Introduction
Coronary artery bypass grafting (CABG) surgery is a
common cardiac surgical procedure. CABG surgery is performed either
with or without cardiopulmonary bypass, referred to as the
traditional method of on-pump CABG and the newer off-pump CABG,
respectively (1). A number of
studies have confirmed that, compared with on-pump CABG, off-pump
CABG has a lower risk of renal damage, myocardial injury, brain
injury, stroke, atrial fibrillation and neurocognitive and organ
dysfunction (2–5). Clinical trials have revealed that
off-pump CABG has no significant benefit regarding mortality,
stroke or myocardial infarction compared with on-pump CABG,
although off-pump CABG may be preferable when patients have
contraindications for cardiopulmonary bypass (6,7).
CABG surgery can lead to ischemic injury due to a transient period
of local ischemia with temporary occlusion of the target vessel,
particularly in patients with poor cardiac contractile function
(8–10). Certain cytokines, including
interleukin (IL)-6, interferon-γ, high sensitivity C-reactive
protein and granulocyte colony-stimulating factor, are released in
off-pump CABG surgery to a similar or larger extent than in on-pump
CABG surgery (11,12). Therefore, perioperative management
for off-pump CABG surgery requires improvement to reduce myocardial
ischemic-reperfusion damage.
Various interventions, including certain anesthetic
agents, prior to and following myocardial ischemia have the
potential, to a certain extent, to reduce myocardial ischemic
damage and subsequent reperfusion injury (13,14).
Sevoflurane is a type of inhalational anaesthetic agent, which
significantly reduces the size of infarcts and Ca2+
loading to protect the myocardium against reperfusion injury
(14,15) and has myocardial protective
properties for low risk patients, who are undergoing CABG surgery
(16). It has been demonstrated
that preconditioning by sevoflurane downregulates
platelet-endothelial cell adhesion molecule-1 and upregulates
catalase in atrial biopsies from patients undergoing CABG surgery
(17). The levels of IL-6 and IL-8
in the serum are suppressed by sevoflurane, while the IL-10 and
IL-1 receptor antagonist, remain significantly increased in
patients undergoing CABG (18). In
addition to volatile anesthetics, intravenous anesthetics,
including propofol, may also reduce myocardial reperfusion injury
in patients undergoing CABG surgery (19). It has been observed that propofol
reduces lipid peroxidation, which is mediated by free radicals, and
systemic inflammation in patients with impaired left ventricular
function undergoing CABG surgery, with no differences in urinary
isoprostane concentrations or leucocyte function (20). The activities of nitric oxide
synthase and phosphoinositide-3-kinase/Akt, which are maintained by
propofol, may be partly responsible for reduced
ischemic-reperfusion injury (21,22).
Kottenberg et al (23)
revealed that propofol attenuates the effects of remote ischemic
preconditioning in patients undergoing CABG surgery (23) and, with a chemical structure
similar to that of free-radical scavengers, interferes with remote
ischemic preconditioning, since the release of free radicals is
necessary to evoke isoflurane-induced preconditioning (24–26).
There is increasing evidence that sevoflurane and propofol have
effective cardioprotective effects, however, the underlying
mechanisms of these anaesthetic agents for cardioprotection remain
to be elucidated (27).
Lucchinetti et al (28)
analyzed the differentially regulated pathways in sevoflurane- and
propofol-treated patients by gene expression profiling using a gene
set enrichment analysis method (28). In the present study, common
differentially expressed genes (DEGs), propofol-specific and
sevoflurane-specific DEGs, were identified. Subsequently, in
addition to performing functional annotation of these DEGs,
functional analysis of the same DEGs in the selected interactive
functional modules was performed. The results of the present study
aimed to reveal specific novel mechanisms underlying the
cardioprotective effects of sevoflurane and propofol.
Materials and methods
Samples
The expression profiling of GSE4386 produced by
Lucchinetti et al (28) and
including 40 atrial samples, was downloaded from the National
Center of Biotechnology Information Gene Expression Omnibus
(http://www.ncbi.nlm.nih.gov/geo/), which
was based on the Affymetrix Human Genome U133 Plus 2.0 Array
platform (Affymetrix, Santa Clara, CA, USA).
The 40 atrial samples were collected at the
beginning and at the end of the off-pump CABG surgery and included
20 atrial samples from 10 patients receiving the anesthetic gas
sevoflurane (Sevorane; Abbott, Baar, Switzerland) and 20 atrial
samples from 10 patients receiving the intravenous anesthetic
propofol (Diprivan 2%; AstraZeneca, Zug, Switzerland). The
sevoflurane and propofol were adjusted to maintain the blood
pressure and heart rate within 20% of the baseline values (28). The downloaded expression profiling
for the 40 atrial samples were further analyzed to identify
DEGs.
Data pre-treatment and DEG
identification
The platform annotation file in txt format, provided
by Affymetrix, was used to map the association between the probes
and the gene symbols. Subsequently, quartile data normalization was
performed using the Affy package in the R language (29) and the Multtest package
(Bioconductor, Fred Hutchinson Cancer Research Center, Seattle, WA,
USA; 30) was used to identify the genes differentially expressed
between the samples at the beginning and at the end of surgery in
the sevoflurane and propofol groups. The Benjamini-Hochberg
procedure (31) was used to adjust
the raw P-values into false discovery rate (FDR). FDR<0.05 and
|logFC|>1 were used as cut-off criteria in the DEG
identification, where FC stands for fold change.
Intergroup comparison of DEGs
Subsequent to obtaining the DEGs in the sevoflurane
and propofol groups, the sevoflurane specific, propofol-specific
and common DEGs between the two groups were selected. The
expression levels of the sevoflurane-specific, propofol-specific
and common DEGs in the samples at the beginning of surgery were
then compared with those in the samples at the end of surgery using
a t-test. P<0.05 was considered to indicate a statistically
significant difference.
Functional enrichment analysis for
DEGs
Gene ontology (GO) biological process enrichment
analyses for the screened DEGs in the sevoflurane and propofol
groups were performed using the Database for Annotation,
Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/), containing
analytical tools and bioinformatic resources for the systematic
extraction of biological functions (32). FDR<0.05 was selected as a
cut-off criterion. The annotated protein sequences of the DEGs were
compared with the proteins in clusters of orthologous groups of
proteins (COG; http://www.ncbi.nlm.nih.gov/COG) (33) database using the Basic Local
Alignment Search tool (BLASTX) (34) with a cut-off threshold of E<1 ×
10−5. The common DEGs, propofol-specific DEGs and
sevoflurane-specific DEGs were then classified into different
functional annotations and COG categories, including cellular
component (CC), molecular function (MF) and biological process
(BP).
Analysis of interactive function modules
for DEGs
WebGestalt software (Vanderbilt University,
Nashville, TN, USA; http://bioinfo.vanderbilt.edu/webgestalt/; 35,36)
was used to search for the interactive function modules of DEGs
within the cut-off criterion of FDR<0.05. Analysis of the GO
biological processes associated with the DEGs in the modules was
then performed using Expression Analysis Systematic Explorer
software (version 2.0, http://david.abcc.ncifcrf.gov/ease/ease.jsp) using the
threshold of P<0.05.
Results
DEG screening
The normalized expression profiles were analyzed to
identify the DEGs with FDR<0.05 and |logFC|>1. A total of 879
and 290 DEGs were selected in the sevoflurane and propofol groups,
respectively.
DEG comparison
The DEGs in the sevoflurane group were compared with
those in the propofol group. There were 275 common DEGs, 604
sevoflurane-specific DEGs and 15 propofol-specific DEGs (Fig. 1A). The percentages of the common
DEGs were 31.3 (275/879) and 94.8% (275/290) in the sevoflurane and
propofol groups, respectively.
The differences in the expression levels were in
accordance with the DEG identification (Fig. 1B). The sevoflurane-specific DEGs
were differentially expressed in the sevoflurane group (P=0.01239)
compared with the propofol group (P=0.214) and the
propofol-specific DEGs were differentially expressed in the
propofol group (P=0.02206) compared with the sevoflurane group
(P=0.2243). Furthermore, the common DEGs were differentially
expressed in the sevoflurane group (P=2.2 × 10−16) and
propofol group (P=2.98 × 10−14).
Functional categories for DEGs
The ‘response to wounding’ function, involving 72
DEGs, was identified as the most significant GO term of the 879
DEGs in the sevoflurane group and the ‘response to organic
substance’ function, involving 45 DEGs, was the most significant GO
term of the 290 DEGs in the propofol group (Fig. 2). The common DEGs,
sevoflurane-specific DEGs and propofol-specific DEGs were analyzed
using BLASTX by comparing with the COG database with the E<1×
10−5. The results demonstrated that the functional
categories in CC, MF and BP for the common, sevoflurane-specific
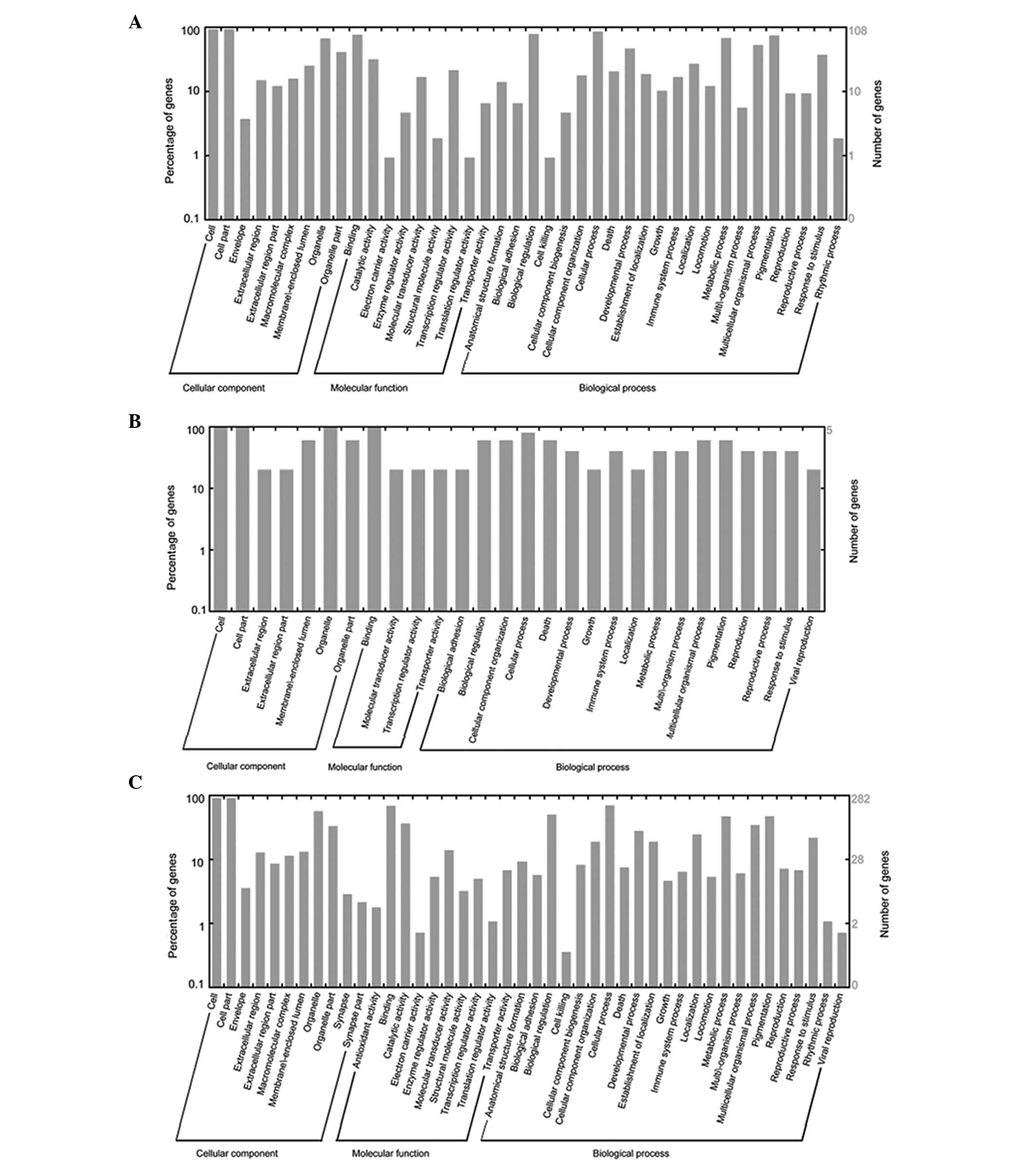
and propofol-specific DEGs were similar (Fig. 3).
Analysis of modules
Total functional modules of the common DEGs (two
total), propofol-specific DEGs and sevoflurane-specific DEGs, with
an FDR<0.05, were identified (Fig.
4). There were 10 and 18 DEGs in the two functional modules for
the common DEGs, including activating transcription factor 3
(ATF3), jun D proto-oncogene (JUND), mitogen-activated protein
kinase (MAPK)3, FBJ murine osteosarcoma viral oncogene homolog B
(FOSB), jun B proto-oncogene (JUNB) and tumor necrosis factor
α-induced protein 3 (TNFαIP3). A total of two upregulated DEGs,
including CD93 and leucine rich repeat containing 32 (LRRC32) were
involved in the functional modules for propofol-specific DEGs.
Eight upregulated DEGs, including cytokine inducible SH2-containing
protein (CISH), protein tyrosine phosphatase, non-receptor type 1
(PTPN1), suppressor of cytokine signaling 1 (SOCS1), interleukin 15
receptor α (IL15Rα), regulator of calcineurin 1 (RCAN1),
interleukin 6 receptor (IL6R), dual specificity phosphatase 4
(DUSP4) and signal transducer and activator of transcription 3
(STAT3) were involved in the functional modules for
sevoflurane-specific DEGs. In addition, seven downregulated DEGs,
including carboxypeptidase X (M14 family), member 1 (CPXM1),
fibroblast growth factor receptor 2 (FGFR2), fibronectin leucine
rich transmembrane protein 3 (FLRT3), interleukin 17 receptor D
(IL17RD), insulin receptor substrate 1 (IRS1), IL7 and colony
stimulating factor 1 receptor (CSF1R) were also observed in the
functional modules for sevoflurane-specific DEGs.
The biological processes associated with the DEGs in
the modules with P<0.05 were selected (Table I). The DEGs in these modules were
involved in cellular processes, including ‘regulation of
transcription’ and ‘regulation of cellular process’, which were
similar to the functional annotations of the DEGs.
 | Table IBiological processes associated with
the differentially expressed genes (DEGs) in the modules for common
DEGs, propofol-specific DEGs and sevoflurane-specific DEGs. |
Table I
Biological processes associated with
the differentially expressed genes (DEGs) in the modules for common
DEGs, propofol-specific DEGs and sevoflurane-specific DEGs.
| Module | GO-ID | P-value | Count | Description |
|---|
| Common 1 | 6355 | 1.668 ×
10−7 | 10 | Regulation of
transcription, DNA-dependent |
| 51252 | 1.668 ×
10−7 | 10 | Regulation of RNA
metabolic processes |
| 45449 | 3.545 ×
10−6 | 10 | Regulation of
transcription |
| 10556 | 5.286 ×
10−6 | 10 | Regulation of
macromolecule biosynthetic process |
| 10468 | 5.286 ×
10−6 | 10 | Regulation of gene
expression |
| 19219 | 5.286 ×
10−6 | 10 | Regulation of
nucleobase, nucleoside, nucleotide and nucleic acid metabolic
process |
| 31326 | 5.286 ×
10−6 | 10 | Regulation of
cellular biosynthetic process |
| 51171 | 5.286 ×
10−6 | 10 | Regulation of
nitrogen compound metabolic process |
| Common 2 | 50794 | 0.018 | 12 | Regulation of
cellular process |
| 50789 | 0.025 | 12 | Regulation of
biological process |
| 65007 | 0.035 | 12 | Biological
regulation |
| 48518 | 0.002 | 9 | Positive regulation
of biological process |
|
Propofol-specific | 42116 | 0.027 | 1 | Macrophage
activation |
| 2274 | 0.027 | 1 | Myeloid leukocyte
activation |
| 6909 | 0.027 | 1 | Phagocytosis |
|
Sevoflurane-specific | 23052 | 1.590 ×
10−4 | 12 | Signaling |
| 50794 | 0.017 | 12 | Regulation of
cellular process |
| 50789 | 0.024 | 12 | Regulation of
biological process |
| 65007 | 0.033 | 12 | Biological
regulation |
| 7165 | 0.001 | 9 | Signal
transduction |
| 23033 | 0.002 | 9 | Signaling
pathway |
| 23060 | 0.002 | 9 | Signal
transmission |
| 23046 | 0.002 | 9 | Signaling
process |
Discussion
Compared with on-pump CABG, the levels of TNFα,
heart-type fatty acid-binding protein and creatine kinase-MB are
significantly lower in off-pump CABG, suggesting a decreased
systemic inflammatory response and reduced myocardial damage
(8,37). However, off-pump CABG surgery can
also lead to ischemic injury (8).
Several studies have identified that, to a certain extent,
sevoflurane and propofol are effective cardioprotective anesthetic
agents (27,38). In the present study, 275 common,
604 sevoflurane-specific and 15 propofol-specific DEGs were
identified from expression profiles of atrial samples, which were
obtained from patients receiving either the anesthetic gas
sevoflurane or the intravenous anesthetic propofol prior to and
following off-pump CABG surgery. Functional analysis of the modules
for the common, sevoflurane-specific and propofol-specific DEGs
revealed that the DEGs in the modules involved with cellular
processes, including ‘regulation of transcription’ and ‘regulation
of cellular process’, were similar to the functional annotations of
the DEGs.
A total of 10 and 18 DEGs were present in the first
and second functional modules for the common DEGs in the
sevoflurane and propofol groups, including ATF3, JUND, JUNB, FOSB,
MAP2K3 and TNFαIP3. ATF3 is a member of the activation
transcription factor family of transcription factors. It has been
demonstrated that ATF3 protects cardiac myocytes against
doxorubicin-induced apoptosis (39). In addition, ATF3 promotes neurite
outgrowth in injured neurons (40)
and is important in promoting neuronal survival (41). ATF3 has been observed to interact
with JUND, which may protect cells from p53-dependent senescence
and apoptosis (42,43). Bergman et al (44) demonstrated that nuclear extracts of
cardiac fibroblasts from hypoxic rats contained Fos-related antigen
1, JUNB and FOSB, which significantly increase the transcriptional
activities of cardiac fibroblasts (44). Furthermore, discrete activator
protein-1 components, including JUNB and FOSB are induced by
oxidative stress and mediate the transcription and translation of
matrix metalloproteinase 2, which is important in the response of
the heart following cardiac ischemic-reperfusion injury (45). MAP2K3 belongs to the MAP kinase
family and is involved in the MAPK-mediated signaling cascade,
which is possibly important in the pathogenesis of cardiac and
vascular disease (46,47). Transfection experiments have
confirmed that p38-MAPK is involved in cardiac myocyte apoptosis
(48). Previous studies have
demonstrated that single nucleotide polymorphisms in TNFαIP3 are
associated with an increased risk of coronary artery disease in
type 2 diabetes and of left ventricular hypertrophy in hypertensive
patients (49,50). The activation of nuclear factor-κB
can be inhibited by overexpression of TNFαIP3 to attenuate cardiac
myocyte hypertrophy (51,52).
In the functional modules for propofol-specific
DEGs, two upregulated DEGs, CD93 and LRRC32, were involved. A total
of eight upregulated DEGs (CISH, PTPN1, SOCS1, IL15Rα, RCAN1, IL6R,
DUSP4 and STAT3) and seven downregulated DEGs (CPXM1, FGFR2, FLRT3,
IL17RD, IRS1, IL7 and CSF1R) were identified in the functional
modules for sevoflurane-specific DEGs. A study by Lucchinetti et
al (28), STAT3 was found to
be associated with the granulocyte-colony stimulating factor
survival pathway. In the present study, the function of ‘response
to wounding’, which involved 72 DEGs, was identified as the most
significant GO term of the 879 DEGs in the sevoflurane group and
the ‘response to organic substance’ function, involving 45 DEGs,
was the most significant GO term of the 290 DEGs in the propofol
group. The sevoflurane-specific DEGs in the interactive functional
modules were also associated significantly with the ‘regulation of
cellular process’ Go term.
However, the genes involved in the molecular
mechanisms underlying sevoflurane and propofol cardioprotective
effects requires further confirmation at the gene and protein
levels using reverse transcription quantitative polymerase chain
reaction and western blot analyses. Therefore, subsequent
investigations aim to include a cell culture system or animal
models to confirm the results of the present study.
In conclusion, sevoflurane and propofol may
synergistically reduce myocardial reperfusion injury in patients
undergoing CABG surgery, since similarity was observed in the
changes in expression induced by sevoflurane and propofol. However,
the underlying molecular mechanisms for the effects of anaesthetic
agents in cardioprotection require further investigation and
confirmation.
References
|
1
|
Shekar PS: Cardiology patient page.
On-pump and off-pump coronary artery bypass grafting. Circulation.
113:e51–e52. 2006. View Article : Google Scholar
|
|
2
|
Ehsan A, Shekar P and Aranki S: Innovative
surgical strategies: Minimally invasive CABG and off-pump CABG.
Curr Treat Options Cardiovasc Med. 6:43–51. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Palmerini T, Biondi-Zoccai G, Riva DD, et
al: Risk of stroke with percutaneous coronary intervention compared
with on-pump and off-pump coronary artery bypass graft surgery:
Evidence from a comprehensive network meta-analysis. Am Heart J.
165:910–917. 2013. View Article : Google Scholar
|
|
4
|
Harskamp RE, Lopes RD, Baisden CE, De
Winter RJ and Alexander JH: Saphenous vein graft failure after
coronary artery bypass surgery: pathophysiology, management and
future directions. Ann Surg. 257:824–833. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bakaeen FG, Chu D, Kelly RF, et al:
Performing Coronary Artery Bypass Grafting Off-Pump May Compromise
Long-Term Survival in a Veteran Population. Ann Thorac Surg.
95:1952–1958. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Moller CH, Penninga L, Wetterslev J,
Steinbruchel DA and Gluud C: Off-pump versus on-pump coronary
artery bypass grafting for ischaemic heart disease. Cochrane
Database Syst Rev. 3:CD0072242012.PubMed/NCBI
|
|
7
|
Nathoe HM, Van Dijk D, Jansen EW, et al: A
comparison of on-pump and off-pump coronary bypass surgery in
low-risk patients. N Engl J Med. 348:394–402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Orhan G, Sargin M, Senay S, et al:
Systemic and myocardial inflammation in traditional and off-pump
cardiac surgery. Tex Heart Inst J. 34:160–165. 2007.PubMed/NCBI
|
|
9
|
Selvanayagam JB, Petersen SE, Francis JM,
et al: Effects of off-pump versus on-pump coronary surgery on
reversible and irreversible myocardial injury: a randomized trial
using cardiovascular magnetic resonanceimaging and biochemical
markers. Circulation. 109:345–350. 2004. View Article : Google Scholar
|
|
10
|
Menasché P: The systemic factor: the
comparative roles of cardiopulmonary bypass and off-pump surgery in
the genesis of patient injury during and following cardiac surgery.
Ann Thorac Surg. 72:S2260–S2265; discussion S2265–S2266,
S2267–S2270. 2001.PubMed/NCBI
|
|
11
|
Tomic V, Russwurm S, Moller E, et al:
Transcriptomic and proteomic patterns of systemic inflammation in
on-pump and off-pump coronary artery bypass grafting. Circulation.
112:2912–2920. 2005.
|
|
12
|
Brown J, Hernandez F, Beaulieu P, Clough
R, Whited C, et al: Off-pump coronary artery bypass does not
influence biomarkers of brain injury, but does exacerbate the
systemic inflammatory response. J Clin Cell Immunik.
S2:0012011.
|
|
13
|
Frassdorf J, De Hert S and Schlack W:
Anaesthesia and myocardial ischaemia/reperfusion injury. Br J
Anaesth. 103:89–98. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Varadarajan SG, An J, Novalija E and Stowe
DF: Sevoflurane before or after ischemia improves contractile and
metabolic function while reducing myoplasmic Ca(2+)
loading in intact hearts. Anesthesiology. 96:125–133. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Preckel B, Schlack W, Comfère T, Obal D,
Barthel H and Thämer V: Effects of enflurane, isoflurane,
sevoflurane and desflurane on reperfusion injury after regional
myocardial ischaemia in the rabbit heart in vivo. Br J Anaesth.
81:905–912. 1998. View Article : Google Scholar
|
|
16
|
Lin E and Symons JA: Volatile anaesthetic
myocardial protection: a review of the current literature. HSR Proc
Intensive Care Cardiovasc Anesth. 2:105–109. 2010.
|
|
17
|
Garcia C, Julier K, Bestmann L, et al:
Preconditioning with sevoflurane decreases PECAM-1 expression and
improves one-year cardiovascular outcome in coronary artery bypass
graft surgery. Br J Anaesth. 94:159–165. 2005.PubMed/NCBI
|
|
18
|
Kawamura T, Kadosaki M, Nara N, et al:
Effects of sevoflurane on cytokine balance in patients undergoing
coronary artery bypass graft surgery. J Cardiothorac Vasc Anesth.
20:503–508. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang Z, Zhong X, Irwin MG, et al: Synergy
of isoflurane preconditioning and propofol postconditioning reduces
myocardial reperfusion injury in patients. Clin Sci (Lond).
121:57–69. 2011. View Article : Google Scholar
|
|
20
|
Corcoran TB, Engel A, Sakamoto H, O’shea
A, O’callaghan-Enright S and Shorten GD: The effects of propofol on
neutrophil function, lipid peroxidation and inflammatory response
during elective coronary artery bypass grafting in patients with
impaired ventricular function. Br J Anaesth. 97:825–831. 2006.
View Article : Google Scholar
|
|
21
|
Sun HY, Xue FS, Xu YC, et al: Propofol
improves cardiac functional recovery after ischemia-reperfusion by
upregulating nitric oxide synthase activity in the isolated rat
hearts. Chin Med J (Engl). 122:3048–3054. 2009.
|
|
22
|
Wang HY, Wang GL, Yu YH and Wang Y: The
role of phosphoinositide-3-kinase/Akt pathway in propofol-induced
postconditioning against focal cerebral ischemia-reperfusion injury
in rats. Brain Res. 1297:177–184. 2009. View Article : Google Scholar
|
|
23
|
Kottenberg E, Thielmann M, Bergmann L, et
al: Protection by remote ischemic preconditioning during coronary
artery bypass graft surgery with isoflurane but not propofol - a
clinical trial. Acta Anaesthesiol Scand. 56:30–38. 2012. View Article : Google Scholar
|
|
24
|
Zaugg M, Lucchinetti E, Spahn DR, Pasch T,
Garcia C and Schaub MC: Differential effects of anesthetics on
mitochondrial K(ATP) channel activity and cardiomyocyte protection.
Anesthesiology. 97:15–23. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Müllenheim J, Ebel D, Frässdorf J, Preckel
B, Thämer V and Schlack W: Isoflurane preconditions myocardium
against infarction via release of free radicals. Anesthesiology.
96:934–940. 2002.PubMed/NCBI
|
|
26
|
Heusch G, Boengler K and Schulz R:
Cardioprotection: nitric oxide, protein kinases and mitochondria.
Circulation. 118:1915–1919. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He W, Zhang FJ, Wang SP, Chen G, Chen CC
and Yan M: Postconditioning of sevoflurane and propofol is
associated with mitochondrial permeability transition pore. J
Zhejiang Univ Sci B. 9:100–108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lucchinetti E, Hofer C, Bestmann L, et al:
Gene regulatory control of myocardial energy metabolism predicts
postoperative cardiac function in patients undergoing off-pump
coronary artery bypass graft surgery: inhalational versus
intravenous anesthetics. Anesthesiology. 106:444–457. 2007.
View Article : Google Scholar
|
|
29
|
Fujita A, Sato JR, de Rodrigues LO,
Ferreira CE and Sogayar MC: Evaluating different methods of
microarray data normalization. BMC Bioinformatic. 7:4692006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Smyth GK: Limma: linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Gentleman R, Carey V, Dudoit S, Irazarry
R and Huber W: Springer; New York, NY: pp. 397–420. 2005,
View Article : Google Scholar
|
|
31
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: a practical and powerful approach to
multiple testing. J R Stat Soc B. 1:289–300. 1995.
|
|
32
|
Huang Da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
Bioinformatics Resources. Nature Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
33
|
Tatusov RL, Natale DA, Garkavtsev IV, et
al: The COG database: new developments in phylogenetic
classification of proteins from complete genomes. Nucleic Acids
Res. 29:22–28. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Altschul SF, Gish W, Miller W, Myers EW
and Lipman DJ: Basic local alignment search tool. J Mol Biol.
215:403–410. 1990. View Article : Google Scholar
|
|
35
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33:W741–W748. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shi G, Zhang L and Jiang T: MSOAR 2.0:
Incorporating tandem duplications into ortholog assignment based on
genome rearrangement. BMC Bioinformatics. 11:102010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Malik V, Kale SC, Chowdhury UK,
Ramakrishnan L, Chauhan S and Kiran U: Myocardial injury in
coronary artery bypass grafting: On-pump versus off-pump comparison
by measuring heart-type fatty-acid-binding protein release. Tex
Heart Inst J. 33:321–327. 2006.
|
|
38
|
Mathur S, Farhangkhgoee P and Karmazyn M:
Cardioprotective effects of propofol and sevoflurane in ischemic
and reperfused rat hearts: role of K(ATP) channels and interaction
with the sodium-hydrogen exchange inhibitor HOE 642 (cariporide).
Anesthesiology. 91:1349–1360. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nobori K, Ito H, Tamamori-Adachi M, et al:
ATF3 inhibits doxorubicin-induced apoptosis in cardiac myocytes: a
novel cardioprotective role of ATF3. J Mol Cell Cardiol.
34:1387–1397. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Seijffers R, Allchorne AJ and Woolf CJ:
The transcription factor ATF-3 promotes neurite outgrowth. Mol Cell
Neurosci. 32:143–154. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Francis JS, Dragunow M and During MJ: Over
expression of ATF-3 protects rat hippocampal neurons from in vivo
injection of kainic acid. Brain Res Mol Brain Res. 124:199–203.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chu HM, Tan Y, Kobierski LA, Balsam LB and
Comb MJ: Activating transcription factor-3 stimulates 3′,5′-cyclic
adenosine monophosphate-dependent gene expression. Mol Endocrinol.
8:59–68. 1994.
|
|
43
|
Weitzman JB, Fiette L, Matsuo K and Yaniv
M: JunD protects cells from p53-dependent senescence and apoptosis.
Mol Cell. 6:1109–1119. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bergman MR, Cheng S, Honbo N, Piacentini
L, Karliner JS and Lovett DH: A functional activating protein 1
(AP-1) site regulates matrix metalloproteinase 2 (MMP-2)
transcription by cardiac cells through interactions with JunB-Fra1
and JunB-FosB heterodimers. Biochem J. 369:485–496. 2003.
View Article : Google Scholar
|
|
45
|
Alfonso-Jaume MA, Bergman MR, Mahimkar R,
et al: Cardiac ischemia-reperfusion injury induces matrix
metalloproteinase-2 expression through the AP-1 components FosB and
JunB. Am J Physiol Heart Circ Physiol. 291:H1838–H1846. 2006.
View Article : Google Scholar
|
|
46
|
Kumar S, Boehm J and Lee JC: p38 MAP
kinases: key signalling molecules as therapeutic targets for
inflammatory diseases. Nat Rev Drug Discov. 2:717–726. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Muslin AJ: MAPK signalling in
cardiovascular health and disease: molecular mechanisms and
therapeutic targets. Clin Sci (Lond). 115:203–218. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Huang S, Sah VP, et al: Cardiac
muscle cell hypertrophy and apoptosis induced by distinct members
of the p38 mitogen-activated protein kinase family. J Biol Chem.
273:2161–2168. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Boonyasrisawat W, Eberle D, Bacci S, et
al: Tag polymorphisms at the A20 (TNFAIP3) locus are associated
with lower gene expression and increased risk of coronary artery
disease in type 2 diabetes. Diabetes. 56:499–505. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Xue H, Wang SX, Wang XJ, et al: Variants
of tumor necrosis factor-induced protein 3 gene are associated with
left ventricular hypertrophy in hypertensive patients. Chin Med J
(Engl). 124:1498–1503. 2011.
|
|
51
|
De Keulenaer GW, Wang Y, Feng Y, et al:
Identification of IEX-1 as a biomechanically controlled nuclear
factor-kappaB target gene that inhibits cardiomyocyte hypertrophy.
Circ Res. 90:690–696. 2002.
|
|
52
|
Hirotani S, Otsu K, Nishida K, et al:
Involvement of nuclear factor-kappaB and apoptosis
signal-regulating kinase 1 in G-protein-coupled receptor
agonist-induced cardiomyocyte hypertrophy. Circulation.
105:509–515. 2002. View Article : Google Scholar
|