Introduction
Renal cell carcinoma (RCC) is a prevalent malignancy
with ~64,000 novel cases diagnosed and 13,500 mortalities per year
(1). Although major progression
has been made in the therapeutic management of kidney cancer in the
past decade, RCC remains the third most common type of urological
cancer and is responsible for 3% of adult neoplasia (2).
Aplasia Ras homologue member I (ARHI) is a
tumor suppressor gene localized to 1p31 which spans ~8 kb and
contains two exons and one intron. ARHI encodes a 26-kDa
guanosine triphosphatase (GTPase) with 55–62% homology to Ras and
Rap, though its function may differ markedly from that of Ras and
Rap (3,4). Downregulation of ARHI expression in
breast, ovarian and stomach cancer may occur through loss of
heterozygosity, DNA methylation or transcriptional regulation
(5–8). Loss of ARHI expression is associated
with tumor progression and poor prognosis (7,8).
Evidence has suggested that overexpression of ARHI is able to
inhibit cancer-cell growth in ovarian and breast cancers (9). However, the roles of ARHI in renal
cancer remain to be elucidated.
In the present study, pathological and functional
studies were performed in order to determine the role of ARHI in
the suppression of renal cancer in vivo and in vitro.
The expression levels of ARHI mRNA and protein in renal cancer were
detected using polymerase chain reaction (PCR) and western-blot
analyses. Further elucidation of the functions of ARHI may lead to
an improved understanding of the pathogenesis of renal cancer and
provide a novel therapeutic target.
Materials and methods
Patients
Surgical specimens of 52 renal cancers were obtained
from the Department of Urology, Institute of Urology, The First
Affiliated Hospital of China Medical University (Shenyang, China)
between January 2008 and December 2012. None of the patients
underwent radiotherapy or chemotherapy prior to operation. Patients
gave their consent for the use of tumor tissues for clinical
research and The First Affiliated Hospital of China Medical
University Ethics Committee (Shenyang, China) approved the research
protocols.
Cell lines
The human renal cancer cell line OS-RC-2 was
purchased from the Shanghai Cell Bank of Type Culture Collection
(Chinese Academy of Sciences, Shanghai, China) and maintained in
RPMI-1640 medium (Gibco-BRL, Invitrogen Life Technologies,
Carlsbad, CA, USA) containing 10% heat-inactivated fetal bovine
serum (FBS; HyClone, GE Healthcare, Little Chalfont, UK), 100 U/ml
penicillin G and 100 μg/ml streptomycin (Invitrogen, Carsbad, CA,
USA) in a humidified 5% CO2 incubator at 37°C.
Transfection
The pcDNA3.1-ARHI vector (provided by Dr Jin Ji,
China Medical University, Shenyang, China) was transfected into
OS-RC-2 cells using Lipofectamine 2000 (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
instructions. Following 48 h of incubation at 37°C, transfected
cells were trypsinized and seeded into 60-mm dishes.
RNA isolation and reverse transcription
quantitative (q)PCR
Total RNA was isolated from cells and tissues by
using a TRIzol reagent (Invitrogen Life Technologies) according to
the manufacturer’s instructions. cDNA was synthesized from 2 μg
total RNA. Oligo (dT) 16 and SuperScript II reverse transcriptase
(Invitrogen Life Technologies) were used for the reverse
transcription reaction, which was performed according to the
manufacturer’s instructions. The ARHI primers were as
follows: forward, 5′-CAGCTGGTTTCTTACCACGTAT-3′ and reverse,
5′-GCACAAGTTCTCCCACACTTAG-3′. To further quantify ARHI gene
expression levels in renal cancer cells, the relative ARHI
mRNA expression levels were measured by qPCR. A housekeeping gene,
GAPDH was used as an endogenous control. The GAPDH primers were as
follows: forward, 5′-AGAAGGCTGGGGCTCATTTG-3′ and reverse,
5′-AGGGGCCATCCACAGTCTTC-3′. Relative quantification was calculated
using the ΔΔCt method.
Immunofluorescence
Transfected cells were washed with
phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde,
permeabilized in 1% Triton X-100 for 5 min and blocked with 5%
bovine serum albumin in PBS containing 0.5% Triton X-100 for 1 h
(Beyotime, Beijing, China). ARHI expression was detected
using goat polyclonal immunoglobulin G (IgG) anti-ARHI antibody,
dilution 1:100, (sc-30321, Santa Cruz Biotechnology, Inc., Dallas,
TX, USA) for 1 h at room temperature. Cells were subsequently
washed with PBS and incubated with Alexa Fluor® 594
donkey anti-goat IgG (heavy and light) (A11058, dilution 1:100;
Invitrogen Life Technologies) for 1 h at room temperature, washed
with PBS and mounted using SlowFade® Gold Antifade
reagent (S36936; Invitrogen Life Technologies).
MTT assay
The proliferation rates of ARHI-transfected
and control cells were measured by MTT assay.
ARHI-transfected cells or control cells were plated at a
density of 1×103/well in 96-well plates and incubated
for 48 h in complete culture medium containing 0.5 mg/ml MTT
(Sigma-Aldrich, St. Louis, MO, USA). Following four hours of
incubation, the medium was replaced with 100 μl dimethylsulfoxide
(Sigma-Aldrich) and agitated for 10 min to dissolve the crystals.
Absorbance optical density of each well was determined at 490 nm
wavelength, including the subtraction of baseline reading (Bio-Rad,
Hercules, CA, USA).
Cell cycle and apoptosis analysis
Cells (3×105/well) were plated and
incubated overnight. Cells were trypsinized, collected in PBS and
fixed on ice with 1% paraformaldehyde, followed by 70% cold
ethanol. Following treatment with 10 μg/ml RNase (Beyotime), cells
were stained with 50 μg/ml propidium iodide (PI; Sigma-Aldrich) for
15 min at room temperature for cell cycle analysis. Apoptotic cells
were detected using Annexin V-fluorescein isothiocyanate (FITC)/PI
double staining. The cells were trypsinized and stained with
Annexin V-FITC and PI (KenGen, Nanjing, China). The stained cells
were immediately analyzed using a FACSCalibur machine (BD
Biosciences, Franklin Lakes, NJ, USA).
Xenograft assays
All experiments with animals were performed
according to the guidelines of the China Medical University Ethical
Committee. NU/NU Nude mice that were six to eight weeks-old were
purchased from Vital River (Beijing, China). The mice were kept at
a 12-h light/dark cycle (6:30 a.m. to 6:30 p.m.) at 22ºC with free
access tofood and water. OS-RC-2 cells (2×107 in 200 μl)
were subcutaneously injected into the axilla of each mouse. Once
the tumor diameters reached 3–5 mm, the mice were divided randomly
into three groups (untreated, mock or transfection) and received a
100 μl intratumoral injection of PBS, pCDNA3.1 or pCDNA3.1-ARHI,
respectively. Three injections were administered at 9am, 3pm and
9pm every three days. Tumor growth was subsequently monitored for
30 days. Every five days until the end of the experiment, one mouse
from each group was randomly selected to be anesthetized,
photographed (Olympus CX31, Olympus, Tokyo, Japan) and sacrificed
using spinal dislocation method. For each tumor, measurements were
taken using calipers and tumor volumes were calculated as follows:
length × width2 × 0.52. Survival was monitored until the
experiments were terminated due to heavy tumor burden.
Preparation of nuclear and cytoplasmic
protein extracts
Nuclear and cytoplasmic protein fractions were
isolated from cell lines at the time-points indicated in the
manufacturer’s instructions with the CelLytic™ NuCLEAR™ Extraction
kit (Sigma-Aldrich). Protein concentrations were determined by
bicinchoninic acid protein assay using bovine serum albumin as a
standard (Pierce Biotechnology, Inc., Rockford, IL, USA).
Western blot analysis
Protein from cells and tissues (30 μg) was denatured
in 6X loading buffer at 95°C for 5 min, separated by SDS-PAGE and
transferred onto a nitrocellulose membrane (Beyotime). The
membranes were subsequently incubated in 5% milk for 2 h at room
temperature and then incubated with the primary antibody overnight
at 4°C. Primary antibodies included: goat polyclonal immunoglobulin
G anti-ARHI (sc-30321, Santa Cruz Biotechnology, Inc.), rabbit
polyclonal immunoglobulin G anti-low-density lipoprotein
receptor-related protein 6 (LRP6) (sc-15399, Santa Cruz
Biotechnology, Inc.), mouse polyclonal immunoglobulin G
anti-phosphorylated-LRP6 (Ser1490) (#2568, Cell Signaling
Technology, Beverly, MA, USA), rabbit monoclonal immunoglobulin G
anti-Axin2 (#5863, Cell Signaling Technology), mouse monoclonal
immunoglobulin G anti-glycogen synthase kinase 3β (GSK-3β;
sc-81462, Santa Cruz Biotechnology, Inc.), mouse monoclonal
immunoglobulin G anti-β-catenin (#610154, BD Biosciences), and
mouse monoclonal immunoglobulin G anti-β-tubulin (T5201,
Sigma-Aldrich). The following day, membranes were washed three
times with PBS and subsequently incubated with the secondary
antibody for 2 h at room temperature. The signals were detected
using an enhanced chemiluminescence kit (GE Healthcare).
Immunohistochemical (IHC) staining
Immunohistochemistry was used to detect the
expression of ARHI protein in renal cancer tissue samples.
Immunohistochemical staining was performed on 4-μm sections
obtained from formalin-fixed, paraffin-embedded blocks. Endogenous
peroxidase activity was inhibited with 3% hydrogen peroxide for 30
min. Antigen retrieval was carried out in citrate buffer (10 mM, pH
6.0) for 30 min at 95°C in a pressure cooker (Beyotime). The
polyclonal antibody used was anti-ARHI (1:500; Santa Cruz
Biotechnology, Inc.), which was applied and incubated overnight at
4°C. Subsequently, sections were incubated with a biotinylated
secondary antibody and then exposed to a streptavidin complex
(horseradish peroxidase). Positive reactions were visualized with
3,3′-diaminobenzidine tetrahydrochloride (Sigma-Aldrich), followed
by counterstaining with hematoxylin. Normal tissue was used as a
positive control. Sections treated without primary antibodies were
used as negative controls. The positive percentage of counted cells
was graded semi-quantitatively according to a four-tier scoring
system: Negative (−), 0–5%; weakly positive (+), 6–25%; moderately
positive (++), 26–50%; and strongly positive (+++), 51–100%.
Statistical analysis
Values are expressed as the mean ± standard
deviation of three independent experiments, each performed in
triplicate with GraphPad Prism 5 (GraphPad Software, La Jolla, CA,
USA). Kaplan-Meier survival plots were generated and comparisons
between survival curves were made with the log-rank statistic.
Differences between groups were assessed by unpaired, two-tailed
Student’s t-test. P<0.05 was considered to indicate a
significant difference between values.
Results
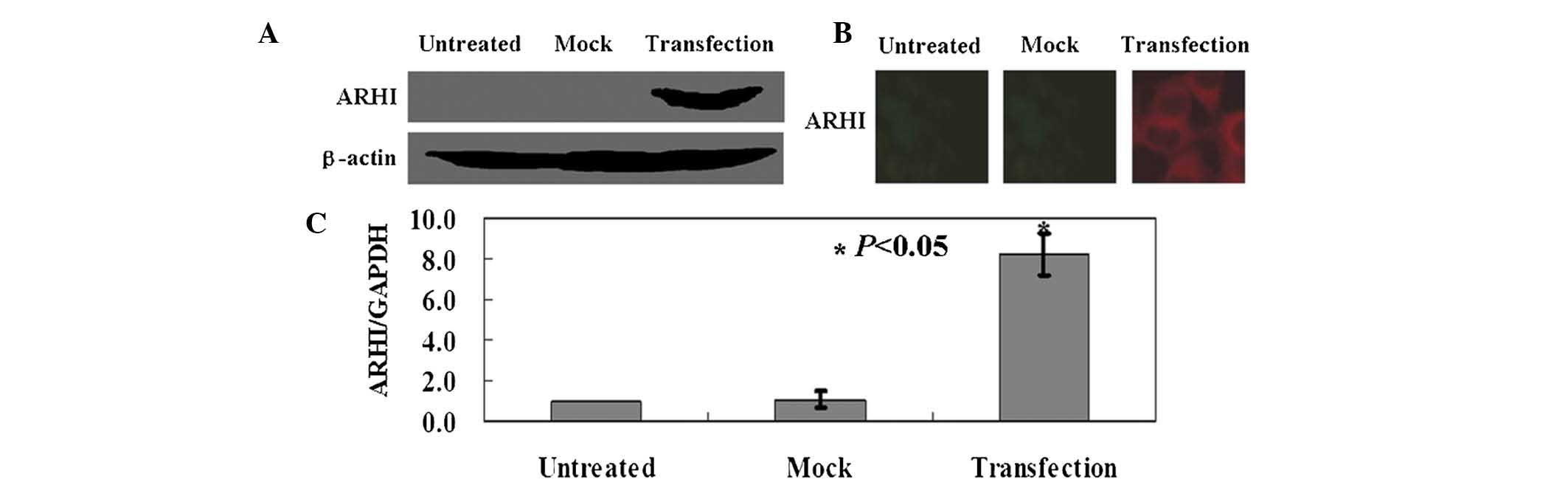
ARHI is exogenously expressed in
transfected OS-RC-2 cells
OS-RC-2 cells were transfected with the
pcDNA3.1-ARHI expression vector, and the expression levels
of ARHI protein and mRNA were measured by western blot
analysis (Fig. 1A),
immunofluorescence assay (Fig. 1B)
and qPCR (Fig. 1C). As indicated
in Fig. 1, the results confirmed
exogenous expression of ARHI in OS-RC-2 cells following
transfection.
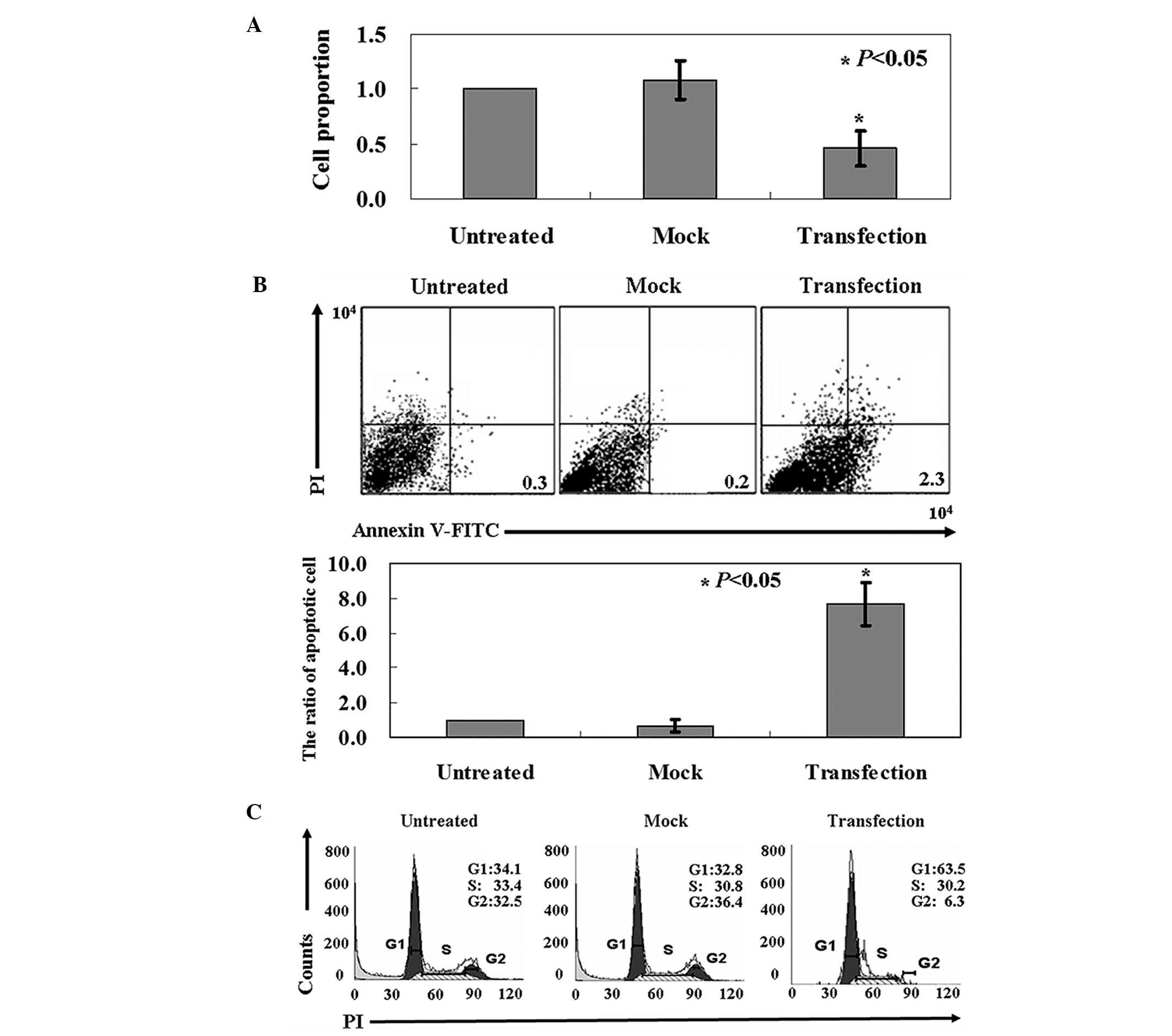
Anti-tumor activity of ARHI in OS-RC-2
cells
MTT assay indicated a significantly lower growth
rate in the ARHI-expressing OS-RC-2 cells than that in the
untreated cells (P<0.05; Fig.
2A). Annexin-V/PI double staining demonstrated a higher level
of apoptosis in ARHI-expressing OS-RC-2 cells vs mock cells or
untransfected cells (P<0.05; Fig.
2B). PI staining of cells revealed that ARHI-expressing OS-RC-2
cells were arrested in the G1 phase (Fig. 2C).
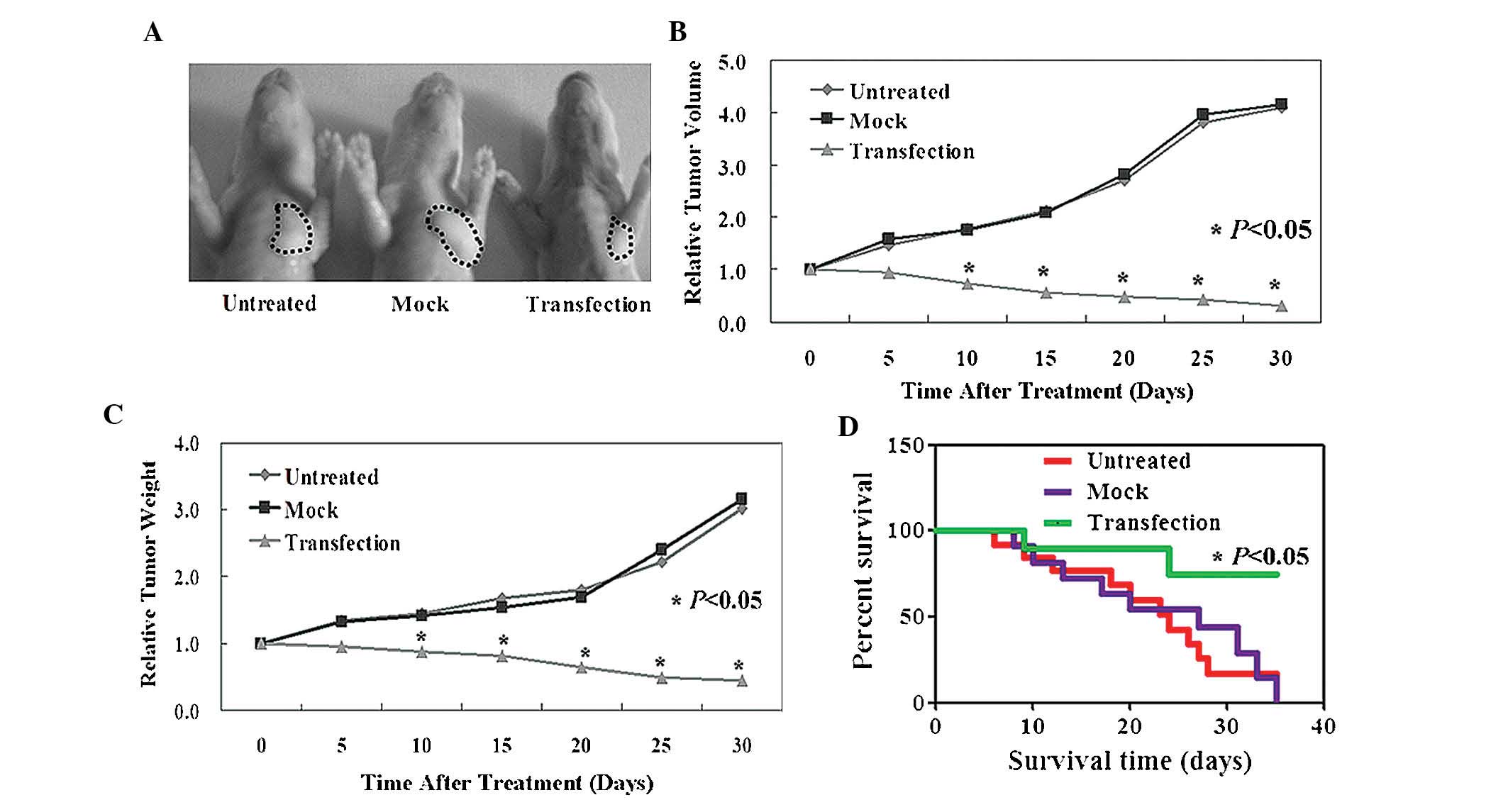
ARHI inhibits tumor growth and promotes
survival rate in vivo
Established xenograft tumor models were used to
determine whether ARHI exhibits antitumor properties in
vivo. As indicated in Fig. 3A and
B, the tumor volumes in ARHI-treated mice were significantly
reduced in comparison to those of the PBS- or pCDNA-3.1-treated
mice (P<0.05). Similarly, tumor weights in the ARHI-treated
group were also significantly reduced compared to those of the PBS-
and pCDNA-3.1-treated groups (P<0.05; Fig. 3C). It was additionally revealed
that ARHI-treated mice displayed an improved survival rate in
comparison with that of the PBS- and mock-treated mice (P<0.05;
Fig. 3D).
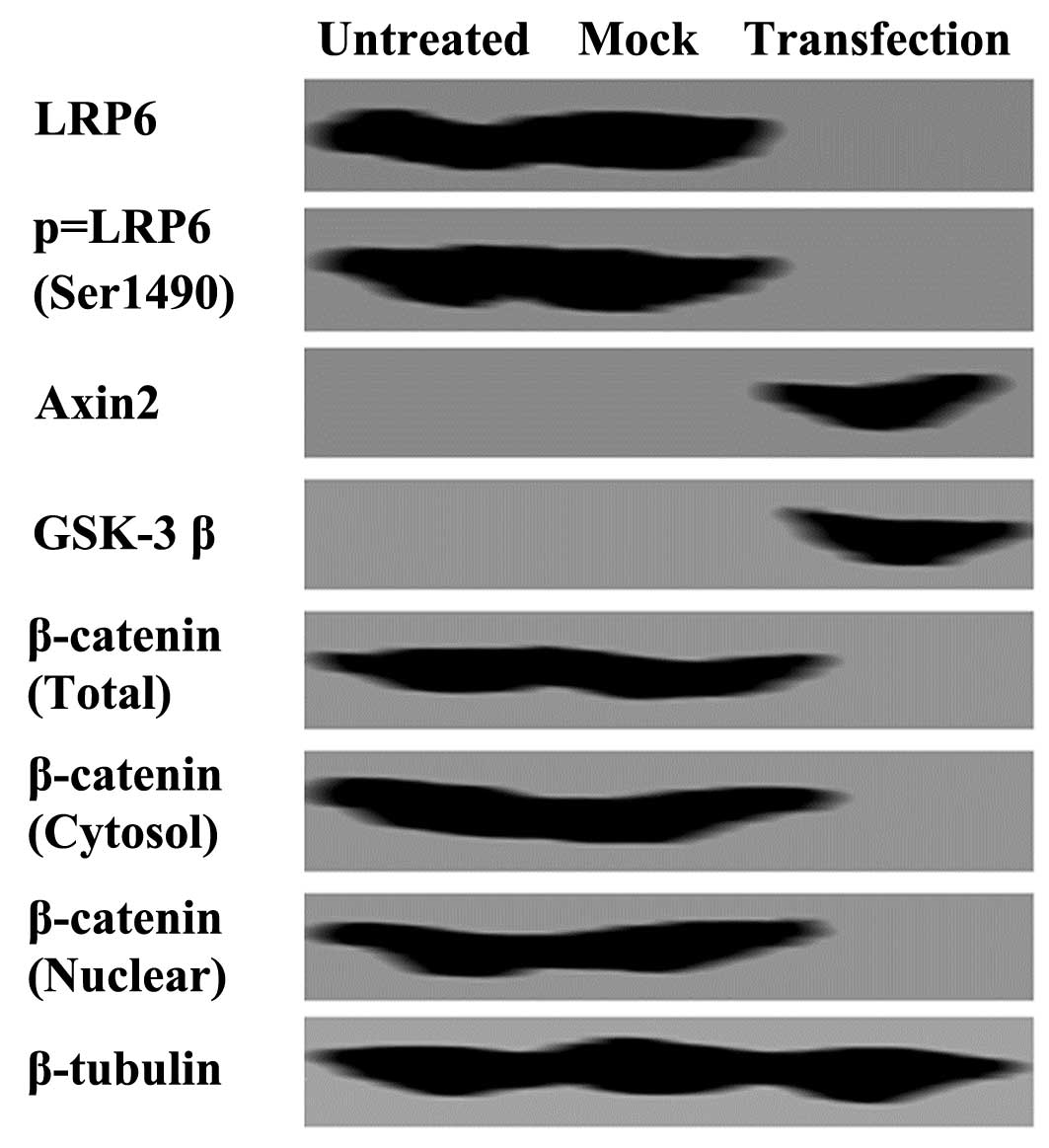
ARHI blocks the β-catenin signaling
pathway in OS-RC-2 cells
To examine the mechanisms induced by ARHI in OS-RC-2
cells, western blot analysis was performed. Cytoplasmic and nuclear
β-catenin expression levels were significantly reduced following
transfection with ARHI compared with those of the controls
(Fig. 4). To determine the
mechanism responsible for the decrease in β-catenin expression
levels, the expression levels of LRP6 were examined. Western blot
analysis detected a significant inhibition of LRP6 expression and
phosphorylation following transfection with ARHI (Fig. 4). An increase in Axin2 and GSK-3β
expression levels were also detected in ARHI-expressing cells
compared with those of the controls (Fig. 4).
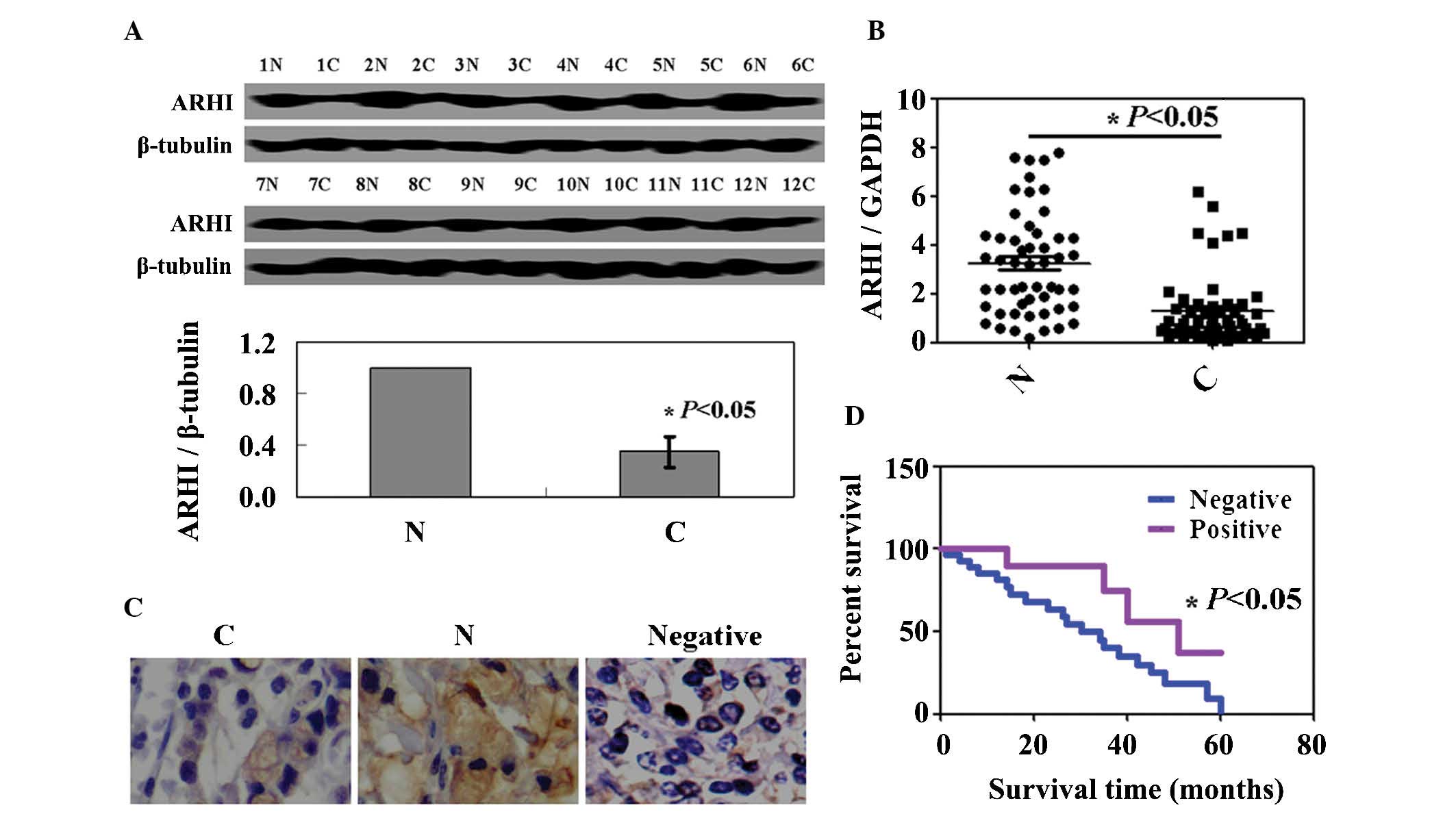
ARHI expression is reduced in renal
cancer
ARHI expression in renal cancer specimens from 52
patients was decreased compared with those in matched normal
tissues (Fig. 5A). As shown in
Fig. 5B, the levels of ARHI
mRNA expression in renal cancer specimens were lower than those in
normal tissues. ARHI protein and mRNA expression levels were
coincident. Immunostaining for ARHI was only localized in the
cytoplasm. ARHI protein was markedly expressed in non-tumor parts
of specimens (Fig. 5C). The
association between the expression levels of ARHI and the
clinicopathological characteristics of these patients is summarized
in Table I. No correlation was
found between gender, age, differentiation, histological grade,
lymph node metastasis and tumor size (P>0.05). However, ARHI
expression was significantly associated with T stage and distant
metastasis (P<0.05). The cumulative survival rate of patients
with ARHI expression was higher than that in those without ARHI
expression (P<0.05; Fig.
5D).
 | Table IAssociation between ARHI expression
and the clinicopathological parameters of renal cancer. |
Table I
Association between ARHI expression
and the clinicopathological parameters of renal cancer.
| | ARHI expression |
|---|
| |
|
|---|
| Clinicopathological
feature | n | − | + | ++ | +++ | PR (%) | χ2 | P |
|---|
| Gender | | | | | | | 0.219 | 0.975 |
| Female | 18 | 15 | 1 | 1 | 1 | 16.7 | | |
| Male | 34 | 29 | 2 | 2 | 1 | 14.7 | | |
| Age (years) | | | | | | | 2.873 | 0.412 |
| <65 | 15 | 11 | 1 | 2 | 1 | 26.7 | | |
| ≥65 | 37 | 33 | 2 | 1 | 1 | 10.8 | | |
| Differentiation | | | | | | | 0.912 | 0.823 |
| Differentiated | 22 | 18 | 1 | 2 | 1 | 18.2 | | |
|
Undifferentiated | 30 | 26 | 2 | 1 | 1 | 13.3 | | |
| Histological
grade | | | | | | | 4.597 | 0.204 |
| I–II | 32 | 29 | 2 | 1 | 0 | 9.3 | | |
| III–IV | 20 | 15 | 1 | 2 | 2 | 25.0 | | |
| T stage | | | | | | | 9.683 | 0.022 |
| T1–T2 | 38 | 35 | 2 | 0 | 1 | 7.9 | | |
| T3–T4 | 14 | 9 | 1 | 3 | 1 | 35.7 | | |
| Lymph node
metastasis | | | | | | | 4.041 | 0.257 |
| − | 35 | 29 | 1 | 3 | 2 | 17.1 | | |
| + | 17 | 15 | 2 | 0 | 0 | 11.8 | | |
| Tumor size | | | | | | | 4.762 | 0.190 |
| < 4 cm | 11 | 8 | 2 | 1 | 0 | 27.3 | | |
| ≥ 4 cm | 41 | 36 | 1 | 2 | 2 | 12.2 | | |
| Distant
metastasis | | | | | | | 9.117 | 0.028 |
| Absent | 15 | 11 | 0 | 2 | 2 | 26.7 | | |
| Present | 37 | 33 | 3 | 1 | 0 | 10.8 | | |
Discussion
ARHI has been demonstrated to act as a suppressor in
numerous types of cancer (3,9,10).
Previous studies have additionally indicated that ARHI inhibited
cell proliferation and promoted apoptosis (9,11,10).
Consistent with previous studies, the results of the present study
confirmed the anti-tumor activity of ARHI in renal cancer cells.
Additionally, ARHI inhibited tumor growth in vivo. To the
best of our knowledge, the present study was the first to
demonstrate the association between ARHI and renal cancer.
Numerous studies have demonstrated the mechanism of
ARHI in cancer cells. ARHI inhibits the Ras/mitogen activated
protein pathway, induces p21 (WAF1/CIP1) and downregulates cyclin
D1 in cancer cells (3,5). A study revealed that ARHI regulates
autophagy in ovarian cancer cells and leads to autophagic mortality
in cell culture through inhibition of the mammalian target of
rapamycin pathway (12). The
results of the present study indicated that ARHI induced apoptosis
via the β-catenin signaling pathway. In order to verify this
molecular mechanism, all associated proteins in the β-catenin
signaling pathway were detected. LRP6 is a Wnt co-receptor which
functions in the Wnt/β-catenin signaling pathway, and its
phosphorylation is required for the activation of Wnt/β-catenin
signaling (13). LRP6
phosphorylation is followed by the formation of the axin-GSK-3β
complex (14). In the absence of
Wnt signaling, GSK-3β has been shown to be active (15), which leads to the degradation of
cytoplasmic β-catenin and an inhibition of nuclear translocation
(16). Cytoplasmic and nuclear
β-catenin localization is considered an indicator of activated Wnt
signaling (17,18). In the present study, the expression
levels of GSK-3β and axin, as well as cytoplasmic and nuclear
β-catenin expression levels detected in ARHI-expressing OS-RC-2
cells were consistent with the results of previous studies and
confirmed that ARHI had the capacity to inhibit the proliferation
of OS-RC-2 cells by suppression of the β-catenin signaling
pathway.
In conclusion, to the best of our knowledge, the
present study was the first to determine the effects of ARHI on
human renal cancer cells in vivo and in vitro. The
results demonstrated that ARHI had a tumor suppressor role in
OS-RC-2 cells, exerting its effects via the β-catenin signaling
pathway. These results provide a novel insight into the possible
therapeutic applications of ARHI in renal cancer.
References
|
1
|
Siegel R, DeSantis C, Virgo K, Stein K,
Mariotto A, Smith T, Cooper D, Gansler T, Lerro C, Fedewa S, et al:
Cancer treatment and survivorship statistics, 2012. CA Cancer J
Clin. 62:220–241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kelley JR and Duggan JM: Gastric cancer
epidemiology and risk factors. J Clin Epidemiol. 56:1–9. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Luo RZ, Fang X, Marquez R, Liu SY, Mills
GB, Liao WS, Yu Y and Bast RC: ARHI is a Ras-related small
G-protein with a novel N-terminal extension that inhibits growth of
ovarian and breast cancers. Oncogene. 22:2897–2909. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hisatomi H, Nagao K, Wakita K and Kohno N:
ARHI/NOEY2 inactivation may be important in breast cancer
pathogenesis. Oncology. 62:136–140. 2002. View Article : Google Scholar
|
|
5
|
Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y,
Cuevas B, Kuo WL, Gray JW, Siciliano M, et al: NOEY2 (ARHI), an
imprinted putative tumor suppressor gene in ovarian and breast
carcinomas. Proc Natl Acad Sci USA. 96:214–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang W, Bu XM, Wang J, Zhang N and Zhao
CH: The expression of ARHI in pT2a and pT2b stage gastric cancer
and its clinical significance. Oncol Rep. 27:1953–1959.
2012.PubMed/NCBI
|
|
7
|
Rosen DG, Wang L, Jain AN, Lu KH, Luo RZ,
Yu Y, Liu J and Bast RC Jr: Expression of the tumor suppressor gene
ARHI in epithelial ovarian cancer is associated with increased
expression of p21 WAF1/CIP1 and prolonged progression-free
survival. Clin Cancer Res. 10:6559–6566. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang L, Hoque A, Luo RZ, Yuan J, Lu Z,
Nishimoto A, Liu J, Sahin AA, Lippman SM, Bast RC Jr and Yu Y: Loss
of the expression of the tumor suppressor gene ARHI is associated
with progression of breast cancer. Clin Cancer Res. 9(10 Pt 1):
3660–3666. 2003.PubMed/NCBI
|
|
9
|
Bao JJ, Le XF, Wang RY, Yuan J, Wang L,
Atkinson EN, LaPushin R, Andreeff M, Fang B, Yu Y and Bast RC Jr:
Reexpression of the tumor suppressor gene ARHI induces apoptosis in
ovarian and breast cancer cells through a caspase-independent
calpain-dependent pathway. Cancer Res. 62:7264–7272.
2002.PubMed/NCBI
|
|
10
|
Tang HL, Hu YQ, Qin XP, Jazag A, Yang H,
Yang YX, Yang XN, Liu JJ, Chen JM, Guleng B and Ren JL: Aplasia ras
homolog member I is downregulated in gastric cancer and silencing
its expression promotes cell growth in vitro. J Gastroenterol
Hepatol. 27:1395–1404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Huang S, Chang IS, Lin W, Ye W, Luo RZ, Lu
Z, Lu Y, Zhang K, Liao WS, Tao T, Bast RC Jr, et al: ARHI (DIRAS3),
an imprinted tumour suppressor gene, binds to importins and blocks
nuclear import of cargo proteins. Biosci Rep. 30:159–168. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare
S, Kondo S, Kondo Y, Yu Y, Mills GB, et al: The tumor suppressor
gene ARHI regulates autophagy and tumor dormancy in human ovarian
cancer cells. J Clin Invest. 118:3917–3929. 2008.PubMed/NCBI
|
|
13
|
Zeng X, Huang H, Tamai K, Zhang X, Harada
Y, Yokota C, Almeida K, Wang J, Doble B, Woodgett J, et al:
Initiation of wnt signaling: control of Wnt coreceptor Lrp6
phosphorylation/activation via frizzled, dishevelled and axin
functions. Development. 135:367–375. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bache KG, Slagsvold T and Stenmark H:
Defective downregulation of receptor tyrosine kinases in cancer.
EMBO J. 23:2707–2712. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ikeda S, Kishida S, Yamamoto H, Murai H,
Koyama S and Kikuchi A: Axin, a negative regulator of the Wnt
signaling pathway, forms a complex with GSK-3beta and beta-catenin
and promotes GSK-3beta-dependent phosphorylation of beta-catenin.
EMBO J. 17:1371–1384. 1998. View Article : Google Scholar
|
|
16
|
Aberle H, Bauer A, Stappert J, Kispert A
and Kemler R: beta-catenin is a target for the ubiquitin-proteosome
pathway. EMBO J. 16:3797–3804. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Anderson CB, Neufeld KL and White RL:
Subcellular distribution of Wnt pathway proteins in normal and
neoplastic colon. Proc Natl Acad Sci USA. 99:8683–8688. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jin Z, Han YX and Han XR: Degraded
iota-carrageenan can induce apoptosis in human osteosarcoma cells
via the Wnt/β-catenin signaling pathway. Nutr Cancer. 65:126–131.
2013.PubMed/NCBI
|