Introduction
According to the National Center for Health
Statistics, presbycusis is the third most common disease in seniors
over the age of 65, following arthritis and high blood pressure
(1). The World Health Organization
predicted that when there will be ~100 million people aged >60
years, 70–80% of those suffering from presbycusis (2). Presbycusis is characterized by the
progressive, bilaterally symmetrical, sensorineural and chronic
loss of hearing. Although the mechanism of presbycusis is unclear,
Markaryan A et al (3) demonstrated
histopathologically, that degeneration of cochlea tissues,
including the spiral ganglion, stria vascularis and hair cells, was
linearly associated with hearing loss (3).
Numerous studies have reported that deletions of
mitochondrial DNA (mtDNA) have a vital role in aging and acquired
impairment in various human organs, including the brain, heart,
liver, skin, skeletal muscle and cornea (4–6).
Cochlea tissue is rich in mtDNA, which have a high probability of
deletion. The deletions in mtDNA in cochlea tissue have been shown
to include mtDNA 13162, 10422, 7663, 7436, 4989 and 4977 bp
deletions, with the mtDNACD4977 being the most common in
cochlea tissue (7–12).
Fischel-Ghodsian et al (13) found that mtDNACD4977 was
a determining factor in the occurrence and development of
presbycusis through polymerase chain reaction (PCR) analysis of
temporal bone specimens. In addition, Bai et al (14) reported that mtDNACD4977
was positive in all patients with presbycusis of the same family.
mtDNACD4977 in the temporal bone is also associated with
the blood supply to the cochlea (15). The diameter of the vessels
supplying the internal auditory meatus in patients with presbycusis
and mtDNACD4977 has been shown to be less than that
observed in patients with presbycusis without
mtDNACD4977 (15).
Previously, Yamasoba et al (16)
demonstrated that mtDNACD4977 aggravates presbycusis in
PolgD257A gene knockout mice (16). The rate of mtDNACD4977
increased in PolgD257A gene knockout mice with
age-related hearing loss. The acceleration of cochlea degeneration
therefore appears to be coupled with mtDNACD4977
(16). These studies indicated
that mtDNACD4977 exhibited a close correlation with
presbycusis, but the degree of mtDNACD4977 in the hair
shaft of patients with presbycusis has not yet been
investigated.
The hair shaft expresses a large amount of mtDNA and
obtaining the hair shaft is a relatively non-invasive procedure.
Analysis of the level and function of mtDNA in the hair shaft may
be used to predict the degree of hearing loss.
The present study investigated the significance of
mtDNACD4977 expression in the hair shaft in patients
with presbycusis and the correlation between mtDNACD4977
and the severity of hearing loss.
Materials and methods
Case selection
The cases selected for the present study were based
on strict audiometric criteria. The criteria were as follows: (i)
All of the patients were >60 years old with bilateral
sensorineural hearing loss and a downward sloping audiometric
pattern; (ii) the hearing loss of all of the patients was
symmetrical within 10 dB at each octave; (iii) all of the
audiograms in the downward sloping high frequency component had a
minimum progression of decreased hearing acuity of 10 dB difference
between each successive octave; (iv) hearing loss was defined as a
sensorineural threshold ≥30 dB; (v) all of the tympanograms were
normal; (vi) the past history was investigated and patients with an
identified disease causing hearing loss were excluded (3). A total of 87 cases of presbycusis
(between 60 and 83 years of age) who met these strict criteria were
selected. Audiograms of 43 cases indicated mild-to-moderate hearing
loss. Audiograms of 31 cases indicated moderate-to-severe, severe
hearing loss. A total of 13 cases indicated profound deafness. A
total of 95 normal hearing individuals were used as control
subjects (between 63 and 81 years of age). Between 0.25 and 8 kHz,
individuals with normal hearing had bone conduction audiometric
thresholds of ≤25 dB. The pure tone averages were analyzed from the
bone conduction thresholds at 0.25, 0.5, 1, 2, 4 and 8 kHz. Six
hair shafts were collected from each individual (17). Informed consent was obtained from
all the patients included in the study, and ethical approval was
granted by the Institutional Review Board of Shandong University
(Shandong, China).
DNA extraction
Approximately 2 cm hair shaft was cut into pieces
and washed three times in 70% alcohol and sterile distilled water.
A total of 215 μl 20% Chelex, 10 μl 1 m DTT and 25 μl Proteinase K
(18 mg/μl) (Sigma-Aldrich Co. Ltd., Poole, Dorset, UK) was added
and the hair shaft was incubated overnight at 56°C until the hair
fragments became invisible to the naked eye. The samples were
boiled for 8 min, centrifuged at 13,000 × g for 3 min and the
supernatant was transferred to a sterile tube and stored at
−20°C.
PCR amplication
The PCR reaction contained 1 μl extracted DNA (40
ng/μl), 0.5 μl each primer (primers 2 and 3, 20 u/μl), 2 μl dNTP
(2.5 mm), 0.25 μl Taq enzyme (5 U/μl; Takara, Dalian, China), 2.5
μl buffer (10 mm Tris-HCl, 50 mm KCl, 1.5 mm MgCl) and 18.25 μl
sterile distilled H2O in a volume of 25 μl. The PCR
assays were performed on an Mastercycler® gradient
(Eppendorf, New York, NY, USA). The PCR reaction condition was as
follows: Initial denaturation at 95°C for 5 min, followed by 40
cycles of denaturation at 95°C for 60 sec, annealing at 54°C for 60
sec and extension at 72°C for 60 sec. The PCR reaction was extended
at 72°C for 7 min. The primers were synthesized by Sangon Biotech
(Shanghai, China) (Table I). The
products of the PCR amplication was separated by electrophoresis on
a 1.0% agarose gel and were detected following staining with
ethidium bromide. The images were captured using an Alpha
Imager® EC System (Alpha Innotech, San Leandro, CA,
USA).
 | Table IPrimer sequences for
mtDNACD4977 amplification. |
Table I
Primer sequences for
mtDNACD4977 amplification.
| Primer | Site (nt) | Sequence (5′-3′) |
|---|
| 1 | 8292–8312 |
GCCCACTGTAAAGCTAACTTA |
| 2 | 13450–13469 |
TCCCTCACCATTGGCAGCCT |
| 3 | 13644–13664 |
CTTCCCCACCCTTACTAACAT |
| 4 | 8251–8270 |
GCCCGTATTTACCCTATAGC |
| 5 | 13845–13826 |
GTCTAGGGCTGTTAGAAGTC |
| 6 | 8406–8425 |
CCCCCATACTCCTTACACTA |
| 7 | 13525–13506 |
CGATGATGTGGTCTTTGGAG |
Nested PCR
The nested PCR assays were performed on an
Mastercycler® gradient (Eppendorf). The first-step PCR
reaction contained 3 μl extracted DNA sample (40 ng/μl), 0.5 μl
each primer (primers 4 and 5, 20 u/μl), 2 μl dNTP (2.5 mm), 0.25 μl
Taq enzyme (5 U/μl), 2.5 μl buffer (10 mm Tris-HCl, 50 mm KCl, 1.5
mm MgCl), 16.25 μl sterile distilled H2O, in a volume of
25 μl. The PCR reaction conditions were performed as previously
described. A total of 3 μl of the product from the first-step PCR
was used as the template in the nested PCR, which used primer 1 and
3 and the same PCR cycling conditions. The products of the nested
PCR was separated by electrophoresis on a 1% agarose gel and were
detected following staining with ethidium bromide. The images were
captured using an Alpha Imager® EC System (Alpha
Innotech).
Sequencing
The amplified fragments were purified and sequenced
by BGI-Shenzhen (Shenzhen, Guangzhou, China). The sequenced results
was analyzed by Basic Local Alignment Search Tool.
Quantitative (q)PCR
The presence of the mtDNACD4977 was
confirmed by sequencing the previously generated PCR product. The
PCR assays were performed on a Mastercycler ep realplex
(Eppendorf). The PCR reaction contained 3 μl extracted DNA sample
(40 ng/μl), 0.5 μl each primer (primers 2 and 3 or primers 6 and 7,
10 u/μl), 12.5 μl SYBR™ Green, 8.5 μl sterile distilled
H2O in a volume of 25 μl. The PCR reaction condition was
as follows: initial denaturation at 94°C for 2 min, followed by 40
cycles of denaturation at 94°C for 15 sec, annealing at 55°C for 15
sec and extension at 68°C for 30 sec. The melting curve was
analyzed at 95°C for 15 sec, followed by 60°C for 20 min and
terminated at 95°C for 15 sec. The quantitative variations of
mtDNACD4977 were evaluated using the relative cycle
threshold (Ct) quantification method (ΔΔ Ct). The relative amount
of mRNA was calculated as the calibrator normalized ratio using
LightCycler® 480 Software 1.5 (Roche Diagnostics,
Mannheim, Germany). The calibrator normalized ratio was measured as
the following formula: RQ = 2−ΔΔCt.
Statistical analysis
SPSS version 17.0 was used for the statistical
analysis (SPSS, Inc., Chicago, IL, USA). The data are presented as
the means ± standard deviation. Analysis of variance (ANOVA) was
used to determine whether the differences in the presence of
mtDNACD4977 between the different groups were
significant. The Pearson correlation coefficient analysis was used
to determine the significance of the differences between the
presence of mtDNACD4977 and hearing loss. A two-sided
P<0.05 was considered to indicate a statistically significant
difference.
Results
PCR analysis of mtDNA
The amount of DNA extracted from the hair shaft was
in the range of 40–75 ng/μl. In all of the samples of each group,
the 135-bp PCR product was detected (Fig. 1A), which indicated the effective
extraction of mtDNA from the hair shaft. Through nested PCR, a
product of 396-bp was detected in certain samples, which indicated
positive expression of mtDNACD4977. In the age-matched
individuals with normal hearing, 8/95 cases (8.42%) were found to
have mtDNACD4977 expression by nested PCR. In the
presbycusis group, a total of 59/87 cases (67.82%) were detected to
have the positive mtDNACD4977 expressed in the hair
shaft. Of those 87 cases, the expression of mtDNACD4977
was found in 22/43 cases (51.16%) with mild-to-moderate hearing
loss. A total of 25/31 cases (80.65%) with moderate-to-severe,
severe hearing loss were indicated to express
mtDNACD4977. A total of 12/13 cases (92.31%) with
profound deafness were indicated to express mtDNACD4977
(Fig. 1B). In total, 67.82% of the
hair shafts were positive in mtDNACD4977. Using Pearson
correlation coefficients analysis, a statistically significant
difference was identified between the presence of
mtDNACD4977 and hearing loss (r=0.858; P<0.001). By
the use of ANOVA, statistically significant differences of
mtDNACD4977 were indicated among all of the groups
(F=47.145; P<0.001).
Sequencing of the nested PCR
products
The results of the sequencing of the nested PCR
products were matched with National Center for Biotechnology
Information reference sequence NC_012920 (http://www.ncbi.nlm.nih.gov/nuccore/251831106) in
the gene library, which confirmed that the deleted fragment was
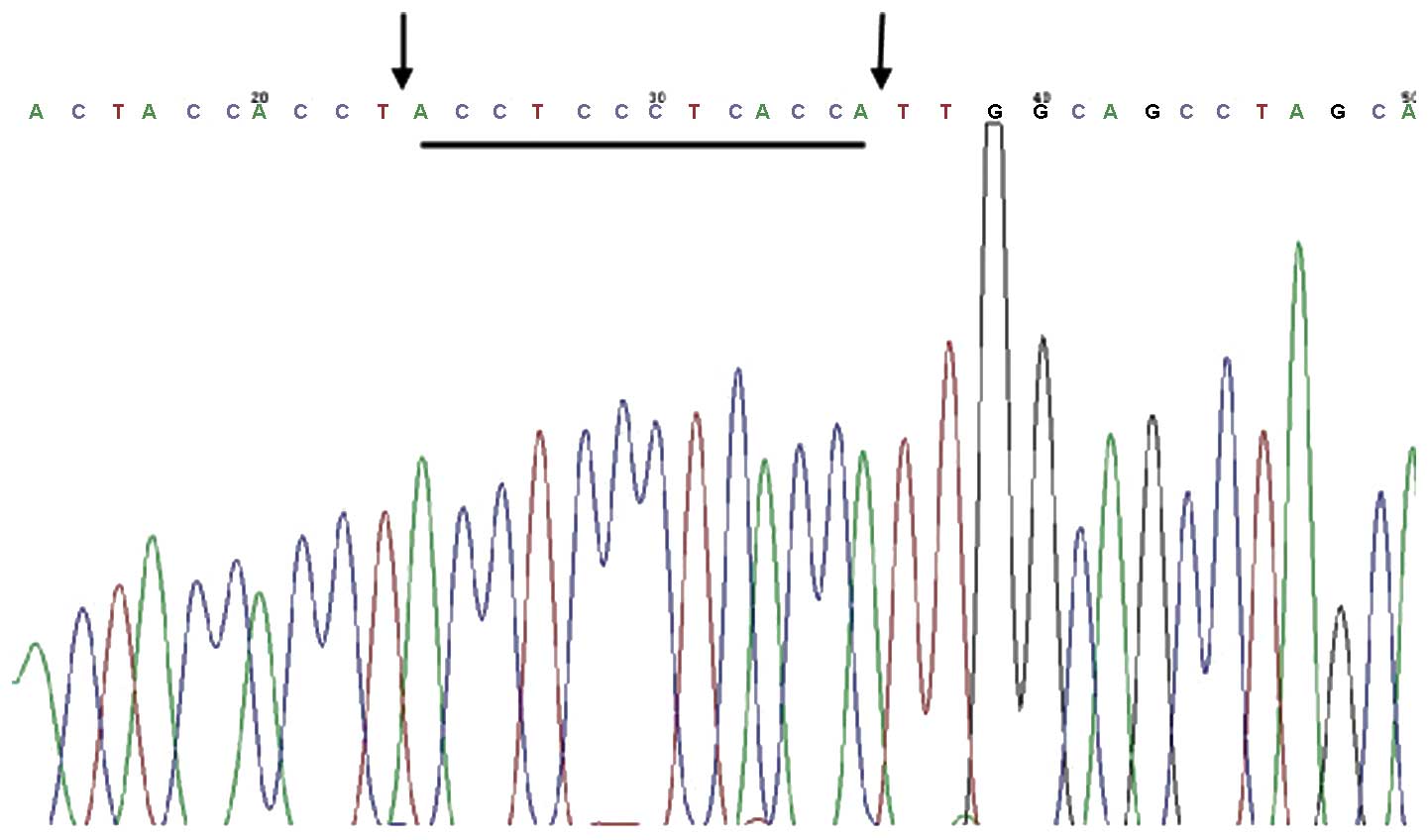
mtDNACD4977 (Fig. 2).
The deleted fragment was located to nucleotides 8470–13447.
qPCR assay
The presence of the mtDNACD4977 was
identified by sequencing the generated nested PCR products. The
mean level of the common deletion (CD) in the specimens and
auditory thresholds of these cases are listed in Table II. A mean CD level of 11.97±4.12,
19.75±5.29, 33.68±10.30 and 4.91±4.16, was detected in groups 1–4,
respectively. This difference in CD levels reached statistical
significance in all groups (P<0.001). A trend toward increasing
levels of the CD with more severe hearing loss was also observed
(P<0.001). Furthermore, there was evidence for a significant
association between the CD level and hearing loss based on the
audiometric thresholds at 8 kHz (r=0.778, P<0.001) and all
ranges of frequency (r=0.858, P<0.001).
 | Table IIAudiometric thresholds and levels of
common deletion across four groups with varying degrees of hearing
loss. |
Table II
Audiometric thresholds and levels of
common deletion across four groups with varying degrees of hearing
loss.
| Group | Group number, n | Mean age, y | Mean PTA, dB | Mean HL8, dB | Mean CD % |
|---|
| 1 | 22 | 69±7.3 | 48.68±10.12 | 72.72±11.10 | 11.97±4.12 |
| 2 | 25 | 72±7.9 | 51.72±12.27 | 80.32±15.72 | 19.75±5.29 |
| 3 | 12 | 75±8.1 | 100.00±0.00 | 100.00±0.00 | 33.68±10.30 |
| 4 | 7 | 80±4.2 | 17.50±5.04 | 17.50±5.04 | 4.91±4.16 |
Discussion
Numerous reports have proposed that oxidative stress
may lead to presbycusis. By this hypothesis, oxygen free radicals
(OFRs) induce lipid and protein peroxidation, and are considered to
damage protein and lipid structure and function. Additionally, OFRs
cause deletion and mutation of nuclear and mitochondrial genes,
which may subsequently cause a significant effect in the ageing and
degeneration of cells and tissues (18,19).
Due to a lack in histone protection and defective
repair mechanisms, mis-pairing and deletion of highly repetitive
sequences in mtDNA occurs frequently under conditions of oxidative
stress. The most common mutation or deletion of mtDNA is
mtDNACD4977 (20).
mtDNA encodes 13 genes, including one cytochrome b gene, two ATP
synthetase subunit genes, three cytochrome c genes and seven
respiratory chain dehydrogenase subunit genes. These genes are
involved in the synthesis of oxidative phosphorylation and
mitochondrial function. mtDNACD4977 results in a
deficiency of 12 genes, including five tRNA and seven
protein-coding genes, which therefore blocks the transcription of
complex enzyme I, III, IV and V and oxidative phosphorylation in
the respiratory chain (21–23).
When generation of ATP through oxidative phosphorylation does not
meet the energy requirements of the inner ear, irreversible hearing
loss may occur (24,25).
Dai et al (15)
examined mtDNACD4977 expression in temporal bone
specimens using nested PCR. The authors showed that the percentage
of mtDNACD4977 was 50.0% (17/34) in the presbycusis
group, 21.1% (4/19) in the age-matched control old-aged group and
0.0% (0/14) in the young and middle-aged group. Markaryan et al
(3) studied mtDNACD4977
in cochlear tissues of old-aged individuals. It was found that the
incidence of mtDNACD4977 was 32±14% in the presbycusis
group and 12±2% in the age-matched normal hearing control group.
These studies have therefore demonstrated that
mtDNACD4977 is commonly observed in patients with
presbycusis. The present study examined the correlation between
presbycusis and mtDNACD4977 in the hair shaft. It was
identified that the incidence of mtDNACD4977 in patients
with presbycusis was significantly higher as compared with the
age-matched normal hearing control individuals (67.82% vs. 8.42%,
P=0.000). The presented PCR results are consistent with a previous
study (7). Furthermore, the PCR
products indicated the absence of mtDNACD4977 in
positive individuals verified by nested-PCR amplication (Fig. 2). Notably, the degree of
mtDNACD4977 is correlated to hearing loss. Based on the
qPCR analysis, the degree of mtDNACD4977 in groups 1–4
was 11.97±4.12, 19.75±5.29, 33.68±10.30 and 4.91±4.16, respectively
(Table II). To the best of our
knowledge, this is the first study to demonstrate the an increase
in the percentage of mtDNACD4977 in the hair shaft along
with the severity of hearing loss.
Zheng et al (7),
detected mtDNACD4977 in the hair shaft of 90 selected
cases (age ranges, 5 days-90 years) using PCR. The results
indicated that the incidence of mtDNACD4977 expression
was 98.3% (89/90). It was therefore concluded that the rate of
mtDNACD4977 was correlated with age. The study by Zheng
et al differed in conclusion from the majority of the previous
reports. These conflicting data may be due to differences in the
criteria for selection of the cases, a small sample number and
error of detection. Further studies are therefore required to
better understand these results. Markaryan et al (3) reported the degree of
mtDNACD4977 in the temporal bone was associated with the
severity of hearing loss at 8 kHz (r=0.44, P=0.034; age-adjusted
partial correlation =0.55, P=0.007). The present study reconfirmed
the a significant association between the mtDNACD4977
expression and hearing loss at 8 kHz (r=0.778, P=0.000) and all
ranges of frequency (r=0.858, P<0.001).
The hair shaft has high sensitivity to oxidative
stress. As compared with previous in vivo studies (3,15,26),
analyzing mtDNA expression in peripheral blood and temporal bone
specimens, the present study was non-invasive and effective through
analysis of mtDNACD4977 in the hair shaft. Therefore,
hair shaft detection may be used to predict hearing function.
mtDNACD4977 in the hair shaft may be used as an
indicator for the prevention, diagnosis and monitoring of
presbycusis.
The present study has demonstrated that
mtDNACD4977 in the hair shaft has a close association
with presbycusis. Analysis suggested as a method to simplify and
further standardise the threshold of mtDNACD4977
resulting in presbycusis reporting.
Acknowledgements
The present study was supported by a grant from the
Science and Technology Development Program of Shandong province,
Provincial Hospital Affiliated to Shandong University (Shandong,
China; grant no. 2012G0021838).
References
|
1
|
Mao Z, Zhao L, Pu L, et al: How well can
centenarians hear? PLoS One. 8:e655652013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sprinzl GM and Riechelmann H: Current
trends in treating hearing loss in elderly people: a review of the
technology and treatment options-a mini-review. Gerontology.
56:351–358. 2010. View Article : Google Scholar
|
|
3
|
Markaryan A, Nelson EG and Hinojosa R:
Quantification of the mitochondrial DNA common deletion in
presbycusis. Laryngoscope. 119:1184–1189. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gendron SP, Mallet JD, Bastien N and
Rochette PJ: Mitochondrial DNA common deletion in the human eye: a
relation with corneal aging. Mech Ageing Dev. 133:68–74. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nie H, Shu H, Vartak R, et al:
Mitochondrial common deletion, a potential biomarker for cancer
occurrence, is selected against in cancer background: a
meta-analysis of 38 studies. PLoS One. 8:e679532013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Torrell H, Montaña E, Abasolo N, et al:
Mitochondrial DNA (mtDNA) in brain samples from patients with major
psychiatric disorders: gene expression profiles, mtDNA content and
presence of the mtDNA common deletion. Am J Med Genet B
Neuropsychiatr Genet. 162B:213–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng Y, Luo X, Zhu J, et al:
Mitochondrial DNA 4977 bp deletion is a common phenomenon in hair
and increases with age. Bosn J Basic Med Sci. 12:187–192.
2012.PubMed/NCBI
|
|
8
|
Pesce V, Cormio A, Marangi LC, et al:
Depletion of mitochondrial DNA in the skeletal muscle of two
cirrhotic patients with severe asthenia. Gene. 286:143–148. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Obara-Moszynska M, Maceluch J, Bobkowski
W, et al: A novel mitochondrial DNA deletion in a patient with
Kearns-Sayre syndrome: a late-onset of the fatal cardiac conduction
deficit and cardiomyopathy accompanying long-term rGH treatment.
BMC Pediatr. 13:272013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taylor RW, Schaefer AM, McFarland R,
Maddison P and Turnbull DM: A novel mitochondrial DNA tRNA(Ile)
(A4267G) mutation in a sporadic patient with mitochondrial
myopathy. Neuromuscul Disord. 12:659–664. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Love RL and Bird P: Cochlear implantation
in mitochondrial deafness due to A7445G mutation. Cochlear Implants
Int. 14:28–31. 2013. View Article : Google Scholar
|
|
12
|
Hao H, Bonilla E, Manfredi G, DiMauro S
and Moraes CT: Segregation patterns of a novel mutation in the
mitochondrial tRNA glutamic acid gene associated with myopathy and
diabetes mellitus. Am J Hum Genet. 56:1017–1025. 1995.PubMed/NCBI
|
|
13
|
Fischel-Ghodsian N, Bykhovskaya Y, Taylor
K, et al: Temporal bone analysis of patients with presbycusis
reveals high frequency of mitochondrial mutations. Hear Res.
110:147–154. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bai U and Seidman MD: A specific
mitochondrial DNA deletion (mtDNA4977) is identified in a pedigree
of a family with hearing loss. Hear Res. 154:73–80. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Dai P, Yang W, Jiang S, et al: Correlation
of cochlear blood supply with mitochondrial DNA common deletion in
presbyacusis. Acta Otolaryngol. 124:130–136. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yamasoba T, Someya S, Yamada C, et al:
Role of mitochondrial dysfunction and mitochondrial DNA mutations
in age-related hearing loss. Hear Res. 226:185–193. 2007.
View Article : Google Scholar
|
|
17
|
Bekaert B, Larmuseau MH, Vanhove MP,
Opdekamp A and Decorte R: Automated DNA extraction of single dog
hairs without roots for mitochondrial DNA analysis. Forensic Sci
Int Genet. 6:277–281. 2012. View Article : Google Scholar
|
|
18
|
Harman D: Free radical theory of aging.
Mutat Res. 275:257–266. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mecocci P, MacGarvey U, Kaufman AE, et al:
Oxidative damage to mitochondrial DNA shows marked age-dependent
increases in human brain. Ann Neurol. 34:609–616. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lagouge M and Larsson NG: The role of
mitochondrial DNA mutations and free radicals in disease and
ageing. J Intern Med. 273:529–543. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen B, Zhong Y, Peng W, Sun Y and Kong
WJ: Age-related changes in the central auditory system: comparison
of D-galactose-induced aging rats and naturally aging rats. Brain
Res. 1344:43–53. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhong Y, Hu YJ, Yang Y, et al:
Contribution of common deletion to total deletion burden in
mitochondrial DNA from inner ear of d-galactose-induced aging rats.
Mutat Res. 712:11–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kim EJ, Kim SY, Yun HJ, et al: Detection
and quantification of a radiation-associated mitochondrial DNA
deletion by a nested real-time PCR in human peripheral lymphocytes.
Mutat Res. 749:53–59. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sequeira A, Martin MV, Rollins B, et al:
Mitochondrial mutations and polymorphisms in psychiatric disorders.
Front Genet. 3:1032012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Niu R, Yoshida M and Ling F: Increases in
mitochondrial DNA content and 4977-bp deletion upon ATM/Chk2
checkpoint activation in HeLa cells. PLoS One. 7:e405722012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yuan YY, Dai P, Zhu XH, et al: Etiologic
analysis of severe to profound hearing loss patients from Chifeng
city in Inner Mongolia. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za
Zhi. 44:292–296. 2009.(In Chinese). PubMed/NCBI
|