Introduction
Neuroblastoma (NB) is a solid embryonal tumor of the
autonomic nervous system, derived from primitive sympathetic
neurons. In >50% of all cases of NB, tumors arise in the adrenal
medulla, while the rest originate in the paraspinal sympathetic
ganglia, with a highly variable clinical presentation (1). Between 1975 and 2011, NB occurred in
1 in 7,000 live births worldwide and accounted for 7–10% of all
childhood cancers (2). The annual
age-standardized incidence rate (ASR) of NB in the Children
Oncology Group between 1986 and 2001 was estimated to be 10.53
cases per million individuals in North American populations
(3). In Lebanon, the National
Cancer Registry (NCR) reports NB as a very rare malignancy, with an
ASR of 2 cases per million individuals in 2007 (4).
According to the Surveillance Epidemiology and End
Results (SEER) databases, the clinical outcome for patients with NB
has improved in the last 40 years (5). However, this improvement is primarily
attributed to an increase in cure rates among patients with the
benign form of the disease. The cure rates among children with
malignant high-risk tumors have shown only modest improvement,
despite marked escalations in the intensity of therapy provided
(6). Despite recent therapeutic
advances, no salvage treatment modalities exist to date, and
high-risk NB is still characterized by a low survival rate, with
50–60% of patients showing a relapse (7).
A number of previous studies indicated that genetic
factors may be involved in the clinical outcome and the response to
treatment of NB patients (8).
Tumor-derived genomic information has been used since the 1980s to
predict prognosis, particularly since the MYCN oncogene was
first discovered to be the target of high-level amplifications at
chromosome band 2p24. Due to the influence of MYCN
amplification on the clinical outcome of NB, it is currently used
as a standard biomarker for treatment stratification (9). In 1985, NB patients with an
MYCN gene amplification of 1, 3 and ≥10 gene copies were
estimated to have an 18-month survival rate of 70, 30 and 5%,
respectively (10). In addition to
MYCN amplification, NB stage III or IV, abnormal chromosomal
deletion/translocation and an older age at diagnosis have all been
identified as being associated with poor prognosis (11).
Current treatments for patients with high-risk NB
include induction of remission, consolidation of the remission, and
eradication of the minimal residual disease. The most commonly used
induction chemotherapy includes dose-intensive cycles of cisplatin
and etoposide, alternated with vincristine, doxorubicin,
cyclophosphamide or ifosfamide and topotecan (12,13).
A number of drug-metabolizing enzymes (DMEs) appear to be involved
in the activation or inactivation of these drugs, including
cytochrome P450 (CYP), N-acetyltransferase (NAT) and
glutathione S-transferase (GST) (14). Therefore, individuals possessing a
modified ability to metabolize these drugs may have an increased
risk of relapse or mortality. In a large cohort of Australian
individuals with NB studied over 15 years (n=209), carriers of the
NAT1*11 allele variant were
significantly less likely to relapse or succumb to the disease,
while children who were GSTM1 null were more likely to relapse or
succumb to the disease, after adjusting for MYCN
amplification, stage of the disease and age at diagnosis [hazard
ratio (HR), 1.6, P=0.04] (15).
The phase I oxidation enzymes CYP3A4 and CYP3A5 were also found to
metabolize a number of the drugs used in NB treatment (16–22)
(Table I). Single nucleotide
polymorphisms (SNPs) in these CYP genes may alter their enzymatic
activity, hence affecting clinical outcome. However, despite these
observations, there are no studies on the role of cytochrome P450
in NB clinical outcome to date. In the present study, the potential
association between the genotype and expression levels of
CYP3A4 and CYP3A5 and mortality were examined in a
group of Lebanese patients with NB.
 | Table IChemotherapeutic agents used in the
treatment of Lebanese patients with neuroblastoma and their
corresponding drug-metabolizing enzymes. |
Table I
Chemotherapeutic agents used in the
treatment of Lebanese patients with neuroblastoma and their
corresponding drug-metabolizing enzymes.
| Anti-cancer
agent | Phase I enzyme | Phase II
enzyme |
|---|
| Cisplatin | GSTM1,
GSTP1, GSTT1 | - |
|
Cyclophosphamide | CYP2A6,
CYP2B6, CYP2C8/9/19, CYP3A4,
CYP3A5 | ALDH,
GSTA1 |
| Doxorubicin | - | GSTP1 |
| Etoposide | CYP2C9,
CYP3A4/5 | UGT1A1,
GSTP1 |
| Ifosfamide | CYP2A6,
CYP2B6, CYP2C8/9/19, CYP3A4,
CYP3A5 | GST |
| Topotecan | - | - |
| Vincristine | CYP3A4,
CYP3A5 | GST |
Materials and methods
Study design and participants
The current study employed a retrospective cohort
design. Three major medical centers in the Lebanese capital city of
Beirut participated. Specimens from children with histologically
confirmed stage III or IV NB, diagnosed between 1993 and 2012, were
obtained from the medical archives of Saint George Hospital
University Medical Center, Hotel Dieu De France Hospital and the
University Medical Center - Rizk Hospital (all Beirut, Lebanon) and
provided to the current study anonymously. Patients excluded from
the study were those with stage I or II NB (n=2) and those with
missing archival tumors (n=9). Of the 38 patients identified, 27
were included in the study.
Approval was obtained from the Institutional Review
Board of the University of Balamand prior to conducting the study.
A review of the medical records of all patients was performed
anonymously to obtain the following information: Age at diagnosis,
gender, NB stage, date of diagnosis, relapse state, date of
mortality and administered chemotherapeutic drugs.
Nucleic acid extraction
Sections (10 μm thick) of identified
paraffin-embedded NB tumors were obtained. These were prepared by a
trained pathologist, avoiding necrotic areas to ensure effective
nucleic acid extraction. Sections were subjected to
deparaffinization by xylene and protein digestion by proteinase K
(Qiagen, Valencia, CA, USA). Tumor DNA was extracted from 50% of
the sections using a QIAamp DNA FFPE Tissue kit (Qiagen), while RNA
was extracted from the remaining 50% of sections using an RNeasy
FFPE kit (Qiagen), both according to the manufacturer’s
instructions.
CYPs genotyping analysis
CYP3A4*1B and
CYP3A4*1A alleles were detected by polymerase
chain reaction and restriction fragment length polymorphism
(PCR-RFLP) using 100–150 ng of extracted DNA and the appropriate
primers (Table II) at an
annealing temperature of 55°C as previously described (23). The resulting 334-bp PCR product was
digested by the PstI restriction enzyme (Thermo Scientific,
Waltham, MA, USA) overnight at 37°C. CYP3A5*1 and
CYP3A5*3 alleles were detected using 100–150 ng
of tumor DNA and the appropriate primers (Table II) at an annealing temperature of
55°C as previously described (24). The resulting 293-bp PCR product was
digested by the Ssp1 restriction enzyme (Thermo Scientific)
overnight at 37°C. Digested fragments were then analyzed using
ethidium bromide staining and agarose gel electrophoresis, and
examined under UV light.
 | Table IIPrimers and probes used in
genotyping, mRNA expression and amplification assays. |
Table II
Primers and probes used in
genotyping, mRNA expression and amplification assays.
| Gene and assay | Sequence |
|---|
| CYP3A4
gene |
| Genotyping forward
primer |
5′-GGACAGCCATAGAGACAACTGCA-3′ |
| Genotyping reverse
primer |
5′-CTTTCCTGCCCTGCACAG-3′ |
| mRNA expression
forward primer |
5′-CACAGATCCCCCTGAAATTAAGCTTA-3′ |
| mRNA expression
reverse primer |
5′-AAAATTCAGGCTCCACTTACGGTG-3′ |
| TaqMan probe |
5′-(6FAM)-AGGACTTCTTCAACCAGAAAAACCCGTTGTTCT-(TAMRA)-3′ |
| CYP3A5
gene |
| Genotyping forward
primer |
5′-CATGCTTAGTAGACAGATGAC-3′ |
| Genotyping reverse
primer |
5′-GGTCCAAACAGGGAAGAAATA-3′ |
| mRNA expression
forward primer |
5′-ACAGATCCCCTTGAAATTAGACACG-3′ |
| mRNA expression
reverse primer |
5′-CTTAGGGTTCCATCTCTTGAATCCA-3′ |
| TaqMan probe |
5′-(6FAM)-AAGGACTTCTTCAACCAGAAAAACCCATTGTTCTA-(TAMRA)-3′ |
| MYCN
amplification |
| Forward primer |
5′-CCCCTGGGTCTGCCCCGTTT-3′ |
| Reverse primer |
5′-GCCGAAGTAGAAGTCATCTT-3′ |
| Fluorogenic
probe |
5′-CCCACCCTCTCCGGTGTGTCTGTCGGTT-3′ |
| β-actin
control gene |
| mRNA expression
forward primer |
5′-TCACCCACACTGTGCCCATCTACGA-3′ |
| mRNA expression
reverse primer |
5′-CAGCGGAACCGCTCATTGCCAATGG-3′ |
| TaqMan probe |
5′-(6FAM)-ATGCCCTCCCCCATGCCATCCTGCGT-(TAMRA)-3′ |
| Amplification
forward primer |
5′-TCACCCACACTGTGCCCATCTACGA-3′ |
| Amplification
reverse primer |
5′-CAGCGGAACCGCTCATTGCCAATGG-3′ |
| Fluorogenic
probe |
5′-ATGCCCTCCCCCATGCCATCCTGCGT-3′ |
MYCN amplification
MYCN amplification was assessed for each NB
tumor sample using extracted DNA in a Real-Time PCR TaqMan
Detection assay (Applied Biosystems, Foster City, CA, USA). The
degree of MYCN amplification was derived from the ratio of
the number of copies of the MYCN oncogene to the number of
copies of the reference gene β-actin
(MYCN:β-actin ratio). The 50-μl PCR mixture contained
10 mmol/l Tris-HCl (pH 8.3), 50 mmol/l KCl, 10 mmol/l EDTA, 60
nmol/l passive reference dye ROX, 3.5 mmol MgCl2, 0.2
mmol/l of each dNTP, 300 nmol/l of each primer, 200 nmol/l of each
probe, 0.5 U of AmpliTaq Gold, and 1 U of AmpErase UNG (Applied
Biosystems) (Table II). The two
genes were amplified as follows: 120 sec at 95°C followed by 45
cycles of: 30 sec at 95°C, 30 sec at 60°C, and 30 sec at 72°C.
The concentrations of MYCN and β-actin
were estimated in the same assay via extrapolation on external
calibration curves. The threshold cycle value (CT) for
each sample was used to estimate the target concentration on the
curves. Calibration curves were constructed with reference DNA
extracted from the peripheral blood of two healthy male donors
(aged >50) with no chronic diseases. The DNA concentration was
determined by spectrophotometric measurement (Nanodrop 1000; Thermo
Scientific) and expressed as number of molecules per microliter as
previously described (25). The
DNA was serially diluted to plot the calibration curves and the
degree of amplification of each sample was derived from the
MYCN:β-actin ratio. Only samples in which the
MYCN:β-actin ratio was >2.0 were considered
amplified. In each assay, positive controls and non-amplified
negative controls were used.
cDNA synthesis and quantitative PCR
(qPCR) for CYPs
cDNA synthesis was performed by reverse
transcription of extracted tumor RNA using a RevertAid First Strand
cDNA Synthesis kit (Thermo Scientific). Briefly, the reaction
consisted of RNA, hexamer primers (provided in the RevertAid kit),
reaction buffer, RNase inhibitor, dNTP mix and reverse
transcriptase which were incubated for 5 min at 25°C and then the
reaction was terminated by heating at 70°C for 5 min. qPCR analysis
was performed using a TaqMan system (Applied Biosystems) according
to the manufacturer’s instructions. β-actin was used as the
reference gene. The 50-μl PCR mixture consisted of 20 ng
synthesized cDNA, primers for CYP3A4, CYP3A5 and
β-actin (600 nmol/l each), 250 nmol/l TaqMan probes and PCR
Master mix (Applied Biosystems) (Table II). Amplification and detection
were performed with the MxPro qPCR Agilent system (Agilent
Technologies, Santa Clara, CA, USA) using the following PCR
reaction profile: 95°C for 10 min followed by 40 cycles of 95°C for
20 sec and 62°C for 1 min. In every assay, liver DNA extracted from
paraffin-embedded healthy liver tissue, archived at St George
Hospital University Medical Center (Beirut, Paris) and a non-DNA
template were used as positive and negative controls,
respectively.
CYP mRNA expression calculations
Gene expression was calculated by absolute
quantification as previously described (26). Standard curves for the target genes
CYP3A4 and CYP3A5, as well as the reference gene
β-actin, were constructed by serial dilutions of a
synthesized cDNA obtained from archival liver tissue of a healthy
donor. The threshold cycle values obtained for each gene from the
tumor samples were extrapolated on corresponding constructed
standard curves to obtain the number of copies per gene per sample.
CYP3A4 and CYP3A5 expression levels were then
calculated as a ratio of the number of copies of the target gene to
the number of copies of the reference gene obtained for each
sample.
Statistical analysis
Variant genotypes were grouped for statistical
analyses based on previous studies of enzymatic function or
phenotypic consequence (27–30).
Homozygote carriers of the CYP3A5*3/*3
mutant genotype were compared with
CYP3A5*1/*3 heterozygotes or wild-type
CYP3A5*1/*1 homozygotes. Similarly,
homozygote and heterozygote carriers of CYP3A4*1B
mutant allele (CYP3A4*1B/1B or
CYP3A4*1A/1B) were compared with homozygote
wild-type CYP3A4*1A/1A carriers. CYP3A4
and CYP3A5 gene expression levels were analyzed as
dichotomous variables (above or below the median expression level).
Overall Survival (OS) is a standard measurement of survival defined
as the duration from the time of diagnosis of disease to either
death or date of last contact (31). A Kaplan-Meier survival analysis was
used to estimate the OS in the total group of patients, and the
MYCN amplification subgroups. The difference in survival
between the MYCN amplified and the MYCN non-amplified groups was
compared using the Log rank test (25). A Cox proportional hazard regression
model was used to assess associations between mortality and
CYP3A4 and CYP3A5 polymorphisms and mRNA expression
after adjusting for MYCN amplification. HRs and 95%
confidence intervals (CIs) were calculated. Statistical analyses
were performed using the Statistical Package for Social Sciences,
version 18.0.0 (SPSS, Inc., Chicago, IL, USA).
Results
Demographic and clinical
characteristics
The mean age at NB diagnosis was 2.5 years (±1.1).
The median OS was 3.7 years (range: birth to 11 years). Cases were
more likely to arise in males than in females (gender ratio 5:4).
The median time to mortality was 3.7 years (range: 9 months to 10
years) and 55.6% of the cases had stage IV disease at diagnosis.
MYCN amplification was present in 59% of the cases. All
patients underwent surgery while 59% underwent radiotherapy. Of all
the patients, 22% showed tumor relapse, and 44% had succumbed to
the disease by the end of follow-up (Table III).
 | Table IIIDescriptive data on the studied
population. |
Table III
Descriptive data on the studied
population.
| Characteristic | Patients
(n=27) |
|---|
| Age at diagnosis
(years), mean (SD) | 2.5 (±1.1) |
| OS (years), median
(range) | 3.7 (0–11) |
| Gender, n (%) |
| Male | 15 (55.6) |
| Female | 12 (44.4) |
| Tumor stage, n
(%) |
| III | 12 (44.4) |
| IV | 15 (55.6) |
| CYP3A4
genotype, n (%) |
|
CYP3A4*1A/*1A | 23 (85.2) |
|
CYP3A4*1A/*1B | 3 (11.1) |
|
CYP3A4*1B/*1B | 1 (3.7) |
| CYP3A5
genotype |
|
CYP3A5*1/*1 | 2 (7.4) |
|
CYP3A5*1/*3 | 3 (11.1) |
|
CYP3A5*3/*3 | 22 (81.5) |
| MYCN
amplification, n (%) |
| Yes | 16 (59.3) |
| No | 11 (40.7) |
| Surgery, n (%) |
| Yes | 27 (100.0) |
| No | 0 (0) |
| Radiotherapy, n
(%) |
| Yes | 16 (59.3) |
| No | 11 (40.7) |
| Relapse, n (%) |
| Yes | 6 (22.2) |
| No | 21 (77.8) |
| Mortality, n
(%) |
| Yes | 12 (44.4) |
| No | 15 (55.6) |
Prevalence of CYP3A4 and CYP3A5
genotypes
The majority of the studied patients were carriers
of the homozygote wild-type genotype CYP3A4*1A/1A
(85.2%), and the homozygote mutant genotype
CYP3A5*3/*3 (81.5%) (Table III and Fig 1).
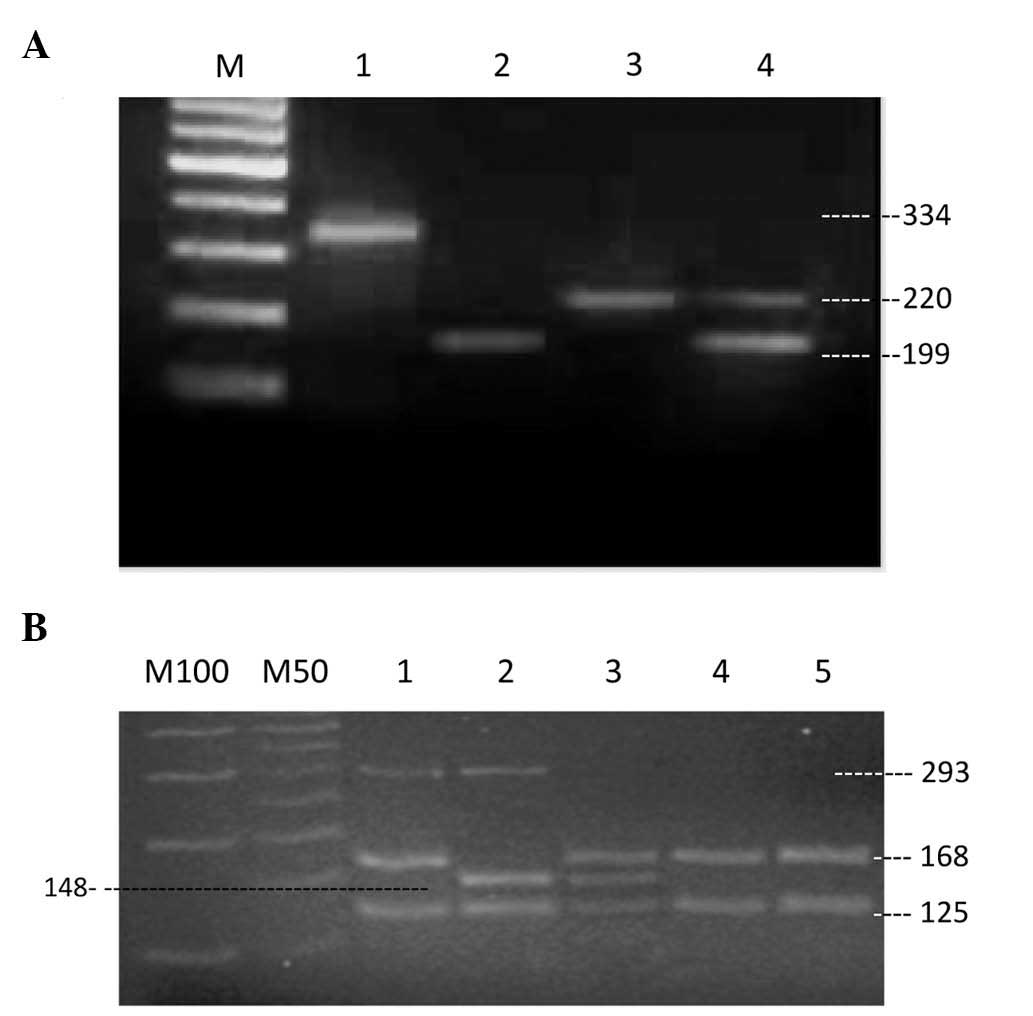 | Figure 1Genotyping gel electrophoresis for
CYP3A4 and CYP3A5 following polymerase chain reaction
and restriction fragment length polymorphism (PCR-RFLP). (A)
CYP3A4. Lane M: 100-bp DNA marker, lane 1: 334-bp PCR
product, lane 2: CYP3A4*1B/*1B
homozygote mutant, lane 3,
CYP3A4*1A/*1A homozygote wild type,
lane 4: CYP3A4*1A/*1B heterozygous
genotype. (B) CYP3A5. Lane M100: 100-bp DNA marker, lane
M50: 50-bp ladder, lanes 1, 4 and 5:
CYP3A5*3/*3 homozygote mutant, lane 2:
CYP3A5*1/*1 homozygote wild type, lane
3: CYP3A5*1/*3 heterozygote
genotype. |
Survival analysis
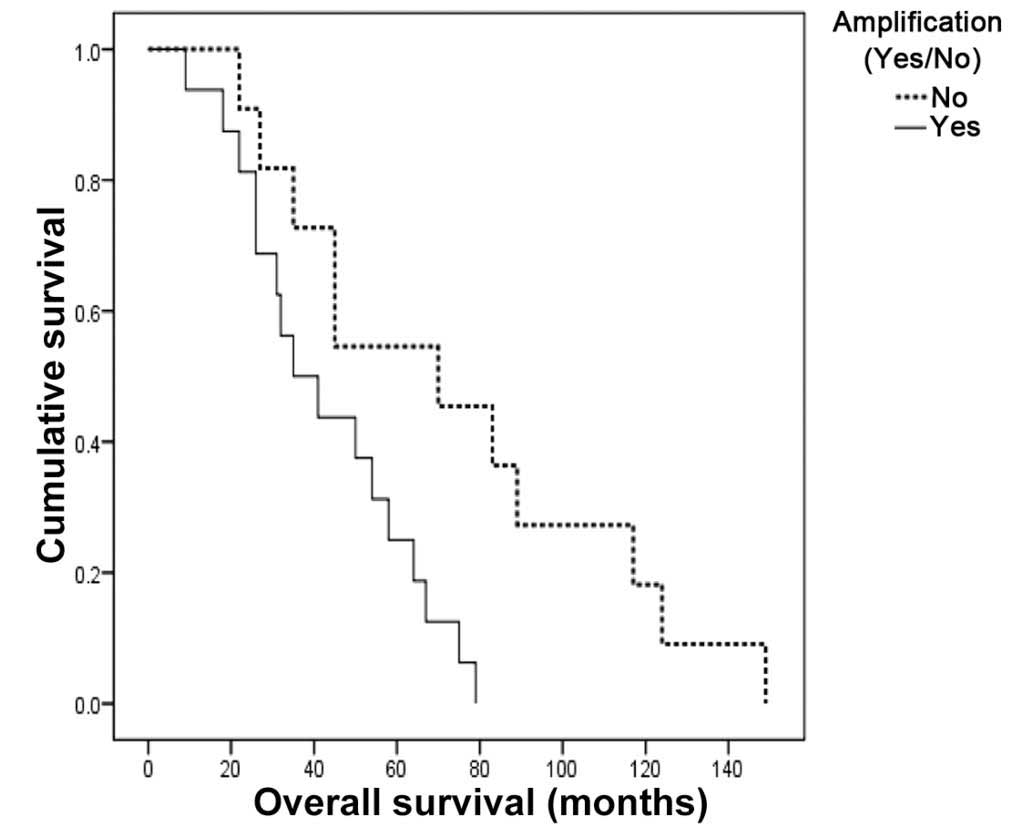
Kaplan-Meier survival analysis revealed that NB
patients with MYCN amplification had a significantly lower
median OS compared with those with no MYCN amplification
(3.1 vs. 5.8 years, P=0.03) (Fig.
2). The associations between the risk of mortality and a number
of genetic factors, calculated using Cox proportional hazard
regression analysis, are shown in Table IV. A 4-fold increased risk of
mortality was associated with MYCN amplification (HR 4.11,
95% CI 1.14–14.80 P<0.02). After adjusting for MYCN
amplification, patients with CYP3A5 expression levels above
the median had a 39% lower mortality risk (HR 0.61, 95% CI
0.21–1.74 P=0.353) while patients with CYP3A4 expression
levels above median had a 2-fold higher mortality risk (HR 2.00,
95% CI 0.67–5.90 P=0.214) compared with that of patients with
expression levels below the median; however, these associations
were not statistically significant.
 | Table IVAssociation of cytochrome P450
expression, genotypes, and MYCN amplification with the risk
of mortality in patients with neuroblastoma. |
Table IV
Association of cytochrome P450
expression, genotypes, and MYCN amplification with the risk
of mortality in patients with neuroblastoma.
| Variable
(reference) | HR (95% CI;
P-value) |
|---|
MYCN
amplification
(No amplification) | 4.11 (1.14–14.80;
0.020) |
CYP3A4
genotypes CYP3A4*1B/1B or
CYP3A4*1A/1B
(CYP3A4*1A/1A) | 0.48 (0.06–3.76;
0.368) |
CYP3A5
genotype CYP3A5*3/*3
(CYP3A5*1/*3 or
CYP3A5*1/*1) | 4.30 (0.56–33.30;
0.511) |
CYP3A4
expression>median
(<median) | 2.00 (0.67–5.90;
0.214) |
CYP3A5
expression>median
(<median) | 0.61 (0.21–1.74;
0.353) |
On the other hand, carriers of the
CYP3A5*3/*3 homozygote mutant genotype
had a 4-fold increased risk of mortality compared with that of the
homozygote wild-type or heterozygote mutant carriers (HR 4.30, 95%
CI 0.56 33.3 P=0.511). Homozygote and heterozygote carriers of the
CYP3A4*1B mutant allele had a 52% lower risk of
mortality compared with that of the non-carriers (HR 0.48, 95% CI
0.06–3.76, P=0.368). These associations were also adjusted for
MYCN amplification and they did not achieve statistical
significance.
Discussion
In the present study, the associations between
survival time and the expression levels or genetic polymorphisms of
CYP3A4 and CYP3A5, as well as MYCN
amplification, were analyzed in Lebanese children diagnosed with
NB. CYP3A4*1B and CYP3A5*3
polymorphisms were specifically targeted as being the most common
among multiple populations, with some insight into their effect on
enzymatic activity and expression (27). In concordance with previous
studies, MYCN amplification was observed to be the strongest
predictor of an unfavorable clinical outcomes in children with NB
in the present study (10). In
addition, after adjusting for MYCN amplification, the
results of the present study demonstrated that individuals with the
CYP3A5*3/*3 mutant genotype and lower
tumor CYP3A5 expression levels demonstrated a higher
mortality risk, while those with the CYP3A4*1B
mutant allele and lower tumor CYP3A4 expression levels
demonstrated a lower mortality risk; however, these associations
did not achieve statistical significance, likely owing to the small
sample size.
CYP3A4 and CYP3A5 are considered to
code for important CYP isoenzymes pertaining to drug metabolism,
due to the diversity of their anti-cancer substrates and their
relative abundance in humans (32). While no previous studies have
examined the association between these enzymes and clinical outcome
in NB, both enzymes have been shown to be important prognostic
modifiers in several common types of malignancy. High protein
expression levels of the CYP3A4/5 genes have previously been
reported to predict metastasis and poor prognosis in osteosarcoma
(33). On the other hand, patients
with breast cancer carrying the CYP3A4*1B and
CYP3A5*1 alleles were found to have a
significantly lower mean survival time compared with that of
carriers of the wild-type alleles (17). Furthermore,
CYP3A5*1 was observed to be associated with an
improved prognosis in a group of Caucasian patients with T-cell
acute lymphocytic leukemia (34)
and CYP3A5*3 was found to modulate
chemotherapy-induced toxicity in a group of Swedish patients with
ovarian cancer (35).
CYP3A5*3 consists of an A to G
transition in intron 3, creating a cryptic splice site (27). Inappropriate splicing leads to a
premature termination codon, resulting in a truncated protein with
101 amino acids (AA) compared with 502 AA in the normal protein
(28). mRNAs with premature stop
codons are more unstable and rapidly degraded, which explains the
lower levels of CYP3A5 mRNA in CYP3A5*3
homozygote carriers compared with those in wild-type carriers
(36).
CYP3A5*3/*3 carriers were determined
to have >50% reduction in catalytic activity for the metabolism
of midazolam compared with that of carriers of the
CYP3A5*1 wild-type allele (37). The results of the present study
regarding CYP3A5 are in agreement with these findings. The
CYP3A5*3/*3 genotype and lower
expression levels of CYP3A5 mRNA were consistent in
predicting poor clinical outcomes in NB patients. As such, it can
be hypothesized that homozygote carriers of the
CYP3A5*3 allele may have much lower levels of
enzymatic activity than carriers of other CYP3A5 variants,
and that the unfavorable outcome observed in the patients with NB
in the present study may be partially due to slower bioactivation
of used chemotherapeutic drugs, particularly etoposide and the
nitrogen mustard drugs, based on the literature (16,19,20,38).
This may be of importance clinically, particularly due to the high
prevalence of the defective allele in the studied population (87%).
The frequency of the allele reported in the Lebanese patients is
almost identical to that reported in Caucasian patients (91%), and
higher than that reported for populations of African descent (50%)
(24,39). The hypothesis proposed in the
present study, that the CYP3A5 genotype and associated
phenotype are part of the prognosis profile of patients with NB,
was based on the reported hazard ratios and supporting molecular
biological evidence from the literature. However, this hypothesis
could not be accepted with confidence as the results were not
statistically significant, possibly due to the small sample size of
the study.
CYP3A4*1B is an A to G transition
within the 5′-flanking region of the gene. This polymorphism is
considered to be associated with enhanced expression due to reduced
binding of a repressor that may affect the transcriptional rate
(29,30). However, no associations have been
found between the CYP3A4*1B allele genotype and
pharmacokinetics of its substrates (40,41).
Regulation of CYP3A4 expression is known to be essentially
pre-translational and its mRNA levels allow a good estimate of its
enzymatic activity (42). The
CYP3A4*1B polymorphism, however, cannot explain
the observed mRNA expression levels (43). In the current study, the possible
association between a higher mortality risk and higher
CYP3A4 expression levels may be more significant than the
reported protective effect for the CYP3A4*1B
polymorphism, particularly since it has a low frequency in the
studied group (8.7%). This is similar to frequencies reported in
Caucasian individuals (3.6–9.6%) and much lower than those reported
in African individuals (53–67%) (44, 45). A potential association between
higher CYP3A4 expression levels and clinical outcome in NB
may be hypothesized, particularly since similar associations have
been previously reported in breast cancer (46), however, the results of the present
study are inconclusive. Phenotypes may vary with tissue and may be
dependent upon other endogenous and environmental factors. Further
studies are required to elucidate the function of CYP3A4 in
the clinical outcome of NB.
There were a number of limitations in the present
study that should be considered when interpreting the results. The
sample size of the study was smaller than desired, due to the low
incidence of NB in the Lebanese population and missing tumor
samples for a number of identified patients. Another limitation of
this study is the possibility that other DMEs may be modifying the
response to drugs received by patients. Additional CYP3A4/5
SNPs, GSTs, N-acetyltransferases, and other CYPs not
investigated by the current study may influence the metabolic
outcome of anticancer drugs most commonly used in the NB setting
(Table I). The effects of these
factors were not adjusted for and may explain why the obtained
results were not significant. In addition, due to the retrospective
nature of the study, serum samples from patients were not available
to conduct direct measurements of drug levels. Furthermore,
investigation of the mechanisms by which CYP3A4/5
polymorphisms and expression levels may act to influence patient
outcome following high-dose therapy was not possible and further
studies should be conducted to address this point. Additionally,
DNA from blood samples was not available to check genotype
concordance between the blood and the tumor.
However, this study showed some strength. Despite
the small sample size, the prognostic value of MYCN
amplification was found to be statistically significant, in
concordance with the literature. Furthermore,
CYP3A5*3/*3 genotype and lower mRNA
expression levels predicted similar clinical outcome, which
reflects the internal validity of the reported results.
The hypothesis that CYP3A5*3
polymorphism and low expression levels may be associated with an
inferior clinical outcome in children diagnosed with NB is the
first of its kind. Although the observed effects did not achieve
statistical significance, the suggested associations may apply for
a substantial proportion of cases given the high prevalence of this
particular polymorphism in numerous populations including Lebanese.
The Lebanese population appears to have a high
CYP3A5*3 mutant genotype frequency, similar to
that of the Caucasian population. The results of the current study
may have an important clinical value. Since CYP3A5
represents at least half the total hepatic CYP3A content, it may be
an important genetic contributor to inter-individual and
interracial differences in clearance and response to NB anticancer
drugs, particularly in CYP3A5 homozygote mutant patients
(33). Characterization of
associations between CYP3A5 polymorphisms and clinical
outcome could be used to genetically define subgroups of patients
with NB who may be more predisposed to therapeutic failures or
adverse drug reactions. However, the prognostic value of functional
polymorphisms of CYP3A5 and CYP3A4 remains to be
fully determined. We recommend building upon the observations of
the current study by conducting cohort studies which incorporate
larger sample sizes. Confirming these hypotheses and generating
supportive mechanistic data will ultimately allow for an
individualized and optimized NB therapy that may improve
survival.
Acknowledgements
The authors thank the Pathology Departments at Saint
George Hospital University Medical Center, Hotel Dieu de France
Hospital, and University Medical Center - Rizk Hospital for
providing the archival tumor sections. This study was supported by
a University of Balamand Research Grant.
References
|
1
|
Maris JM: Recent advances in
neuroblastoma. N Engl J Med. 362:2202–2211. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Howlader N, Noone A, Krapcho M, et al:
SEER Cancer Statistics Review, 1975–2011. National Cancer
Institute; Bethesda, MD: 2012, http://seer.cancer.gov/csr/1975_2011/.
Accessed August 14, 2014
|
|
3
|
London WB, Castleberry RP, Matthay KK, et
al: Evidence for an age cutoff greater than 365 days for
neuroblastoma risk group stratification in the Children’s Oncology
Group. J Clin Oncol. 23:6459–6465. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lebanese Ministry of Public Health and
Epidemiology Surveillance Program. National cancer registry. 2007,
http://www.moph.gov.lb/Prevention/Surveillance/Pages/Cancer.aspx.2010.
Accessed December 9, 2013
|
|
5
|
Ries L, Smith M, Gurney J, Linet M, Tamra
T, Young J and Bunin G: Cancer incidence and survival among
children and adolescents: United States SEER Program 1975–1995. NIH
publication no. 99-4649. National Cancer Institute; Bethesda, MD:
1999
|
|
6
|
Maris JM, Hogarty MD, Bagatell R and Cohn
SL: Neuroblastoma. Lancet. 369:2106–2120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brodeur GM: Neuroblastoma: biological
insights into a clinical enigma. Nat Rev Cancer. 3:203–216. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kushner BH, Gilbert F and Helson L:
Familial neuroblastoma. Case reports, literature review, and
etiologic considerations. Cancer. 57:1887–1893. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brodeur GM, Seeger RC, Schwab M, Varmus HE
and Bishop JM: Amplification of N-myc in untreated human
neuroblastomas correlates with advanced disease stage. Science.
224:1121–1124. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Seeger RC, Brodeur GM, Sather H, Dalton A,
Siegel SE, Wong KY and Hammond D: Association of multiple copies of
the N-myc oncogene with rapid progression of neuroblastomas. N Engl
J Med. 313:1111–1116. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tonini GP and Romani M: Genetic and
epigenetic alterations in neuroblastoma. Cancer Lett. 197:69–73.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kushner BH, LaQuaglia MP, Bonilla MA, et
al: Highly effective induction therapy for stage 4 neuroblastoma in
children over 1 year of age. J Clin Oncol. 12:2607–2613.
1994.PubMed/NCBI
|
|
13
|
Simon T, Längler A, Berthold F, Klingebiel
T and Hero B: Topotecan and etoposide in the treatment of relapsed
high-risk neuroblastoma: results of a phase 2 trial. J Pediatr
Hematol Oncol. 29:101–106. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Daly AK: Pharmacogenetics of the major
polymorphic metabolizing enzymes. Fundam Clin Pharmacol. 17:27–41.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ashton LJ, Murray JE, Haber M, Marshall
GM, Ashley DM and Norris MD: Polymorphisms in genes encoding drug
metabolizing enzymes and their influence on the outcome of children
with neuroblastoma. Pharmacogenet Genomics. 17:709–717. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang Z, Roy P and Waxman DJ: Role of
human liver microsomal CYP3A4 and CYP2B6 in catalyzing
N-dechloroethylation of cyclophosphamide and ifosfamide. Biochem
Pharmacol. 59:961–972. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Petros WP, Hopkins PJ, Spruill S, et al:
Associations between drug metabolism genotype, chemotherapy
pharmacokinetics, and overall survival in patients with breast
cancer. J Clin Oncol. 23:6117–6125. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Walker D, Flinois JP, Monkman SC, et al:
Identification of the major human hepatic cytochrome P450 involved
in activation and N-dechloroethylation of ifosfamide. Biochem
Pharmacol. 47:1157–1163. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dennison JB, Jones DR, Renbarger JL and
Hall SD: Effect of CYP3A5 expression on vincristine metabolism with
human liver microsomes. J Pharmacol Exp Ther. 321:553–563. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Relling MV, McLeod HL, Bowman LC and
Santana VM: Etoposide pharmacokinetics and pharmacodynamics after
acute and chronic exposure to cisplatin. Clin Pharmacol Ther.
56:503–511. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
McCune JS, Risler LJ, Phillips BR, Thummel
KE, Blough D and Shen DD: Contribution of CYP3A5 to hepatic and
renal ifosfamide N-dechloroethylation. Drug Metab Dispos.
33:1074–1081. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bosch TM, Meijerman I, Beijnen JH and
Schellens JH: Genetic polymorphisms of drug-metabolising enzymes
and drug transporters in the chemotherapeutic treatment of cancer.
Clin Pharmacokinet. 45:253–285. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
van Schaik RH, de Wildt SN, van Iperen NM,
Uitterlinden AG, van den Anker JN and Lindemans J: CYP3A4-V
polymorphism detection by PCR-restriction fragment length
polymorphism analysis and its allelic frequency among 199 Dutch
Caucasians. Clin Chem. 46:1834–1836. 2000.PubMed/NCBI
|
|
24
|
van Schaik RH, van der Heiden IP, van den
Anker JN and Lindemans J: CYP3A5 variant allele frequencies in
Dutch Caucasians. Clin Chem. 48:1668–1671. 2002.PubMed/NCBI
|
|
25
|
Raggi CC, Bagnoni ML, Tonini GP, et al:
Real-time quantitative PCR for the measurement of MYCN
amplification in human neuroblastoma with the TaqMan detection
system. Clin Chem. 45:1918–1924. 1999.PubMed/NCBI
|
|
26
|
Peirson SN, Butler JN and Foster RG:
Experimental validation of novel and conventional approaches to
quantitative real-time PCR data analysis. Nucleic Acids Res.
31:e732003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lamba JK, Lin YS, Schuetz EG and Thummel
KE: Genetic contribution to variable human CYP3A-mediated
metabolism. Adv Drug Deliv Rev. 54:1271–1294. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Busi F and Cresteil T:
Phenotyping-genotyping of alternatively spliced genes in one step:
study of CYP3A5*3 polymorphism. Pharmacogenet Genomics.
15:433–439. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sinues B, Vicente J, Fanlo A, et al:
CYP3A5*3 and CYP3A4*1B allele distribution
and genotype combinations: differences between Spaniards and
Central Americans. Ther Drug Monit. 29:412–416. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Amirimani B, Ning B, Deitz AC, Weber BL,
Kadlubar FF and Rebbeck TR: Increased transcriptional activity of
the CYP3A4*1B promoter variant. Environ Mol Mutagen.
42:299–305. 2003. View Article : Google Scholar
|
|
31
|
Lim HJ, Gill S, Speers C, et al: Impact of
irinotecan and oxaliplatin on overall survival in patients with
metastatic colorectal cancer: a population-based study. J Oncol
Pract. 5:153–158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gellner K, Eiselt R, Hustert E, et al:
Genomic organization of the human CYP3A locus: identification of a
new, inducible CYP3A gene. Pharmacogenetics. 11:111–121. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dhaini HR, Thomas DG, Giordano TJ, et al:
Cytochrome P450 CYP3A4/5 expression as a biomarker of outcome in
osteosarcoma. J Clin Oncol. 21:2481–2485. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Borst L, Wallerek S, Dalhoff K, Rasmussen
KK, Wesenberg F, Wehner PS and Schmiegelow K: The impact of
CYP3A5*3 on risk and prognosis in childhood acute
lymphoblastic leukemia. Eur J Haematol. 86:477–483. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gréen H, Khan MS, Jakobsen-Falk I,
Avall-Lundqvist E and Peterson C: Impact of CYP3A5*3 and
CYP2C8-HapC on paclitaxel/carboplatin-induced myelosuppression in
patients with ovarian cancer. J Pharm Sci. Jun 23–2011.(Epub ahead
of print).
|
|
36
|
Maquat LE: When cells stop making sense:
effects of nonsense codons on RNA metabolism in vertebrate cells.
RNA. 1:453–465. 1995.PubMed/NCBI
|
|
37
|
Kuehl P, Zhang J, Lin Y, et al: Sequence
diversity in CYP3A promoters and characterization of the genetic
basis of polymorphic CYP3A5 expression. Nat Genet. 27:383–391.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lewis AD, Lau DH, Durán GE, Wolf CR and
Sikic BI: Role of cytochrome P-450 from the human CYP3A gene family
in the potentiation of morpholino doxorubicin by human liver
microsomes. Cancer Res. 52:4379–4384. 1992.PubMed/NCBI
|
|
39
|
Xie HG, Wood AJ, Kim RB, Stein CM and
Wilkinson GR: Genetic variability in CYP3A5 and its possible
consequences. Pharmacogenomics. 5:243–272. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wandel C, Witte JS, Hall JM, Stein CM,
Wood AJ and Wilkinson GR: CYP3A activity in African American and
European American men: population differences and functional effect
of the CYP3A4*1B5′-promoter region polymorphism. Clin
Pharmacol Ther. 68:82–91. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Westlind A, Löfberg L, Tindberg N,
Andersson TB and Ingelman-Sundberg M: Interindividual differences
in hepatic expression of CYP3A4: relationship to genetic
polymorphism in the 5′-upstream regulatory region. Biochem Biophys
Res Commun. 259:201–205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fisher CD, Lickteig AJ, Augustine LM,
Ranger-Moore J, Jackson JP, Ferguson SS and Cherrington NJ: Hepatic
cytochrome P450 enzyme alterations in humans with progressive
stages of nonalcoholic fatty liver disease. Drug Metab Dispos.
37:2087–2094. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Rodríguez-Antona C, Donato MT, Pareja E,
Gómez-Lechón MJ and Castell JV: Cytochrome P-450 mRNA expression in
human liver and its relationship with enzyme activity. Arch Biochem
Biophys. 393:308–315. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Garcia-Martín E, Martínez C, Pizarro RM,
Garcia-Gamito FJ, Gullsten H, Raunio H and Agúndez JA: CYP3A4
variant alleles in white individuals with low CYP3A4 enzyme
activity. Clin Pharmacol Ther. 71:196–204. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ball SE, Scatina J, Kao J, et al:
Population distribution and effects on drug metabolism of a genetic
variant in the 5′ promoter region of CYP3A4. Clin Pharmacol Ther.
66:288–294. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Miyoshi Y, Ando A, Takamura Y, Taguchi T,
Tamaki Y and Noguchi S: Prediction of response to docetaxel by
CYP3A4 mRNA expression in breast cancer tissues. Int J Cancer.
97:129–132. 2002. View Article : Google Scholar : PubMed/NCBI
|