Introduction
MicroRNAs (miRNAs) are short noncoding RNAs, 18 to
25 nucleotides in length, which regulate gene expression (1,2).
They are essential to a number of biological processes, such as
embryogenesis, development, cell growth, cell differentiation and
cell death (2,3). Recent studies have shown that miRNAs
regulate a large number of oncogenes and tumor suppressor genes
(4,5). Therefore, understanding the
downstream targets of a number of miRNAs is important in diagnostic
therapeutic applications for human cancer.
5-Fluorouracil (5-FU) is a commonly-used
chemotherapeutic agent that is effective for treating a range of
malignant tumors (6). 5-FU is
converted to fluorodeoxyuridine monophosphate, which forms a stable
complex with thymidylate synthase and thus inhibits deoxythymidine
monophosphate production (7,8).
However, a primary cause of treatment failure in advanced colon
cancer is the development of chemoresistance to 5-FU. Currently,
the overall response rate for advanced colorectal cancer to 5-FU
alone remains at only 10–15% (9),
and the combination of 5-FU with other antitumor drugs has improved
the response rates to 40–50% (10). Therefore, a novel therapeutic
strategy is required to target cellular signaling molecules in
order to overcome chemoresistance to colorectal cancer
treatments.
The Warburg effect describes the phenomenon by which
cancer cells exhibit dysregulated aerobic glycolysis regardless of
their oxygen status. It has been intensively investigated and is
recognized as one of the characteristic hallmarks of cancer cells
in the current understanding of cancer cell metabolism (11,12).
Furthermore, cancer cells were found to require high levels of
glucose and to be sensitive to changes in glucose concentration
(13,14). Lactate dehydrogenase A (LDHA) is
one of the primary isoforms of LDH expressed in cancer tissue. It
controls the conversion of pyruvate to lactate during the cellular
glycolytic process. It has been shown that LDHA is involved in
cancer cell glycolysis and growth, and tumor maintenance (15). Furthermore, it has been reported
that LDH activity may be a reliable prognostic marker in certain
types of cancer (16).
The present study investigated the effect of
miR-34a-mediated glucose metabolism inhibition, via targeting of
the 3′ untranslated region (UTR) region of LDHA, on the mechanism
of 5-FU resistance in colon cancer cells.
Materials and methods
Cell lines and culture
Cells from the DLD-1 human colon cancer cell line
were obtained from the American Type Culture Collection (Manassas,
VA, USA). Cells were cultured in RPMI-1640 (Sigma-Aldrich, Hong
Kong, China) supplemented with 10% fetal bovine serum
(Sigma-Aldrich) and 1X penicillin-streptomycin-glutamine
(10378-016; Invitrogen Life Technologies, Carlsbad, CA, USA) at
37°C in a humidified incubator with 95% air and 5%
CO2.
Generation of the 5-FU-resistant cell
line
The 5-FU-resistant cell line from DLD-1 human colon
cancer cells was generated as described previously (17). Briefly, DLD-1 cells were treated
with gradually increasing concentrations of 5-FU in regular cell
culture conditions in order to select resistant cells. Following
successive treatments for a duration of up to three months,
resistant cell clones were pooled and used for all subsequent
experiments in this study. Resistant cells were treated with 5-FU
each month for elimination of the cells which may have regained
sensitivity to 5-FU.
Antibodies and reagents
Rabbit polyclonal LDHA and β-actin antibodies were
obtained from Cell Signaling Technology, Inc., (Danvers, MA, USA;
#2012 and #4967, respectively) and 5-FU antibodies were obtained
from Sigma-Aldrich.
Western blot analysis
Whole cells were lysed in 1X SDS sample buffer,
resolved by electrophoresis using SDS-PAGE and transferred to
nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA, USA).
The membranes were probed with primary antibodies overnight and
incubated with horseradish peroxidase-conjugated polyclonal goat
anti-rabbit immunoglobulin G secondary antibodies (Thermo Fisher
Scientific, Waltham, MA, USA) for 3 h, prior to detection using a
Super Signal Enhanced Chemiluminescence kit (Pierce Biotechnology,
Inc., Rockford, IL, USA). For sequential blotting, membranes were
stripped with Stripping Buffer (Pierce Biotechnology, Inc.) and
re-probed with other primary antibodies.
Cell viability assay
A colorimetric assay using the tetrazolium salt, MTT
(EMD Millipore, Billerica, MA, USA), was used to assess the
cytotoxicity of the anticancer agent, 5-FU. Single-cell suspensions
were prepared and cell density was measured. MTT assays were
performed according to the manufacturer’s instructions. Briefly, an
equal number of cells were added into each well with culture medium
containing normal concentrations of either 5-FU or
phosphate-buffered saline for the untreated control. Following four
days of culture, 0.1 mg MTT was added to each well and was
incubated at 37°C for an additional 4 h. Plates were centrifuged at
450 × g for 5 min at room temperature, and the medium was
discarded. Dimethyl sulfoxide (0.15 ml) was added to each well to
solubilize the crystals, and the plates were immediately read at
540 nm on a scanning multiwell spectrometer (Bio-Tek instruments,
Inc., Burlington, VT, USA). All experiments were performed three
times.
Pre-miRNA transfection
miRNA precursors (pre-miRNAs) and pre-miR negative
controls were purchased from Applied Biosystems Life Technologies
(Foster City, CA, USA). Lipofectamine® 2000 (Invitrogen
Life Technologies) was used for the transfection of pre-miRNAs. At
48 h following transfection, the expression of miR-34a was detected
by reverse transcription-quantitative polymerase chain reaction
(RT-qPCR), and the expression of LDHA, a target of miR-34a, was
measured using western blotting.
Plasmid DNA transfection
Transfection was performed using the Lipofectamine
2000 Transfection reagent (Invitrogen Life Technologies) according
to the manufacturer’s instructions. Overexpression vectors
containing wild type LDHA (Myc-DDK-tagged) were purchased from
OriGene Technologies, Inc. (Rockville, MD, USA). At 48 h following
transfection, cells were collected or whole-cell lysates were
prepared for further analysis.
RT-qPCR
RNA was extracted from cancer cells using TRIzol
reagent (Invitrogen Life Technologies). cDNA synthesis was
performed using a SuperScript First-Standard Synthesis system for
RT-qPCR (Invitrogen Life Technologies) according to the
manufacturer’s instructions. qPCR analyses were performed using
Assay-on-Demand primers and the TaqMan Universal PCR Master Mix
reagent (Applied Biosystems Life Technologies). Samples were
analyzed using an ABI Prism 7700 Sequence Detection system (Applied
Biosystems Life Technologies). The following primers were used:
LDHA, forward 5′-TGGAGTGGAATGAATGTTGC-3′, reverse
5′-ATAGCCCAGGATGTGTAGCC-3′; and β-actin, forward
5′-AGGCACCAGGGCGTGAT-3′, and reverse 5′-GCCCACATAGGAATCCTTCTGAC-3′.
LDHA expression levels were normalized to those of β-actin. For
miRNA expression analysis, RT-qPCR was conducted using the TaqMan
microRNA reverse transcription kit (Applied Biosystems) and TaqMan
microRNA assays kit (Applied Biosystems) according the
manufacturer’s instructions. All reactions were performed in
triplicate. Human U6 served as an internal control. The relative
quantities of mRNA were calculated using the comparative
CT method (17).
Experiments were performed three times.
Lactate production assay
Lactate production was detected using a Lactate
assay kit (BioVision, Inc., Milpitas, CA, USA). Results were
normalized to the quantity of total protein in the control
cells.
LDH activity assay
Total LDH activity in cell lysates was examined
using the LDH cytotoxicity assay kit (BioVision, Inc.) according to
the manufacturer’s instructions. Briefly, 2×105 cells
were seeded in a 24-well plate one day prior to the assay and all
samples were analyzed in triplicate. Cells were collected and
washed, and protein was extracted in order to measure LDH activity.
Results were normalized to the quantity of total protein in the
control cells.
Luciferase assays
The DLD-1 cells were plated at 5×104
cells/well in 24-well plates. The following day, the cells were
co-transfected with luciferase reporter plasmids (pMIR-REPORT™
miRNA Expression Reporter Vector System; Invitrogen Life
Technologies; AM5795); with wild type 3′-UTR or mutant 3′-UTR of
LDHA, and pre-miR-34a or pre-miR-negative (control miR; Applied
Biosystems), using Lipofectamine® 2000 reagent
(Invitrogen Life Technologies; 11668019). Forty-eight hours
post-transfection the cells were harvested and lysed using passive
lysis buffer (Dual-Luciferase® Reporter Assay System;
Promega; E1910). Luciferase (LUC) activity was measured using a
dual luciferase reporter assay (Dual-Luciferase®
Reporter Assay System; Promega; E1910). The pRL-TK vector was used
as an internal control. The results are expressed as relative LUC
activity (firefly LUC/renilla LUC).
Statistical analysis
The data were analyzed using GraphPad 5.0 (GraphPad
Software, Inc., La Jolla, CA, USA). Unpaired Student’s t-test was
used for the data analysis. All data are shown as the mean ±
standard error. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-34a is downregulated in
5-FU-resistant colon cancer cells
Since miR-34a has been reported to act as a tumor
suppressor in a number of tumor types (18) and is associated with 5-FU
resistance (17), the present
study investigated the role of miR-34a in 5-FU resistance in human
colon cancer cells. A 5-FU-resistant cell line was generated using
DLD-1 cells, by selection with gradually increasing concentrations
of 5-FU in a cell culture medium. Following successive treatments
for a duration of three months, several 5-FU-resistant cell clones
were developed and pooled for use in the subsequent experiments. To
verify resistance, parental cells and resistant pool cells were
treated with 5-FU at various concentrations for 72 h. As
hypothesized, cell viability assays demonstrated that DLD-1
5-FU-resistant cells tolerated markedly higher concentrations of
5-FU compared with sensitive cells, which exhibited significant
inhibition of viability at 4, 20 and 50 μM (Fig. 1A). The IC50 was ~5 μM
for 5-FU-sensitive cells and 45 μM for resistant cells. As
hypothesized, the expression of miR-34a was significantly
downregulated in 5-FU-resistant cells compared with sensitive cells
(Fig. 1B), suggesting that miR-34a
may act as a tumor suppressor in human colon cancers and that it is
involved in the development of resistance to 5-FU.
LDHA is a direct target of miR-34a in
colon cancer cells
The initial results showed that miR-34a is
downregulated in 5-FU-resistant cells. Potential targets of miR-34a
were then investigated. An miRNA database (www.targetscan.org) was searched for predicted targets
of miR-125b that may contribute to 5-FU resistance. The public
miRNA database, TargetScan, predicted that LDHA may be a target for
miR-34a, and showed that the 3′-UTR of LDHA contains a highly
conserved binding site for miR-34a (Fig. 2A). To the best of our knowledge,
thus far no publication has reported that LDHA is a direct target
of miR-34a in colon cancer cells. To determine whether miR-34a
targets LDHA in colon cancer cells, pre-miR-34a was transfected
into DLD-1 cells. Overexpression of miR-34a significantly
downregulated expression of the LDHA protein (Fig. 2B). The following experiment sought
to investigate whether miR-34a directly targets the 3′-UTR of LDHA
mRNA. A luciferase reporter analysis was performed by
co-transfecting a vector containing pMIR reporter-luciferase fused
with either the original 3′-UTR sequence or a sequence with a
mutation in the predicted binding site of the 3′-UTR of LDHA mRNA,
and with either pre-miR-34a or control microRNA. Overexpression of
miR-34a decreased the luciferase activity of the reporter
containing the wild type 3′-UTR of LDHA by ~60% in DLD-1 cells
(Fig. 2C). However, no such
inhibitory effects of miR-34a on the activity of the reporter fused
with the 3′-UTR of LDHA with the mutation in the predicted binding
site were detected (Fig. 2C),
These results demonstrate that LDHA is a direct target of miR-34a
in colon cancer cells.
5-FU-resistant cells exhibit increased
expression and activity of LDHA
Previous studies have shown that dysregulated
cellular metabolism is associated with 5-FU resistance in cancer
cells (19). LDHA catalyzes the
final step in the glycolytic pathway, which is the conversion of
pyruvate and nicotinamide adenine dinucleotide dehydrogenase to
lactate and nicotinamide adenine dinucleotide, and is known to be
involved in tumor maintenance. The present study aimed to
investigate whether LDHA is involved in miR-34a-mediated 5-FU
resistance in colon cancer cells. Notably, the expression of LDHA
was upregulated at protein and mRNA levels in 5-FU-resistant cells
(Fig. 3A and B), suggesting that
the downregulation of miR-34a in 5-FU-resistant cells may
contribute to LDHA upregulation. Consistent with this, the activity
of LDH and the levels of lactate were increased in 5-FU-resistant
cells compared with 5-FU-sensitive cells (Fig. 3C and D). These results demonstrate
that LDHA is associated with 5-FU resistance in colon cancer cells
and may be a promising therapeutic target.
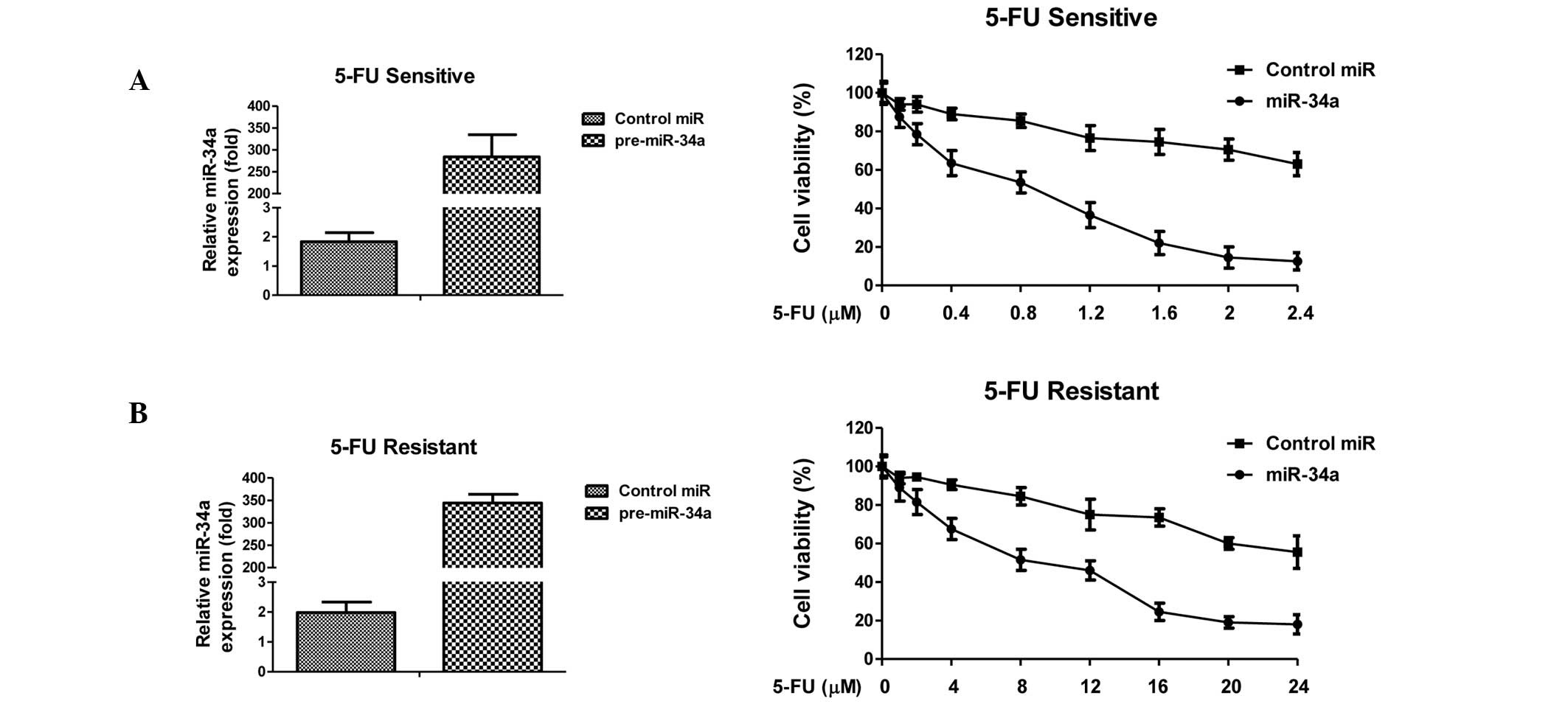
Overexpression of miR-34a sensitizes
5-FU-resistant cells through direct targeting of LDHA
To investigate the mechanism underlying the
association between miR-34a-mediated downregulation of LDHA and
5-FU resistance in colon cancer cells, miR-34a was exogenously
overexpressed in DLD-1 5-FU-sensitive and resistant cells through
transient transfection. These cells were then treated with
increasing concentrations of 5-FU for 72 h. Transfection with
miR-34a significantly inhibited cell viability in sensitive and
resistant cancer cells (Fig. 4A and
B). Compared with control microRNA, overexpression of miR-34a
in 5-FU-sensitive cells led to a decrease in IC50 from 4
to 1 μM. The IC50 of 5-FU-resistant cells in response to
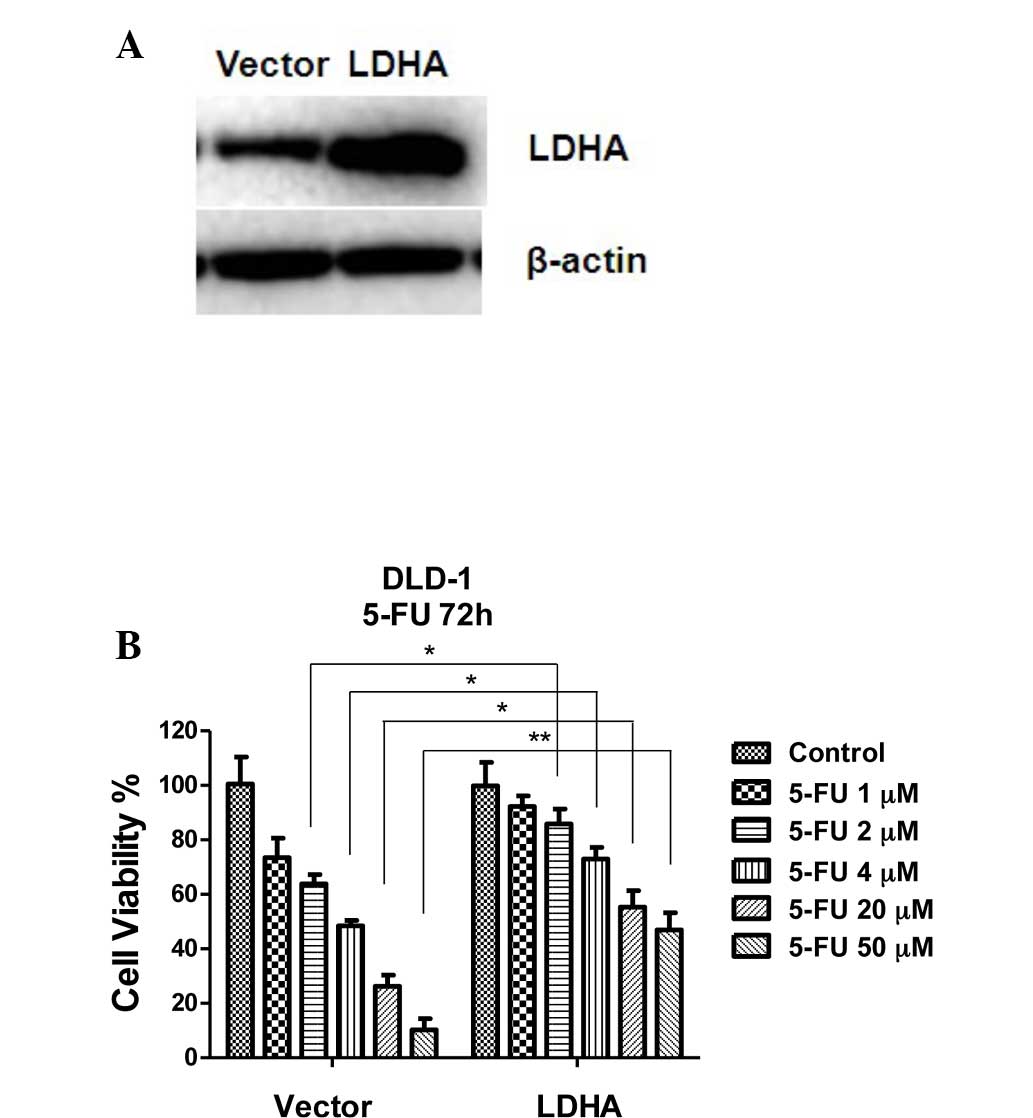
5-FU decreased from 45 to 8 μM. To verify whether overexpression of
miR-34a sensitizes colon cancer cells to 5-FU treatment through
inhibition of LDHA, LDHA was transiently transfected into DLD-1
5-FU-sensitive cells (Fig. 5A) and
the sensitivity to increasing concentrations of 5-FU for 72 h was
measured (Fig. 5B). The results
demonstrated that exogenous overexpression of LDHA rendered DLD-1
cells resistant to 5-FU, indicating that overexpression of miR-34a
results in cells that are susceptive to 5-FU via the inhibition of
LDHA.
Discussion
miRNAs have been shown to be involved in the
regulation of a number of processes that are deregulated in cancer
cells, such as proliferation, differentiation and apoptosis
(1,2). Downregulation of miR-34a has been
reported in a number of cancer types, including colorectal cancer
(20), pancreatic cancer (21), prostate cancer (22) and neuroblastoma (23). To date, numerous targets of the
miR-34 family have been postulated, including
mesenchymal-epithelial transition factor (23), cyclin-dependent kinase 6 (24), c-Myc and N-Myc (25), silent mating type information
regulation 2 homolog (SIRT1) (26)
and Bcl-2 (27). The greatest
level of induction by p53 was observed in miR-34a, which has been
shown to be a direct target gene of p53 (28). Furthermore, ectopic miR-34
expression induces apoptosis, and cell-cycle arrest or senescence
(29). Thus, miR-34a is recognized
as a tumor suppressor. A recent study showed that ectopic
expression of miR-34a in 5-FU-resistant colon cells inhibited cell
growth and attenuated the resistance to 5-FU through the
downregulation of SIRT1 and E2F3 (17), suggesting that miR-34a is involved
in cancer cell resistance to 5-FU. The results of the present study
showed a downregulation of miR-34a expression in 5-FU-resistant
colon cancer cells, which suggests a tumor suppressive function of
this miRNA in colon cancer cells.
In 1956, Warburg observed that the rate of
glycolysis was abnormally high in cancer cells, yet a smaller
fraction of this glucose is broken down by oxidative
phosphorylation. This Warburg effect indicates that the metabolic
properties of cancer cells are different from those of normal
cells. Cancer cells are more dependent than healthy cells on
aerobic glycolysis, fatty acid synthesis and glutaminolysis for
proliferation (12). Therefore,
targeting cancer cell metabolism may be a selective approach by
which to treat cancer patients. Recently, a number of studies have
reported that dysregulated metabolism is associated with drug
resistance. Cancer cells are known to take up glucose avidly and
generate lactate through LDHA, which has been reported to be
involved in tumor maintenance and progression. In addition, LDH
activity is increased in colon cancer, indicating that LDHA may be
of use as a prognostic marker (30). Therefore, as chemoresistant cancer
cells exhibit an abnormal metabolism, this could in itself be a
target for the development of novel therapeutic agents.
The current study demonstrated that LDHA is a direct
target of miR-34a in colon cancer cells. Overexpression of miR-34a
decreased LDHA protein levels, which contributed to the
re-sensitization of 5-FU-resistant cancer cells. The role of LDHA
in acquired 5-FU resistance in human colon cancer cells was
investigated. Compared with parental cells, 5-FU-resistant cells
exhibited an increase in the expression and activity of LDHA.
Overexpression of LDHA resulted in increased resistance to 5-FU. In
addition, the current study found that inhibition of LDHA by
overexpressing miR-34a significantly re-sensitized 5-FU-resistant
cells. This demonstrated the importance of miR-34a-mediated
inhibition of LDHA in overcoming chemoresistance in cancer cells.
Further investigation into other putative targets of miR-34a is
required. This may be facilitated by the use of gene expression
profiling approaches to identify signal pathways regulated by
miR-34a and their association with sensitivity to 5-FU in order to
develop novel therapeutic approaches to overcome 5-FU
resistance.
Acknowledgements
The authors would like to thank the staff and
faculty of the Department of Hematology and Oncology, the 101st
Hospital of the People’s Liberation Army and Dr Haibin Zhao of the
Department of Pathology, the 101st Hospital of the People’s
Liberation Army for his editorial assistance.
References
|
1
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
4
|
Fabbri M, Croce CM and Calin GA:
MicroRNAs. Cancer J. 14:1–6. 2008. View Article : Google Scholar
|
|
5
|
Slaby O, Bienertova-Vasku J, Svoboda M and
Vyzula R: Genetic polymorphisms and microRNAs: new direction in
molecular epidemiology of solid cancer. J Cell Mol Med. 16:8–21.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Longley DB, Harkin DP and Johnston PG:
5-fluorouracil: mechanisms of action and clinical strategies. Nat
Rev Cancer. 3:330–338. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Curtin NJ, Harris AL and Aherne GW:
Mechanism of cell death following thymidylate synthase inhibition:
2′-deoxyuridine-5′-triphosphate accumulation, DNA damage, and
growth inhibition following exposure to CB3717 and dipyridamole.
Cancer Res. 51:2346–2352. 1991.
|
|
8
|
Peters GJ, van Triest B, Backus HH, Kuiper
CM, van der Wilt CL and Pinedo HM: Molecular downstream events and
induction of thymidylate synthase in mutant and wild-type p53 colon
cancer cell lines after treatment with 5-fluorouracil and the
thymidylate synthase inhibitor raltitrexed. Eur J Cancer.
36:916–924. 2000. View Article : Google Scholar
|
|
9
|
Giacchettii S, Perpoint B, Zidani R, Le
Bail N, Faggiuolo R, Focan C, Chollet P, Llory JF, Letourneau Y,
Coudert B, et al: Phase III multicenter randomized trial of
oxaliplatin added to chronomodulated fluorouracil-leucovorin as
first-line treatment of metastatic colorectal cancer. J Clin Oncol.
18:136–147. 2000.
|
|
10
|
Douillard JY, Cunningham D, Roth AD,
Navarro M, James RD, Karasek P, Jandik P, Iveson T, Carmichael J,
Alakl M, et al: Irinotecan combined with fluorouracil compared with
fluorouracil alone as first-line treatment for metastatic
colorectal cancer: a multicentre randomised trial. Lancet.
355:1041–1047. 2000. View Article : Google Scholar
|
|
11
|
Vander Heiden MG, Cantley LC and Thompson
CB: Understanding the Warburg effect: the metabolic requirements of
cell proliferation. Science. 324:1029–1033. 2009.PubMed/NCBI
|
|
12
|
Warburg O: On respiratory impairment in
cancer cells. Science. 124:269–270. 1956.PubMed/NCBI
|
|
13
|
Kim JW and Dang CV: Cancer’s molecular
sweet tooth and the Warburg effect. Cancer Res. 66:8927–8930.
2006.
|
|
14
|
Zhao Y, Butler EB and Tan M: Targeting
cellular metabolism to improve cancer therapeutics. Cell Death Dis.
7:e5322013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Doherty JR and Cleveland JL: Targeting
lactate metabolism for cancer therapeutics. J Clin Invest.
123:3685–3692. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fantin VR, St-Pierre J and Leder P:
Attenuation of LDH-A expression uncovers a link between glycolysis,
mitochondrial physiology, and tumor maintenance. Cancer Cell.
9:425–434. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Akao Y, Noguchi S, Iio A, Kojima K, Takagi
T and Naoe T: Dysregulation of microRNA-34a expression causes
drug-resistance to 5-FU in human colon cancer DLD-1 cells. Cancer
Lett. 300:197–204. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hermeking H: The miR-34 family in cancer
and apoptosis. Cell Death Differ. 17:193–199. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou Y, Tozzi F, Chen J, Fan F, Xia L,
Wang J, et al: Intracellular ATP levels are a pivotal determinant
of chemoresistance in colon cancer cells. Cancer Res. 72:304–314.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Akao Y, Nakagawa I, Hirata I, Iio A, Itoh
T, Kojima K, Nakashima R, Kitade Y and Naoe T: Role of
anti-oncomirs miR-143 and -145 in human colorectal tumors. Cancer
Gene Ther. 17:398–408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chang TC, Wentzel EA, Kent OA,
Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M,
Ferlito M, Lowenstein CJ, et al: Transactivation of miR-34a by p53
broadly influences gene expression and promotes apoptosis. Mol
Cell. 26:745–752. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rokhlin OW, Scheinker VS, Taghiyev AF,
Bumcrot D, Glover RA and Cohen MB: MicroRNA-34 mediates
AR-dependent p53-induced apoptosis in prostate cancer. Cancer Biol
Ther. 7:1288–1296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan D, Zhou X, Chen X, Hu DN, Dong XD,
Wang J, et al: MicroRNA-34a inhibits uveal melanoma cell
proliferation and migration through downregulation of c-Met. Invest
Ophthalmol Vis Sci. 50:1559–1565. 2009. View Article : Google Scholar
|
|
24
|
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R,
et al: Downregulation of CCND1 and CDK6 by miR-34a induces cell
cycle arrest. FEBS Lett. 582:1564–1568. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wei JS, Song YK, Durinck S, Chen QR, Cheuk
AT, Tsang P, et al: The MYCN oncogene is a direct target of
miR-34a. Oncogene. 27:5204–5213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yamakuchi M, Ferlito M and Lowenstein CJ:
miR-34a repression of SIRT1 regulates apoptosis. Proc Natl Acad Sci
USA. 105:13421–13426. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang F, Li QJ, Gong ZB, Zhou L, You N,
Wang S, Li XL, Li JJ, An JZ, Wang DS, et al: MicroRNA-34a targets
Bcl-2 and sensitizes human hepatocellular carcinoma cells to
sorafenib treatment. Technol Cancer Res Treat. 13:77–86.
2014.PubMed/NCBI
|
|
28
|
Ji Q, Hao X, Meng Y, Zhang M, Desano J,
Fan D and Xu L: Restoration of tumor suppressor miR-34 inhibits
human p53-mutant gastric cancer tumorspheres. BMC Cancer.
8:2662008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Welch C, Chen Y and Stallings RL:
MicroRNA-34a functions as a potential tumor suppressor by inducing
apoptosis in neuroblastoma cells. Oncogene. 26:5017–5022. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Koukourakis I, Giatromanolaki A and
Sivridis E: Colorectal cancer: Lactate dehydrogenase (LDH) activity
as a prognostic marker. Methods of Cancer Diagnosis, Therapy, and
Prognosis. Springer; Berlin, Germany: 4. pp. 241–253. 2009
|