroblasts/keratinocytes on 3D matrices of sericin hope cocoons.
PLoS One. 8:e747792013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li H, Chu Y, Zhang Z, et al: Construction
of bilayered tissue-engineered skin with human amniotic mesenchymal
cells and human amniotic epithelial cells. Artif Organs.
36:911–919. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nie X, Zhang JY, Cai KJ, et al: Cosmetic
improvement in various acute skin defects treated with
tissue-engineered skin. Artif Organs. 31:703–710. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang X, Sheng L, Xie F and Zhang Q:
Differentiation of bone marrow-derived mesenchymal stem cells into
chondrocytes using chondrocyte extract. Mol Med Rep. 6:745–749.
2012.PubMed/NCBI
|
|
5
|
Håkelien AM, Landsverk HB, Robl JM,
Skålhegg BS and Collas P: Reprogramming fibroblasts to express
T-cell functions using cell extracts. Nat Biotechnol. 20:460–466.
2002.PubMed/NCBI
|
|
6
|
Gaustad KG, Boquest AC, Anderson BE,
Gerdes AM and Collas P: Differentiation of human adipose tissue
stem cells using extracts of rat cardiomyocytes. Biochem Biophys
Res Commun. 314:420–427. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Walev I, Bhakdi SC, Hofmann F, et al:
Delivery of proteins into living cells by reversible membrane
permeabilization with streptolysin-O. Proc Natl Acad Sci USA.
98:3185–3190. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Collas P and Håkelien AM: Teaching cells
new tricks. Trends Biotechnol. 21:354–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zuk PA, Zhu M, Ashjian P, et al: Human
adipose tissue is a source of multipotent stem cells. Mol Biol
Cell. 13:4279–4295. 2002.PubMed/NCBI
|
|
10
|
Cousin B, André M, Arnaud E, Pénicaud L
and Casteilla L: Reconstitution of lethally irradiated mice by
cells isolated from adipose tissue. Biochem Biophys Res Commun.
301:1016–1022. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Håkelien AM, Gaustad KG and Collas P:
Transient alteration of cell fate using a nuclear and cytoplasmic
extract of an insulinoma cell line. Biochem Biophys Res Commun.
316:834–841. 2004.PubMed/NCBI
|
|
12
|
Wernig M, Meissner A, Foreman R, et al: In
vitro reprogramming of fibroblasts into a pluripotent ES-cell-like
state. Nature. 448:318–324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Herzog EL, Chai L and Krause DS:
Plasticity of marrow-derived stem cells. Blood. 102:3483–3493.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Landsverk HB, Håkelien AM, Küntziger T,
Robl JM, Skålhegg BS and Collas P: Reprogrammed gene expression in
a somatic cell-free extract. EMBO Rep. 3:384–389. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Maison C, Bailly D, Peters AH, et al:
Higher-order structure in pericentric heterochromatin involves a
distinct pattern of histone modification and an RNA component. Nat
Genet. 30:329–334. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sleutels F, Zwart R and Barlow DP: The
non-coding Air RNA is required for silencing autosomal imprinted
genes. Nature. 415:810–813. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Mikkelsen TS, Hanna J, Zhang X, et al:
Dissecting direct reprogramming through integrative genomic
analysis. Nature. 454:49–55. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huangfu D, Osafune K, Maehr R, et al:
Induction of pluripotent stem cells from primary human fibroblasts
with only Oct4 and Sox2. Nat Biotechnol. 26:1269–1275. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huangfu D, Maehr R, Guo W, et al:
Induction of pluripotent stem cells by defined factors is greatly
improved by small-molecule compounds. Nat Biotechnol. 26:795–797.
2008. View
Article : Google Scholar : PubMed/NCBI
|