Introduction
Hepatocellular carcinoma (HCC) is the fifth most
prevalent type of cancer worldwide and, with a five-year survival
rate of <5%, HCC remains one of the most fatal cancers (1). HCCs are malignant tumors notorious
for their resistance to conventional chemotherapy, and few
treatments have proven to be effective. HCC exhibits multidrug
resistance, mediated by the high expression of anti-apoptosis genes
[including B-cell lymphoma 2 (Bcl-2) and B-cell lymphoma extra
large (Bcl-xl)]that molecularly oppose the pro-apoptotic effects of
chemotherapeutic agents (2–6).
The Bcl-2 gene family is involved in the regulation
of cellular apoptosis and tumorigenesis. Bcl-xl, a member of the
Bcl-2 family, plays a critical role in regulating cell survival and
apoptosis, and is frequently overexpressed in various types of
human cancer (7). The inhibition
of this gene is associated with decreased tumorigenesis and
resistance to conventional chemotherapy (8).
Research on the function of microRNAs (miRNAs) in
various cancers has attracted significant attention due to the
regulatory role of miRNAs in a broad range of biological processes,
including embryogenesis, differentiation, proliferation, apoptosis
and carcinogenesis (9). miRNAs are
a class of small (19–23 nucleotides), single-stranded, endogenous
non-coding RNAs that post-transcriptionally regulate gene
expression by binding to the 3′ untranslated region (UTR) of target
mRNAs, thus inhibiting translation and/or inducing mRNA degradation
(10). A previous study
demonstrated that single nucleotide polymorphisms (SNPs) found in
miRNA binding sites can affect the expression of miRNA target genes
and may therefore contribute to the resistance of cancers to
multiple drug treatments (11).
Furthermore, our previous studies have shown that miRNAs can affect
the sensitivity of HCC to drug treatment by regulating the
expression of the Bcl-2 gene family (12,13).
Therefore, we proposed that miRNA could additionally regulate
Bcl-xl and that SNPs in miRNA binding sites could disrupt this
regulation. Using various computational programs, a potential miRNA
binding site was identified for the let-7 miRNA family in the 3′UTR
of the Bcl-xl gene. The miRNAs of the let-7 family share a common
seed sequence (GAUGGAG) and are poorly expressed in HCC as compared
with normal tissues (14).
Materials and methods
Prediction of miRNA targets
Three computational programs were selected to
predict miRNAs that could potentially bind to Bcl-xl mRNA: miRanda
(http://www.microrna.org/), Pictar (http://pictar.mdcberlin.de/) and TargetScan
(http://www.targetscan.org/).
Cell culture
The present study was approved by the ethics
committee of the University of South China (Hengyang, China).
BEL-7402 human HCC cells (Cell Bank of the Chinese Academy of
Sciences, Shanghai, China) were cultured in RPMI-1640 media
(HyClone Laboratories, Logan, UT, USA) containing 10% fetal bovine
serum (HyClone Laboratories) and 1% penicillin/streptomycin sulfate
at 37°C in a humidified atmosphere of 5% CO2. 5-FU was
purchased from Sigma-Aldrich (St. Louis, MO, USA).
Construction of the vector and
dual-luciferase reporter assay
Full-length Bcl-xl cDNA sequence containing the
entire 3′UTR was synthesized and cloned into pcDNA3.1 plasmid and
confirmed by DNA sequencing (Invitrogen Biotechnology Co., Ltd.,
Shanghai, China). Site-directed mutagenesis was achieved using the
Takara MutanBEST kit (Takara Bio, Inc., Shiga, Japan) using the
following primers: Forward primer,
5′-CCCTCAGGCAGGAAGGGCAGGAAGGAG-3′ and reverse primer:
5′-AGGGAAGACCCTGGGGCTCCCATAGCTG-3′. The 3′UTR of Bcl-xl was
synthesized and inserted into the 3′-end of the Renilla luciferase
gene in the dual-luciferase miRNA target expression vector
psi-CHECK2 (Promega Corporation, Madison, WI, USA) between
XhoI and NotI sites. Site-directed mutagenesis was
performed in accordance with the manufacturer’s instructions
(Takara Bio, Inc.). The dual-luciferase reporter plasmids were
co-transfected with let-7b mimics into HEK293 cells. At 48 h
post-transfection, the cells were assayed for luciferase activity
using the Dual-Glo Luciferase Assay System (Promega Corporation)
according to the manufacturer’s instructions. The Renilla
luciferase activities were normalized to firefly luciferase
activity. For each transfection, the luciferase activity was
averaged from three replicates.
Transfection of miRNA mimics and
vectors
The let-7b mimics were purchased from GenePharma
(Shanghai, China). The let-7b mimic (20 μM) or control
oligonucleotide was transfected into BEL-7402 cells using
Lipofactamine™ 2000 (Invitrogen Life Technologies, Carlsbad, CA,
USA) according to the manufacturer’s instructions. Forty-eight
hours after transfection, the cells were harvested and lysed. For
transient co-transfection, control vector, pcDNA3.1-Bcl-xl (A) or
pcDNA3.1-Bcl-xl (C) was co-transfected with let-7b mimic (20 μM) or
control oligonucleotide into BEL-7402 cells using Lipofactamine™
2000. Forty-eight hours after transfection, the cells were
harvested and lysed.
Western blot analysis
At 48 h post-transfection, the cells were harvested
and lysed in radioimmunoprecipitation assay buffer with protease
inhibitors. Proteins were separated on 12% SDS-polyacrylamide gel
and transferred to polyvinylidene fluoride membranes (Merck
Millipore, Darmstadt, Germany). A PageRuler prestained protein
ladder (Pierce Biotechnology Inc., Rockford, IL, USA) was used as a
molecular marker. A single membrane was cut into three parts at the
43, 30 and 20 kD bands and incubated with anti-β-actin, -Bcl-xl and
-Bcl2-associated X protein (Bax) monoclonal primary antibodies
(Cell Signaling Technology, Inc., Danvers, MA, USA), respectively.
The protein was detected using a horseradish peroxidase-conjugated
secondary antibody and a Chemilucent Enhanced Chemiluminescence
Detection system (Millipore, Billerica, MA, USA).
Cell counting kit 8 (CCK8) assay
Cells from the various transfection groups were
seeded into a 96-well plate at a density of 5×103 cells
per well. Forty-eight hours after transfection, various
concentrations of 5-FU (0, 5, 50, 500, 5,000 and 50,000 μM) were
added and the cells were incubated for 24 h. The cells were then
treated with CCK8 (Beyotime Institute of Biotechnology, Shanghai,
China) for 2 h at 37°C. The reaction was optically monitored at 450
nm using a 96-well microtiter plate reader (Amersham Pharmacia
Biotech, Piscataway, NJ, USA).
Statistical analysis
SPSS software (Version 14.0; SPSS, Inc., Chicago,
IL, USA) was used for statistical analysis. One-way analysis of
variance was performed to evaluate the significance of the
difference between the samples. P<0.05 indicates a statistically
significant difference.
Results
The 3′UTR of Bcl-xl contains an miRNA
binding site for let-7b, and SNP rs3208684 A>C disrupts the
binding efficiency
To identify the miRNAs that regulate Bcl-xl
expression, three computational programs (TargetScan, Pictar and
miRanda) were used to search for miRNA binding sites in the 3′UTR
of the Bcl-xl gene. Each program predicted different miRNA binding
sites; however, one common binding site for the let-7b miRNA
(Fig. 1A) was identified.
Subsequent to searching the Ensembl genome browser (http://www.ensembl.org/index.html), an SNP
(rs3208684) was identified in the let-7b binding site of the Bcl-xl
3′UTR. The SNP rs3208684 results in the production of two alleles,
A and C (Fig. 1A and B).
To investigate the effect of SNP rs3208684 A>C
variation on miRNA binding, the wild-type and mutant 3′UTR were
cloned into the dual-luciferase reporter vector psi-CHECK2 and
co-transfected with miRNA mimics into HEK293T cells. The A>C
substitution in the miRNA binding site increased the luciferase
activity of the Bcl-xl 3′UTR, indicating that the A>C
substitution suppressed or disrupted the binding of let-7b to its
target (Fig. 1C).
Let-7b downregulates endogenous Bcl-xl
expression and sensitizes BEL-7402 cells to 5-FU
To examine the effects of let-7b on endogenous
Bcl-xl expression, let-7b mimics and negative control were
transfected into BEL-7402 cells, which are known to express high
levels of Bcl-xl protein. The enhanced expression of let-7b in
BEL-7402 cells significantly decreased the amount of Bcl-xl
protein, compared with mock transfection (Fig. 2A and B), whereas the Bax protein
expression was not significantly different.
 | Figure 2Let-7b downregulates endogenous Bcl-xl
expression and sensitizes BEL-7402 cells to 5-FU. (A and B) Let-7b
mimics and negative control were transfected into BEL-7402 cells,
respectively. At 48 h after transfection, cell lysates were
prepared and subjected to western blot analysis. The data are
presented as the mean ± SD (n=3), *P<0.05 vs. the
negative control group. (C) The let-7b mimics and negative control
were transfected into BEL-7402 cells, respectively. At 48 h after
transfection, various concentrations of 5-FU (0, 5, 50, 500, 5,000,
50,000 μM) were added and the cells were incubated for 24 h and
subjected to cell counting kit 8 assay. The data are presented as
the mean ± SD (n=3), *P<0.05. NC, negative control;
Bcl-xl, B-cell lymphoma-extra large; Bax, Bcl2-associated X
protein; 5-FU, fluorouracil; SD, standard deviation. |
To further investigate whether let-7b sensitizes
BEL-7402 cells to 5-FU, let-7b mimics and negative control were
transfected into BEL-7402 cells. CCK8 assay was performed to
determine the cell survival rate. The results indicated that the
transfection of let-7b mimics significantly reduced cell viability,
as compared with the negative control and mock transfection
(Fig. 2C). These results
demonstrated that let-7b sensitizes BEL-7402 cells to 5-FU by
negatively regulating Bcl-xl expression.
SNP rs3208684 A>C variation in the
Bcl-xl 3′UTR inhibits the binding of let-7b and enhances Bcl-xl
expression
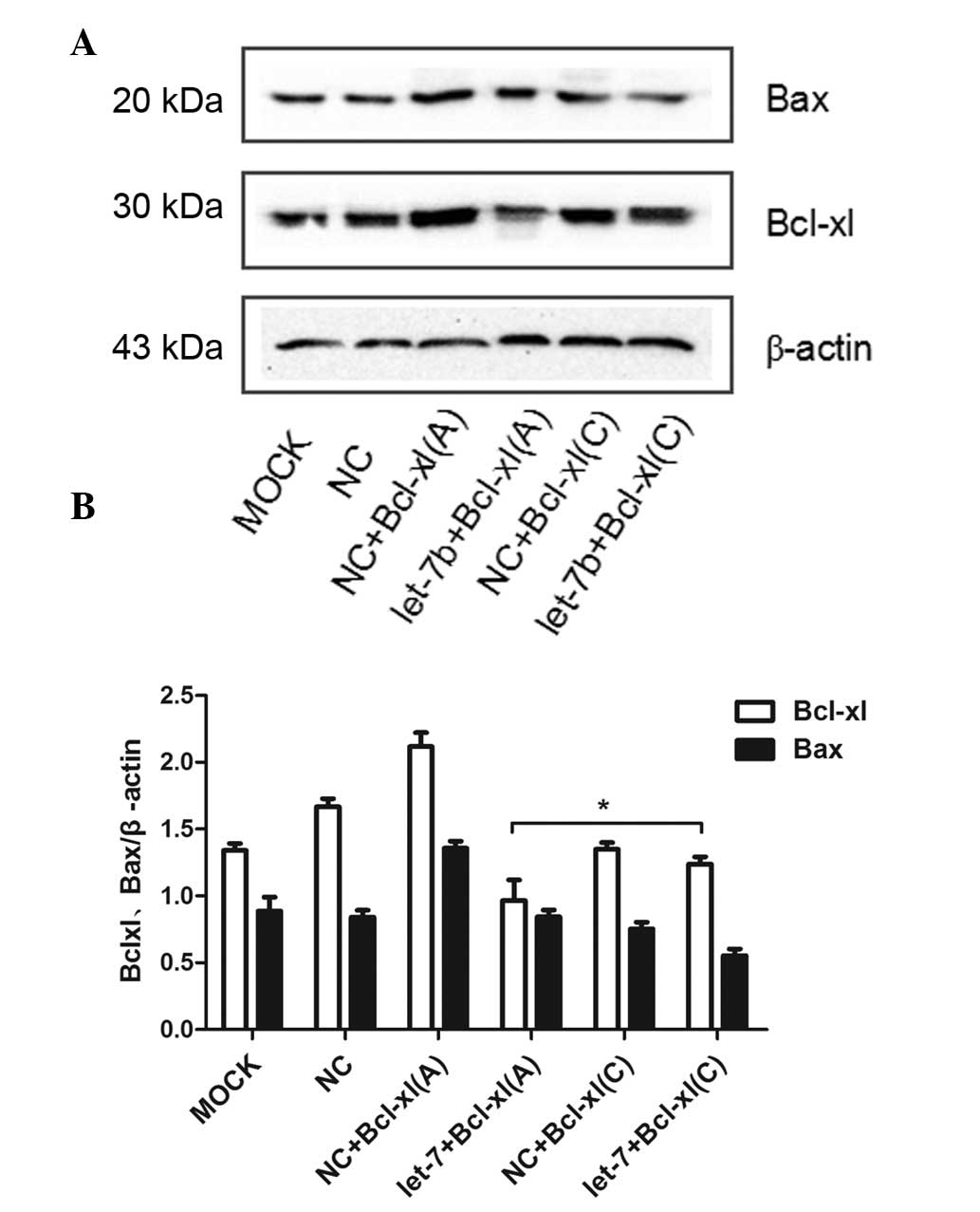
To determine the effects of SNP rs3208684 on the
let-7b-mediated regulation of Bcl-xl, expression constructs of
Bcl-xl containing a wild-type or mutant 3′UTR [pcDNA3.1-Bcl-xl (A)
and pcDNA3.1-Bcl-xl (C), respectively] were generated and
co-transfected with let-7b mimics into BEL-7402 cells. The
overexpression of let-7b decreased the expression of Bcl-xl with a
wild-type 3′UTR but had no effect on the expression of Bcl-xl with
a mutant 3′UTR (Fig. 3A and B).
Taken together, these results indicated that SNP rs3208684 A>C
variation mediated Bcl-xl upregulation through disruption of the
binding of let-7b to the 3′UTR of Bcl-xl.
Let-7b miRNA binding site polymorphism in
the Bcl-xl 3′UTR desensitizes human BEL-7402 cells to 5-FU
Since SNP rs3208684 A>C variation enhances the
expression of Bcl-xl protein, and Bcl-xl overexpression has been
shown to decrease chemosensitivity in tumor cells (15,16),
we analyzed whether SNP rs3208684 affected the response to 5-FU
treatment. BEL-7402 cells were co-transfected with let-7b mimics
and wild-type (A allele) or mutated (C allele) Bcl-xl cDNA
constructs, and treated with various concentrations of 5-FU for 24
h. The result of the CCK8 assay showed that, as compared with cells
in the let-7b mimics plus wild-type-Bcl-xl vector co-transfection
group and the negative control group, the BEL-7402 cells in the
let-7b mimics plus mutant-Bcl-xl vector co-transfection group
exhibited decreased drug sensitivity to 5-FU (Fig. 4). As stated previously, the mutant
Bcl-xl was insensitive to let-7b. Therefore, BEL-7402 cells with
mutant Bcl-xl exhibited higher levels of Bcl-xl protein in the
presence of low levels of let-7b than the cells with wild-type
Bcl-xl. Cells with wild-type Bcl-xl exhibited increased drug
sensitivity. These results indicated that the SNP rs3208684 A>C
decreased 5-FU sensitivity in BEL-7402 cells.
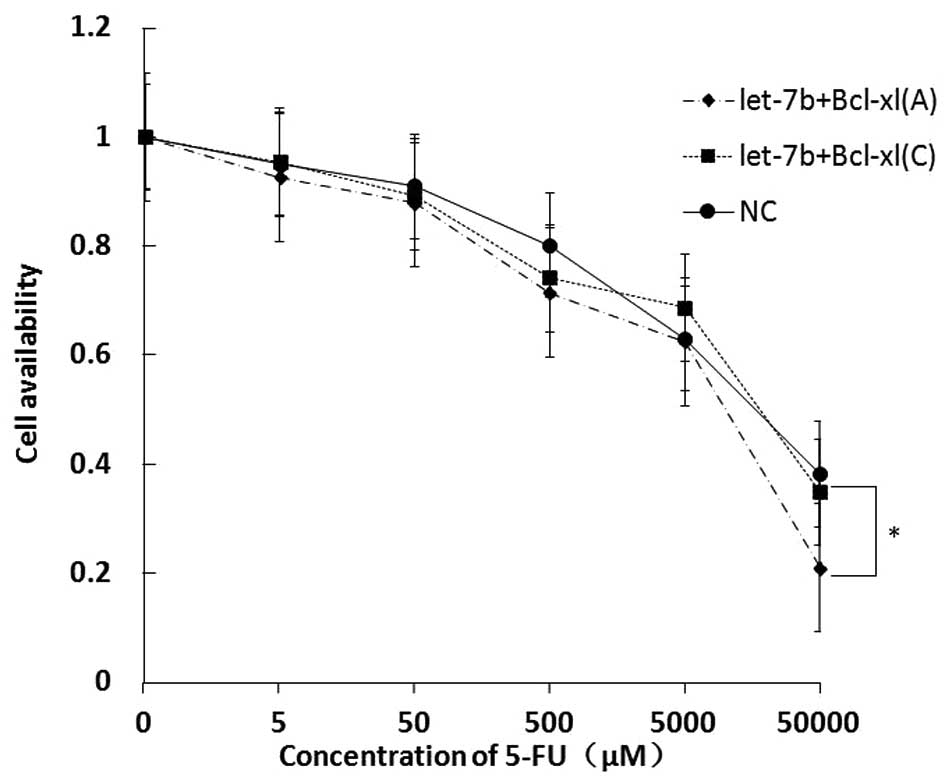 | Figure 4Let-7b microRNA-binding site
polymorphism in the Bcl-xl 3′ untranslated region desensitizes
human BEL-7402 cells to 5-FU. The pcDNA-3.1 expression construct of
full-length Bcl-xl with the A or C allele was co-transfected with
miRNA mimics into BEL-7402 cells. At 48 h after transfection,
various concentrations of 5-FU (0, 5, 50, 500, 5,000 and 50,000 μM)
were added and the cells were incubated for 24 h and subjected to
cell counting kit 8 assay. Data are presented as the mean ±
standard deviation (n=3), *P<0.05. NC, negative
control; Bcl-xl, B-cell lymphoma-extra large; Bax, Bcl2-associated
X protein; 5-FU, fluorouracil. |
Discussion
Let-7, first identified in 2001 (17), is a precursor of human miRNA and
has been shown to be downregulated in several malignancies.
Numerous target genes have been identified for let-7 miRNA,
including Ras (18), Myc (19), HMGA2 (20), CDC25A and CDK6 (21). The major function of these genes is
the promotion of cell proliferation. These genes have been shown to
exhibit oncogenic function in tumor cells; therefore, let-7 miRNA
is considered to function as a tumor suppressor (22). In the present study, we present
evidence that let-7b miRNAs are negative regulators of Bcl-xl,
which is consistent with findings of Shimizu et al (23) and Qin et al (24). Using computational programs, it was
predicted that the 3′UTR of Bcl-xl contained a potential binding
site for let-7b. The binding of let-7b to the 3′UTR of Bcl-xl was
then verified using the luciferase assay. Finally, the effects of
let-7b on the expression of endogenous Bcl-xl were investigated.
Ectopic expression of let-7b suppressed the expression of Bcl-xl in
BEL-7402 cells.
The let-7 family is expressed at lower levels in HCC
than in normal liver tissues, and its low expression is correlated
with 5-FU resistance (25). In
this study, it was shown that Bcl-xl overexpression in HCC BEL-7402
cells also decreased sensitivity of cancer cells to 5-FU. Cells
that were transfected with wild-type Bcl-xl, which contains a 3′UTR
that can bind to let-7b, had significantly higher sensitivity to
5-FU than cells with mutant Bcl-xl. These results suggest that
overexpression of let-7b induced 5-FU resistance by repressing
Bcl-xl expression in BEL-7402 cells.
SNPs are the most common form of genetic variation
that occurs in the human genome. SNPs that are located at or near
an miRNA binding site of a functional gene can affect gene
expression by altering the interaction between miRNA and mRNA
(26,27). Previous studies have shown that
miRNA binding site polymorphisms are associated with tumor
susceptibility and the chemotherapeutic response (11,28–30).
For example, the miR-200b/200c/429 binding site polymorphism in the
3′UTR of the AP-2α gene is associated with cisplatin resistance
(31). Identifying SNPs that are
associated with cancer and chemosensitivity may therefore be useful
in the generation of personalized cancer diagnostic and therapeutic
approaches. Numerous studies have shown that Bcl-xl status is
associated with the chemosensitivity of cancer cells (32,33).
In the present study, it was found that the SNP (rs3208684) A>C
variation in the Bcl-xl 3′UTR disrupted the interaction between the
let-7 precursor miRNA and Bcl-xl, thus upregulating Bcl-xl
expression and Bcl-xl-mediated 5-FU resistance. These results
suggest that SNP rs3208684 may be a potential prognostic marker for
5-FU treatment and that patients with the A allele of SNP rs3208684
may be more sensitive to chemotherapy than those with the mutant C
allele of SNP rs3208684.
Acknowledgements
This study was supported by grants from the Science
and Technology Department Projects of Hunan Province, China
(2012FJ2016), the Construction Projects of Provincial Key
Disciplines, the National Natural Science Foundation of China (no.
81372579) and the Open Fund Based on Innovation Platform of Hunan
Colleges and Universities, China (no. 13K084).
References
|
1
|
Borel F, Han R, Visser A, et al; Réseau
Centre de Ressources Biologiques Foie (French Liver Biobanks
Network), France. Adenosine triphosphate-binding cassette
transporter genes up-regulation in untreated hepatocellular
carcinoma is mediated by cellular microRNAs. Hepatology.
55:821–832. 2012.
|
|
2
|
van Oosterwijk JG, Herpers B, Meijer D, et
al: Restoration of chemosensitivity for doxorubicin and cisplatin
in chondrosarcoma in vitro: BCL-2 family members cause
chemoresistance. Ann Oncol. 23:1617–1626. 2012.PubMed/NCBI
|
|
3
|
Zhu W, Xu H, Zhu D, et al: miR-200bc/429
cluster modulates multidrug resistance of human cancer cell lines
by targeting BCL2 and XIAP. Cancer Chemother Pharmacol. 69:723–731.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu W, Zhu D, Lu S, et al: miR-497
modulates multidrug resistance of human cancer cell lines by
targeting BCL2. Med Oncol. 29:384–391. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lieber J, Kirchner B, Eicher C, Warmann
SW, Seitz G, Fuchs J and Armeanu-Ebinger S: Inhibition of Bcl-2 and
Bcl-X enhances chemotherapy sensitivity in hepatoblastoma cells.
Pediatr Blood Cancer. 55:1089–1095. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wesarg E, Hoffarth S, Wiewrodt R, Kröll M,
Biesterfeld S, Huber C and Schuler M: Targeting BCL-2 family
proteins to overcome drug resistance in non-small cell lung cancer.
Int J Cancer. 121:2387–2394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mallick S, Patil R, Gyanchandani R, Pawar
S, et al: Human oral cancers have altered expression of Bcl-2
family members and increased expression of the anti-apoptotic
splice variant of Mcl-1. J Pathol. 217:398–407. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ma B, Zhang H, Wang J, Zhang B, Xu X and
Cheng B: HIV-1 viral protein R (Vpr) induction of apoptosis and
cell cycle arrest in multidrug-resistant colorectal cancer cells.
Oncol Rep. 28:358–364. 2012.PubMed/NCBI
|
|
9
|
Di Leva G and Croce CM: Roles of small
RNAs in tumor formation. Trends Mol Med. 16:257–267.
2010.PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Slaby O, Bienertova-Vasku J, Svoboda M and
Vyzula R: Genetic polymorphisms and microRNAs: new direction in
molecular epidemiology of solid cancer. J Cell Mol Med. 16:8–21.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yin J, Tang HF, Xiang Q, Yu J, Yang XY, Hu
N and Lei XY: MiR-122 increases sensitivity of drug-resistant
BEL-7402/5-FU cells to 5-fluorouracil via down-regulation of bcl-2
family proteins. Pharmazie. 66:975–981. 2011.PubMed/NCBI
|
|
13
|
Yang X, Yin J, Yu J, et al: miRNA-195
sensitizes human hepatocellular carcinoma cells to 5-FU by
targeting BCL-w. Oncol Rep. 27:250–257. 2012.PubMed/NCBI
|
|
14
|
Hou J, Lin L, Zhou W, et al:
Identification of miRNomes in human liver and hepatocellular
carcinoma reveals miR-199a/b-3p as therapeutic target for
hepatocellular carcinoma. Cancer Cell. 19:232–243. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang J, Cheng C, He CL, Zhou YJ and Cao
Y: The expression of Bcl-XL, Bcl-XS and p27Kip1 in
topotecan-induced apoptosis in hepatoblastoma HepG2 cell line.
Cancer Invest. 26:456–463. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao Y, Zhang CL, Zeng BF, Wu XS, Gao TT
and Oda Y: Enhanced chemosensitivity of drug-resistant osteosarcoma
cells by lentivirus-mediated Bcl-2 silencing. Biochem Biophys Res
Commun. 390:642–647. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson SM, Grosshans H, Shingara J, et
al: RAS is regulated by let-7 microRNA family. Cell. 120:635–647.
2005. View Article : Google Scholar
|
|
19
|
Sampson VB, Rong NH, Han J, et al:
MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth
in Burkitt lymphoma cells. Cancer Res. 67:9762–9770. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Johnson CD, Esquela-Kerscher A, Stefani G,
et al: The let-7 microRNA represses cell proliferation pathways in
human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Büssing I, Slack FJ and Grosshans H: let-7
microRNAs in development, stem cells and cancer. Trend Mol Med.
14:400–409. 2008.
|
|
23
|
Shimizu S, Takehara T, Hikita H, et al:
The let-7 family of microRNAs inhibits Bcl-xL expression and
potentiates sorafenib-induced apoptosis in human hepatocellular
carcinoma. J Hepatol. 52:698–704. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qin B, Xiao B, Liang D, Li Y, Jiang T and
Yang H: MicroRNA let-7c inhibits Bcl-xl expression and regulates
ox-LDL-induced endothelial apoptosis. BMB Rep. 45:464–469. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hummel R, Hussey DJ and Haier J:
MicroRNAs: predictors and modifiers of chemo- and radiotherapy in
different tumour types. Eur J Cancer. 46:298–311. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nicoloso MS, Sun H, Spizzo R, et al:
Single-nucleotide polymorphisms inside microRNA target sites
influence tumor susceptibility. Cancer Res. 70:2789–2798. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mishra PJ and Bertino JR: MicroRNA
polymorphisms: the future of pharmacogenomics, molecular
epidemiology and individualized medicine. Pharmacogenomics.
10:399–416. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Smits KM, Paranjape T, Nallur S, et al: A
let-7 microRNA SNP in the KRAS 3′UTR is prognostic in early-stage
colorectal cancer. Clin Cancer Res. 17:7723–7731. 2011.
|
|
29
|
Zhang L, Liu Y, Song F, et al: Functional
SNP in the microRNA-367 binding site in the 3′UTR of the calcium
channel ryanodine receptor gene3 (RYR3) affects breast cancer risk
and calcification. Proc Natl Acad Sci USA. 108:13653–13658.
2011.PubMed/NCBI
|
|
30
|
Pharoah PD, Palmieri RT, Ramus SJ, et al:
The role of KRAS rs61764370 in invasive epithelial ovarian cancer:
implications for clinical testing. Clin Cancer Res. 17:3742–3750.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu Y, Xiao Y, Ding X, Zhuo Y, Ren P, Zhou
C and Zhou J: A miR-200b/200c/429-binding site polymorphism in the
3′ untranslated region of the AP-2α gene is associated with
cisplatin resistance. PLoS One. 6:e290432011.PubMed/NCBI
|
|
32
|
Peddaboina C, Jupiter D, Fletcher S, et
al: The downregulation of Mcl-1 via USP9X inhibition sensitizes
solid tumors to Bcl-xl inhibition. BMC Cancer. 12:5412012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bansal H, Seifert T, Bachier C, Rao M,
Tomlinson G, Iyer SP and Bansal S: The transcription factor Wilms
tumor 1 confers resistance in myeloid leukemia cells against the
proapoptotic therapeutic agent TRAIL (tumor necrosis factor
α-related apoptosis-inducing ligand) by regulating the
antiapoptotic protein Bcl-xL. J Biol Chem. 287:32875–32880.
2012.PubMed/NCBI
|