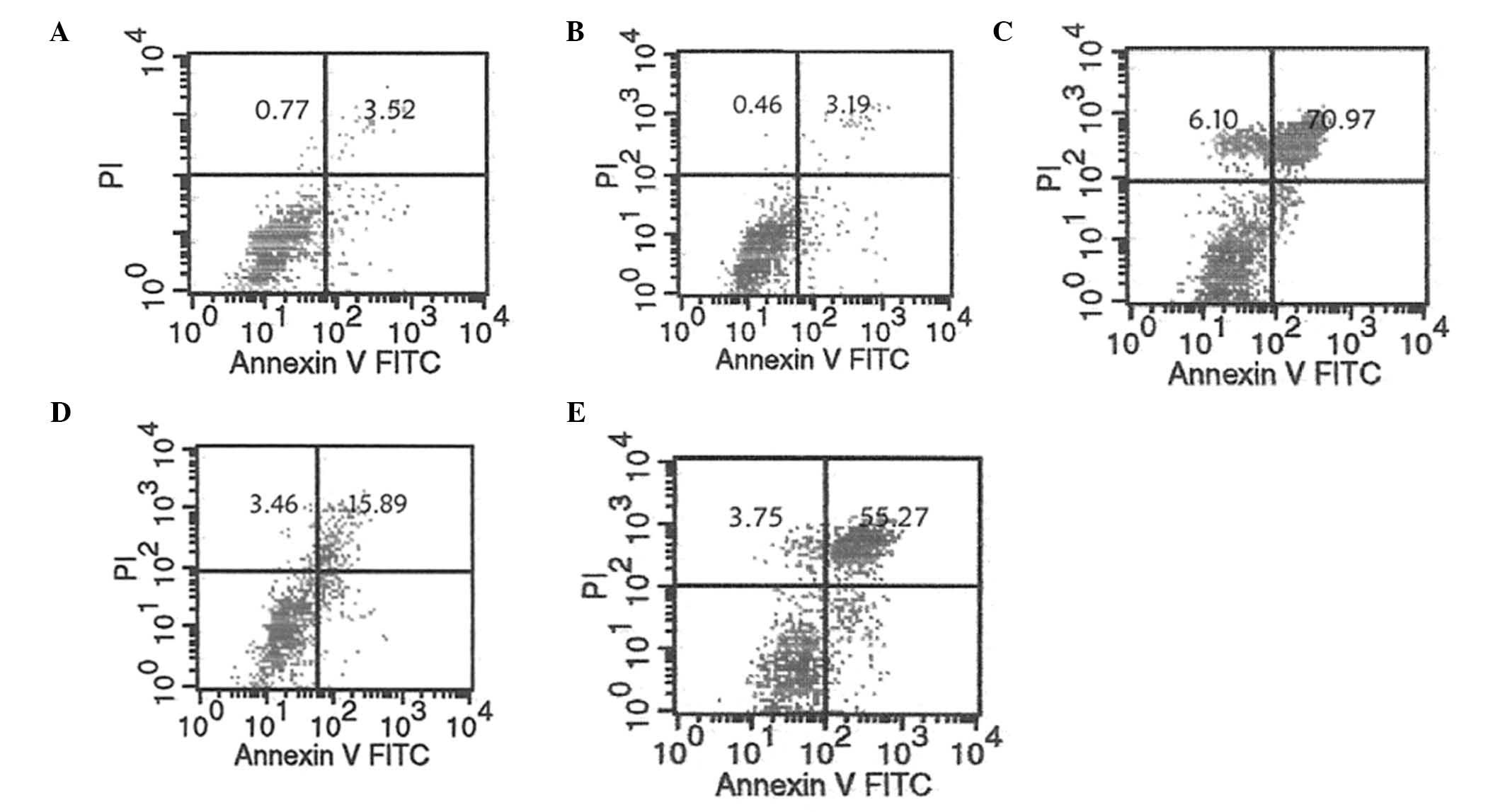
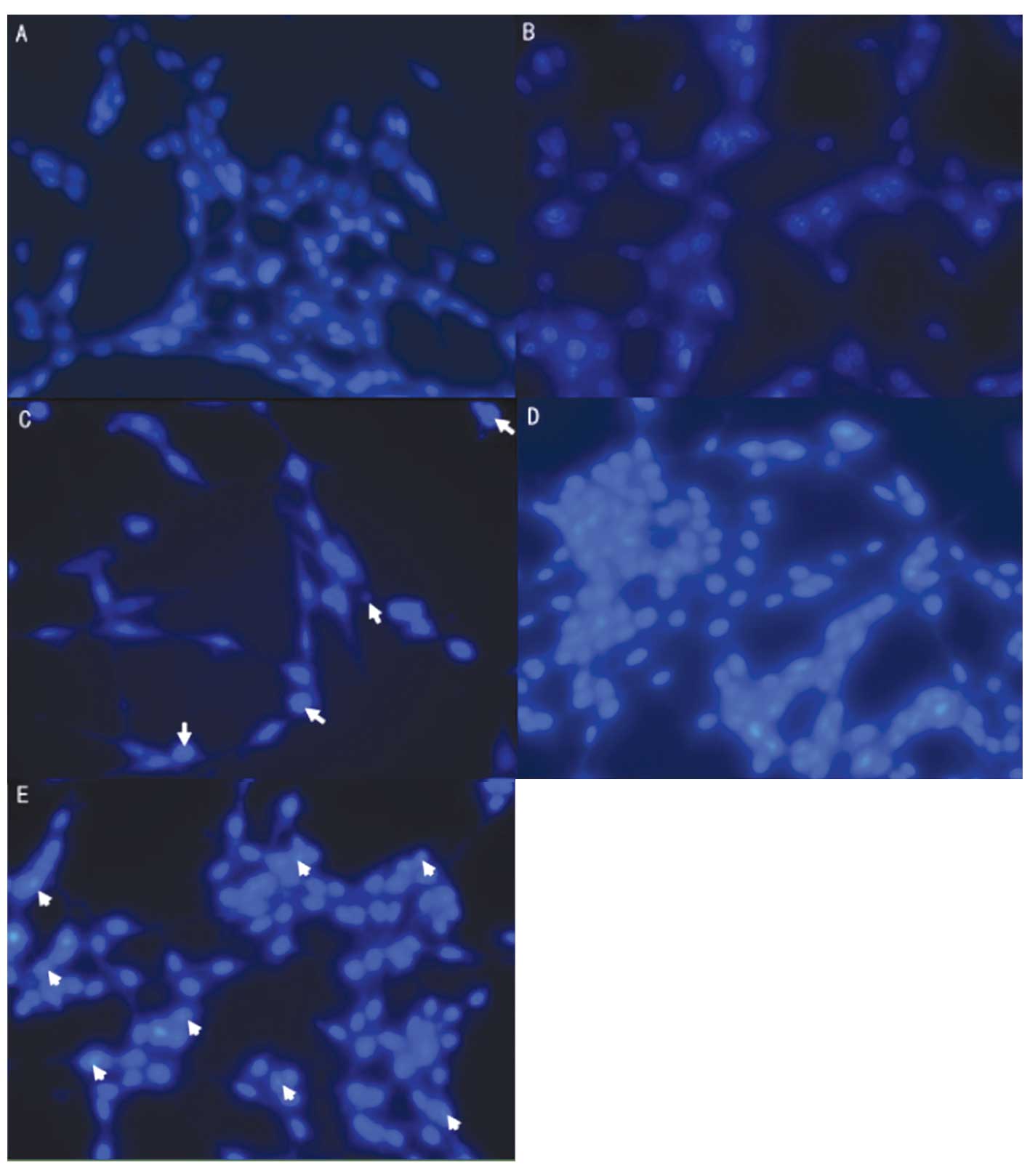
|
1
|
Mamelak AN and Jacoby DB: Targeted
delivery of antitumoral therapy to glioma and other malignancies
with synthetic chlorotoxin (TM-601). Expert Opin Drug Deliv.
4:175–186. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Weiss RB: The anthracyclines: will we ever
find a better doxorubicin? Semin Oncol. 19:670–686. 1992.PubMed/NCBI
|
|
3
|
Minotti G, Menna P, Salvatorelli E, Cairo
G and Gianni L: Anthracyclines: molecular advances and
pharmacologic developments in antitumor activity and
cardiotoxicity. Pharmacol Rev. 56:185–229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng X, Chen B, Lim CC and Sawyer DB: The
cardiotoxicology of anthracycline chemotherapeutics: translating
molecular mechanism into preventative medicine. Mol Interv.
5:163–171. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stewart LA: Chemotherapy in adult
high-grade glioma: a systematic review and meta-analysis of
individual patient data from 12 randomised trials. Lancet.
359:1011–1018. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lawson HC, Sampath P, Bohan E, et al:
Interstitial chemotherapy for malignant gliomas: the Johns Hopkins
experience. J Neurooncol. 83:61–70. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Johannessen T-CA, Bjerkvig R and Tysnes
BB: DNA repair and cancer stem-like cells - potential partners in
glioma drug resistance? Cancer Treat Rev. 34:558–567. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
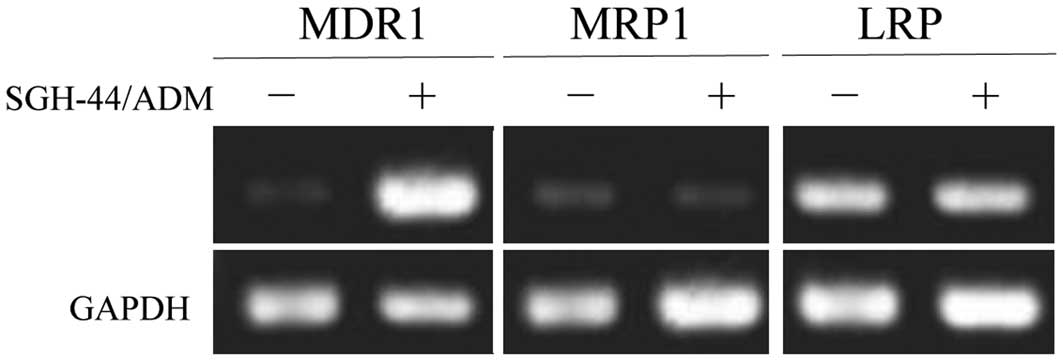
Bronger H, König J, Kopplow K, et al: ABCC
drug efflux pumps and organic anion uptake transporters in human
gliomas and the blood-tumor barrier. Cancer Res. 65:11419–11428.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Calatozzolo C, Gelati M, Ciusani E, et al:
Expression of drug resistance proteins Pgp, MRP1, MRP3, MRP5 and
GST-pi in human glioma. J Neurooncol. 74:113–121. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dalton WS, Grogan TM, Meltzer PS, et al:
Drug-resistance in multiple myeloma and non-Hodgkin’s lymphoma:
detection of P-glycoprotein and potential circumvention by addition
of verapamil to chemotherapy. J Clin Oncol. 7:415–424. 1989.
|
|
11
|
Miller TP, Grogan TM, Dalton WS, et al:
P-glycoprotein expression in malignant lymphoma and reversal of
clinical drug resistance with chemotherapy plus high-dose
verapamil. J Clin Oncol. 9:17–24. 1991.PubMed/NCBI
|
|
12
|
Régina A, Demeule M, Laplante A, et al:
Multidrug resistance in brain tumors: roles of the blood-brain
barrier. Cancer Metastasis Rev. 20:13–25. 2001.PubMed/NCBI
|
|
13
|
Mohri M, Nitta H and Yamashita J:
Expression of multidrug resistance-associated protein (MRP) in
human gliomas. J Neurooncol. 49:105–115. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spiegl-Kreinecker S, Buchroithner J,
Elbling L, et al: Expression and functional activity of the
ABC-transporter proteins P-glycoprotein and multidrug-resistance
protein 1 in human brain tumor cells and astrocytes. J Neurooncol.
57:27–36. 2002. View Article : Google Scholar
|
|
15
|
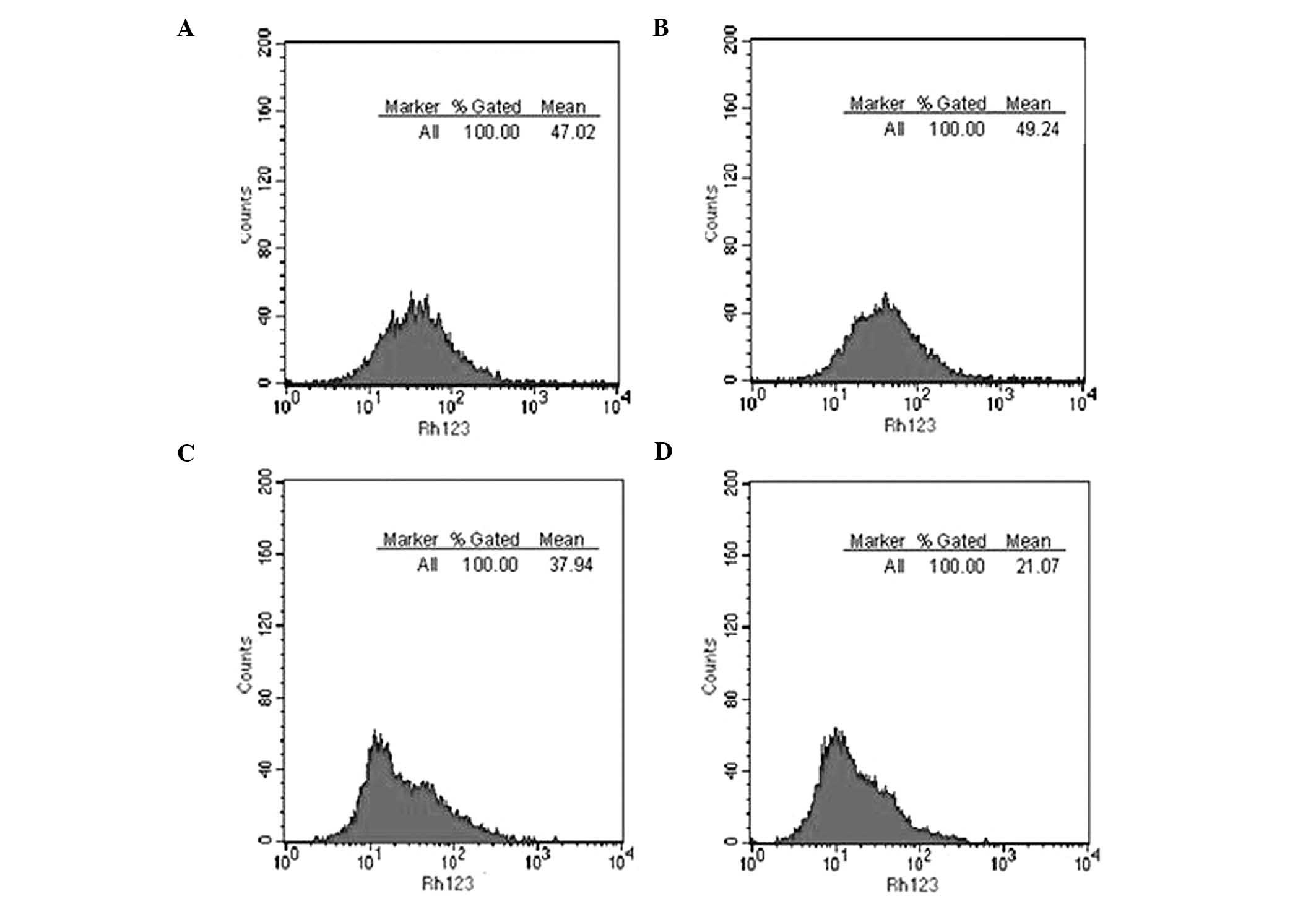
Herz J, Kowal RC, Goldstein JL and Brown
MS: Proteolytic processing of the 600 kd low density lipoprotein
receptor-related protein (LRP) occurs in a trans-Golgi compartment.
EMBO J. 9:1769–1776. 1990.PubMed/NCBI
|
|
16
|
Pinzón-Daza M, Garzón R, Courard P, et al:
The association of statins plus LDL receptor-targeted
liposome-encapsulated doxorubicin increases in vitro drug delivery
across blood-brain barrier cells. Br J Pharmacol. 167:1431–1447.
2012.
|
|
17
|
Ratnasinghe D, Daschner PJ, Anver MR, et
al: Cyclooxygenase-2, P-glycoprotein-170 and drug resistance; is
chemoprevention against multidrug resistance possible? Anticancer
Res. 21:2141–2147. 2001.PubMed/NCBI
|
|
18
|
Ali-Osman F, Caughlan J and Gray GS:
Decreased DNA interstrand cross-linking and cytotoxicity induced in
human brain tumor cells by 1,3-bis (2-chloroethyl)-1-nitrosourea
after in vitro reaction with glutathione. Cancer Res.
49:5954–5958. 1989.PubMed/NCBI
|
|
19
|
Chen C, Taniguchi T and D’Andrea A: The
Fanconi anemia (FA) pathway confers glioma resistance to DNA
alkylating agents. J Mol Med. 85:497–509. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wu XG, Peng SB and Huang Q:
Transcriptional regulation of breast cancer resistance protein. Yi
Chuan. 34:1529–1536. 2012.(In Chinese).
|
|
21
|
Candelaria M, de la Cruz-Hernandez E,
Taja-Chayeb L, et al: DNA methylation-independent reversion of
gemcitabine resistance by hydralazine in cervical cancer cells.
PLoS ONE. 7:e291812012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogawa T, Liggett TE, Melnikov AA, et al:
Methylation of death-associated protein kinase is associated with
cetuximab and erlotinib resistance. Cell Cycle. 11:1656–1663. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He J: Expression of glioma stem cell
marker CD133 and O6-methylguanine-DNA methyltransferase is
associated with resistance to radiotherapy in gliomas. Oncol Rep.
26:1305–1313. 2011.PubMed/NCBI
|
|
24
|
Wang B, Li Y, Tan Y, et al: Low-dose Cd
induces hepatic gene hypermethylation, along with the persistent
reduction of cell death and increase of cell proliferation in rats
and mice. PLoS One. 7:e338532012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
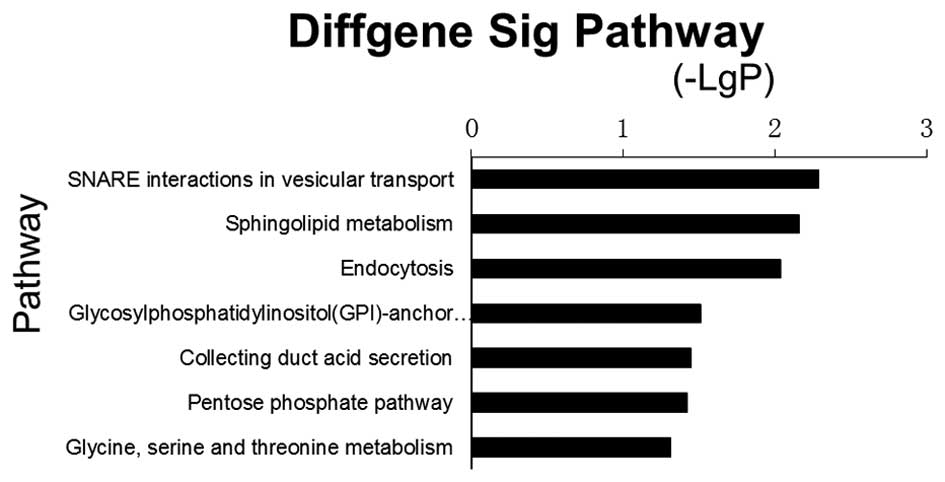
Kanehisa M, Goto S, Kawashima S, Okuno Y
and Hattori M: The KEGG resource for deciphering the genome.
Nucleic Acids Res. 32:D277–D280. 2004.PubMed/NCBI
|
|
26
|
Yi M, Horton JD, Cohen JC, Hobbs HH and
Stephens RM: WholePathwayScope: a comprehensive pathway-based
analysis tool for high-throughput data. BMC Bioinformatics.
7:302006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Draghici S, Khatri P, Tarca AL, et al: A
systems biology approach for pathway level analysis. Genome res.
17:1537–1545. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gene Ontology Consortium. The Gene
Ontology (GO) project in 2006. Nucleic Acids Res. 34:D322–D326.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ashburner M, Ball CA, Blake JA, et al:
Gene ontology: tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dupuy D, Bertin N, Hidalgo CA, et al:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Schlitt T, Palin K, Rung J, et al: From
gene networks to gene function. Genome Res. 13:2568–2576. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim Y-J, Sah RLY, Doong J-YH and
Grodzinsky AJ: Fluorometric assay of DNA in cartilage explants
using Hoechst 33258. Anal Biochem. 174:168–176. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W7748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang J, Duncan D, Shi Z and Zhang B:
WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013.
Nucleic Acids Res. 41:W77–W7783. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Denecke J, Fiedler K, Hacker-Klom U, et
al: Multiple drug-resistant C6 glioma cells cross-resistant to
irradiation. Anticancer Res. 17:4531–4534. 1997.PubMed/NCBI
|
|
36
|
Shi L, Chen J, Yang J, et al: MiR-21
protected human glioblastoma U87MG cells from chemotherapeutic drug
temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and
caspase-3 activity. Brain Res. 1352:255–264. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang L, Xiong Y, Sun Y, et al: HLungDB: an
integrated database of human lung cancer research. Nucleic Acids
Res. 38:D665–D669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kang MK and Kang SK: Tumorigenesis of
chemotherapeutic drug-resistant cancer stem-like cells in brain
glioma. Stem Cells Dev. 16:837–847. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
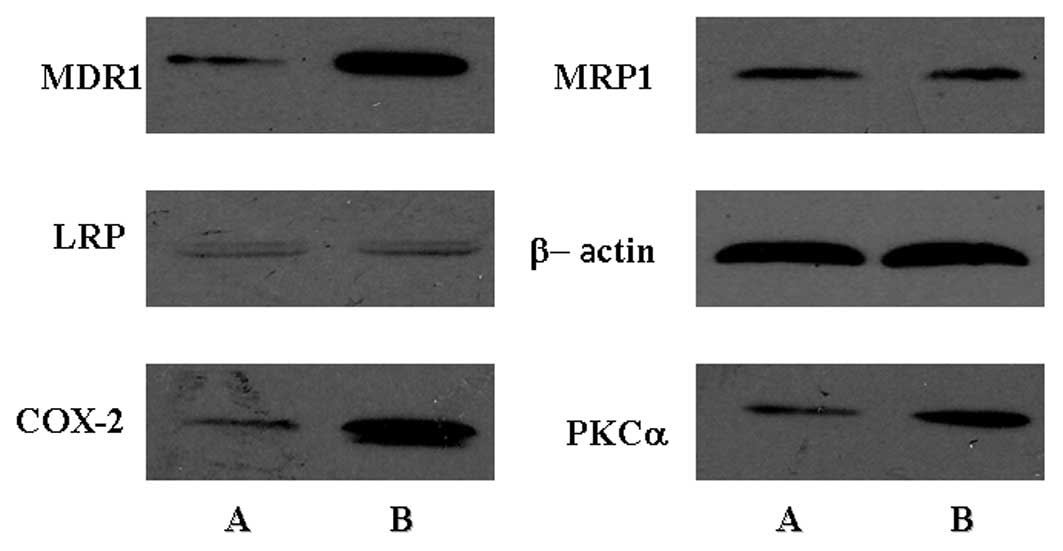
Sorokin A: Cyclooxygenase-2: potential
role in regulation of drug efflux and multidrug resistance
phenotype. Current Pharmaceutical Design. 10:647–657. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fan Q-W, Cheng C, Knight ZA, et al: EGFR
signals to mTOR through PKC and independently of Akt in glioma. Sci
Signal. 2:ra42009.PubMed/NCBI
|
|
41
|
Basu A: PKC and resistance to
chemotherapeutic agents. Protein Kinase C in Cancer Signaling and
Therapy. Kazanietz MG: Humana Press; pp. 409–429. 2010, View Article : Google Scholar
|
|
42
|
Leirdal M and Sioud M: Ribozyme inhibition
of the protein kinase C alpha triggers apoptosis in glioma cells.
Br J Cancer. 80:1558–1564. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fujita H, Yamanaka M, Imamura K, et al: A
dominant negative form of the AAA ATPase SKD1/VPS4 impairs membrane
trafficking out of endosomal/lysosomal compartments: class E vps
phenotype in mammalian cells. J Cell Sci. 116:401–414. 2003.
View Article : Google Scholar
|
|
44
|
Schnitzer JE, Liu J and Oh P: Endothelial
caveolae have the molecular transport machinery for vesicle
budding, docking, and fusion including VAMP, NSF, SNAP, annexins,
and GTPases. J Biol Chem. 270:14399–14404. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kawano M, Kumagai K, Nishijima M and
Hanada K: Efficient trafficking of ceramide from the endoplasmic
reticulum to the Golgi apparatus requires a VAMP-associated
protein-interacting FFAT motif of CERT. J Biol Chem.
281:30279–30288. 2006. View Article : Google Scholar
|
|
46
|
Guizetti J and Gerlich DW: ESCRT-III
polymers in membrane neck constriction. Trends Cell Biol.
22:133–140. 2012. View Article : Google Scholar : PubMed/NCBI
|