Introduction
Cartilage injuries remain a major clinical problem
due to the poor self-healing ability of articular cartilage
(1). Despite recent
cell-engineering advances, the treatment of cartilage injuries has
remained challenging. As chondrocytes have a limited capability of
regeneration, it is necessary to obtain a sufficient number of
chondrocytes in vitro for repairing traumatic joint surface
lesions. Although stem cell transplantation therapy using
mesenchymal stem cells (MSCs) has been considered a prominent
strategy, the major problem of limited proliferative capacity of
autologous cells remains to be addressed (2).
It is well-established that the successful
reprogramming of differentiated human somatic cells into a
pluripotent state would allow for the creation of patient- and
disease-specific stem cells. Since Takahashi et al (3) demonstrated that fibroblasts were
reprogrammed into induced pluripotent stem cells (iPSCs) by defined
transcription factors, numerous differentiation strategies have
been applied to reprogram mature cells into a single- or
pluripotent stem cells, including hematopoietic cells (4), cardiomyocytes (5), neural progenitors (6), liver cells (7) and hepatic stem cells (8). A number of studies have suggested
iPSCs as alternative candidates for the treatment of numerous
disorders (9–11). Previously, Teramura et al
(2) demonstrated that the
properties of iPSCs render them suitable alternative candidates for
the treatment of articular disorders and suggested an effective
approach for preparing chondrocytes from iPSCs.
Additionally, current strategies of virus-mediated
formation of iPSCs are markedly limited in their clinical
application (12,13). Other approaches, including a
plasmid transfection method, are able to obtain mouse iPSCs
directly without exogenous gene modification (12). However, the efficiency of such
strategies is markedly low and with poor operability, due to the
low probability of its successful integration into the cellular
genome (12). Previously, Yusa
et al (13) described an
efficient piggyBac transposon-based approach to generate
integration-free iPSCs. The transposons carrying 2A peptide-linked
reprogramming factors induced reprogramming of mouse embryonic
fibroblasts (MEF) with equivalent efficiencies to retroviral
transduction. To date, there are a limited number of studies using
this piggyBac transposon-based approach to reprogram rat embryonic
fibroblasts (REF) to iPSCs without any genetic alteration (13).
Therefore, the aim of the present study was to apply
a piggyBac transposon-based approach to reprogram REF into iPSCs,
and subsequently to investigate its capacity of chondrocyte
differentiation in vitro.
Materials and methods
Reprogramming of REF using transposon
vectors
The present study was approved by the Ethics
Committee of Fudan University (Shanghai, China). REF were isolated
from embryonic day (E)14.5 Sprague-Dawley rat embryos. The pregnant
rats were obtained from Shanghai Laboratory Animal Center, CAS
(Shanghai, China). REFs were cultured in Dulbecco’s modified
Eagle’s medium (DMEM; Hyclone, Logan, UT, USA) supplemented with
10% fetal bovine serum (FBS; Gibco-BRL, Carlsbad, CA, USA). For the
generation of iPSCs, REF were plated onto a 35-mm dish
(5×105 cells per well) 1 day prior to transfection. The
next day, 3 μg pCMV-mPBase and plasmids pPB-CAG.OSKM-puΔtk or
pPB-CAG.OSKML-puΔtk (donated by the Wellcome Trust Sanger
Institute, Hinxton, UK) containing the piggyBac transposon were
transfected using Lipofectamine 2000 (Invitrogen Life Technologies,
Carlsbad CA, USA) according to the manufacturer’s instructions.
Following one day, the REF were trypsinized and re-plated onto
L-wnt3a feeder layers at a ratio of 1:10 in REF medium. The medium
was replaced every other day. Following seven days, the medium was
changed to N2B27 medium [1:1 mixture of N2 medium (DMEM/F12, N2
supplement) and B27 medium (Neurobasal medium, B27 supplement)]
supplemented with 0.1 mM 2-mercaptoethanol (Invitrogen Life
Technologies), 2 mM l-glutamine (Invitrogen Life Technologies), 1×
non-essential amino acids (EMD Millipore, Billerica, MA, USA) and
1,000 U/ml hLIF 1 mM MEK1/2 (mitogen activated protein kinase
kinase 1/2) inhibitor PD0325901 (Selleck Chemicals, Shanghai,
China). Following 14 days, the colonies were stained using the
alkaline phosphatase (AP) detection kit (Sigma-Aldrich, St. Louis,
MO, USA) and counted, selected and further expanded.
Immunostaining
The cells were fixed in 4% paraformaldehyde in
phosphate-buffered saline (PBS) for 20 min at room temperature and
permeabilized in 0.1% Triton-X 100 in PBS for 20 min, then blocked
with 5% FBS in PBS for 40 min at room temperature. The cells were
washed with PBS and incubated with the following primary
antibodies: Goat polyclonal octamer-binding transcription factor 4
(Oct4; 1:100; sc-8628; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA), rabbit polyclonal Nanog (1:300; ab80892; Abcam,
Cambridge, UK), mouse monoclonal stage-specific embryonic antigen 1
(SSEA1; 1:200; sc-21702; Santa Cruz Biotechnology, Inc.) and
His-tag mouse monoclonal immunoglobulin (Ig)G (1:200; ab18184;
Abcam) overnight at 4°C, followed by goat anti-mouse immunoglobulin
IgM-fluorescein isothiocyanate (FITC) (1:400; Beyotime Institute of
Biotechnology, Shanghai, China), donkey anti-goat IgG-FITC (1:40;
Beyotime Institute of Biotechnology), goat anti-mouse IgG-FITC
(1:400; Beyotime Institute of Biotechnology), goat anti-rabbit
IgG-FITC or Cy3 (1:400; Beyotime Institute of Biotechnology) as
secondary antibodies. Nuclei were stained with Hoechst 33258
(Sigma-Aldrich).
Quantitative polymerase chain reaction
(qPCR) and western blot analysis
Total RNA was extracted by using TRIzol reagent
(Invitrogen Life Technologies). RNA was reverse-transcribed with
random primer using SuperScript III (Takara Bio Inc., Shiga, Japan)
according to the manufacturer’s instructions. qPCR was performed
using SYBRgreen PCR Master Mix (Takara Bio Inc.) on the ABI7500
system (Applied Biosystems Life Technologies, Foster City, CA, USA)
according to the manufacturer’s instructions; the primers are
listed in Table I. The expanded
colonies were collected and suspended in radioimmunoprecipitation
buffer (Sangon Biotech Co., Ltd., Shanghai, China) and the proteins
were separated on electrophoresis gels. The protein blots were
analyzed using antibodies against Oct4 (sc-8628; Santa Cruz
Biotechnology, Inc.), Nanog (ab80892; Abcam) or to GAPDH (mouse
monoclonal; Sigma-Aldrich).
 | Table IPrimer sequences used for polymerase
chain reaction. |
Table I
Primer sequences used for polymerase
chain reaction.
| Primer | Sequence |
|---|
| Oct4-F |
5′-CCCCATTTCACCACACTCTACTC-3′ |
| Oct4-R |
5′-GTGACAGGAACAGAGGGAAAGG-3′ |
| Nanog-F |
5′-GACTAGCAACGGCCTGACTCA-3′ |
| Nanog-R |
5′-CTGCAATGGATGCTGGGATA-3′ |
| Sox2-F |
5′-CTGTTTTTTCATCCCAATTGCA-3′ |
| Sox2-R |
5′-CGGAGATCTGGCGGAGAATA-3′ |
| GAPDH-F |
5′-ATCACTGCCACTCAGAAG-3′ |
| GAPDH-R |
5′-AAGTCACAGGAGACAACC-3′ |
| IRx3-F |
5′-GCTCAATGAACACCGCAAGA-3′ |
| IRx3-R |
5′-GTGATGATGGCCAACATGATCT-3′ |
| Maf- F |
5′-CAGACGCTCCTCCTGAGCTT-3′ |
| Maf- R |
5′-CATTCGTTGGGCACCAAAG-3′ |
| Ramp2- F |
5′-CATCTGCCTCATCCCTTTCC-3′ |
| Ramp2-R |
5′-TGCGCATCGCCGTCTT-3′ |
| Sal3- F |
5′-CCAGCTGTCTCCGACCAGTT-3′ |
| Sal3- R |
5′-TGACGTTTGCATAGAGTCAAGCA-3′ |
| Hoxa5-F |
5′-CCCGGACTACCAGTTGCATAA-3′ |
| Hoxa5-R |
5′-CGCCGAGTCCCTGAATTGT-3′ |
Protein transduction and transposon
removal from primary iPSCs
The expression vector PCDH-cPP-mPB-HIS contains a
nuclear localization sequence and protein transduction domain
consisting of nine arginine residues (9R) cPP at the N-terminus and
a HIS tag at the C-terminus of the transposase (mPB) protein. This
expression cassette was assembled by standard PCR methods (13). Expression in 293T and purification
of PCDH-cPP-mPB-HIS was performed as described previously (14). Protein transduction was performed
with rat iPSCs on L-wnt3a feeder cells using 2 mM recombinant
protein for 6 h. Approximately 6×105 cells were then
seeded into 10-cm dishes containing feeder cells following three
days of culture. The next day, 0.2 μM fialuridine (FIAU) was added
and the selection was continued for 10 days. The resulting colonies
were selected and expanded, and then the transposon was removed.
Transposon removal was identified by genomic PCR. Primers for PCR
are listed in Table II.
 | Table IIPrimer sequences. |
Table II
Primer sequences.
| Gene | Sequence |
|---|
| L (Sox2-Klf4
junction)-F |
5′-GCAACGGCAGCTACAGCATGATGCAG-3′ |
| L (Sox2-Klf4
junction)-R |
5′-CAGGAGGTCGTTGAACTCCTCGGTCTC-3′ |
| R (PGK
promoter)-F |
5′-GCGTCGACCGACTGTGCCTTCTAGTTGCCAGCC-3′ |
| R (PGK
promoter)-R |
5′-GTTGGCGCCTACCGGTGGATGTGGAATGTG-3′ |
| Endogenous
control-F |
5′-GGGGCCTTCTGGGGGTAAAGTTCAGAACAC-3′ |
| Endogenous
control-R |
5′-TGGCTGCCTGAGGGCAAGAGGGAAAGAATC-3′ |
Differentiation of embryoid bodys
(EBs)
Aliquots of 25 μl differentiation medium (10% FBS;
Invitrogen Life Technologies) containing 1,000 cells were
cultivated in ‘hanging drops’ for 2 days and subsequently in
suspension in petri dishes for an additional three days. The
combination of bone morphogenetic protein-2 (BMP-2; 15 ng/ml;
Invitrogen Life Technologies) and tumor growth factor-1 (TGF-1; 2
ng/ml; Invitrogen Life Technologies) which enhanced chondrogenic
differentiation efficiency was added to the medium during the
suspension phase. The EBs were plated onto 0.1% gelatin-coated
24-well plates for Alcian blue staining or immunostaining, and onto
35-mm dishes for total RNA isolation. Alcian blue staining was
performed as described previously (15). Immunostaining and qPCR were
performed as aforementioned. The primers for PCR are listed in
Table III.
 | Table IIIPrimer sequences. |
Table III
Primer sequences.
| Gene | Sequence |
|---|
| Aggrecan-F |
5′-GAAGTGGCGTCCAAACCAAC-3′ |
| Aggrecan-R |
5′-AGCTGGTAATTGCAGGGGAC-3′ |
| Sox9-F |
5′-TCCCCGCAACAGATCTCCTA-3′ |
| Sox9-R |
5′-AGCTGTGTGTAGACGGGTTG-3′ |
| Col2a1-F |
5′-GCTTCTGGTAACCCAGGGAC-3′ |
| Col2a1-R |
5′-TTGGGGCCTTGTTCACCTTT-3′ |
| GAPDH-F |
5′-ATCACTGCCACTCAGAAG-3′ |
| GAPDH-R |
5′-AAGTCACAGGAGACAACC-3′ |
Results
Formation of iPSCs
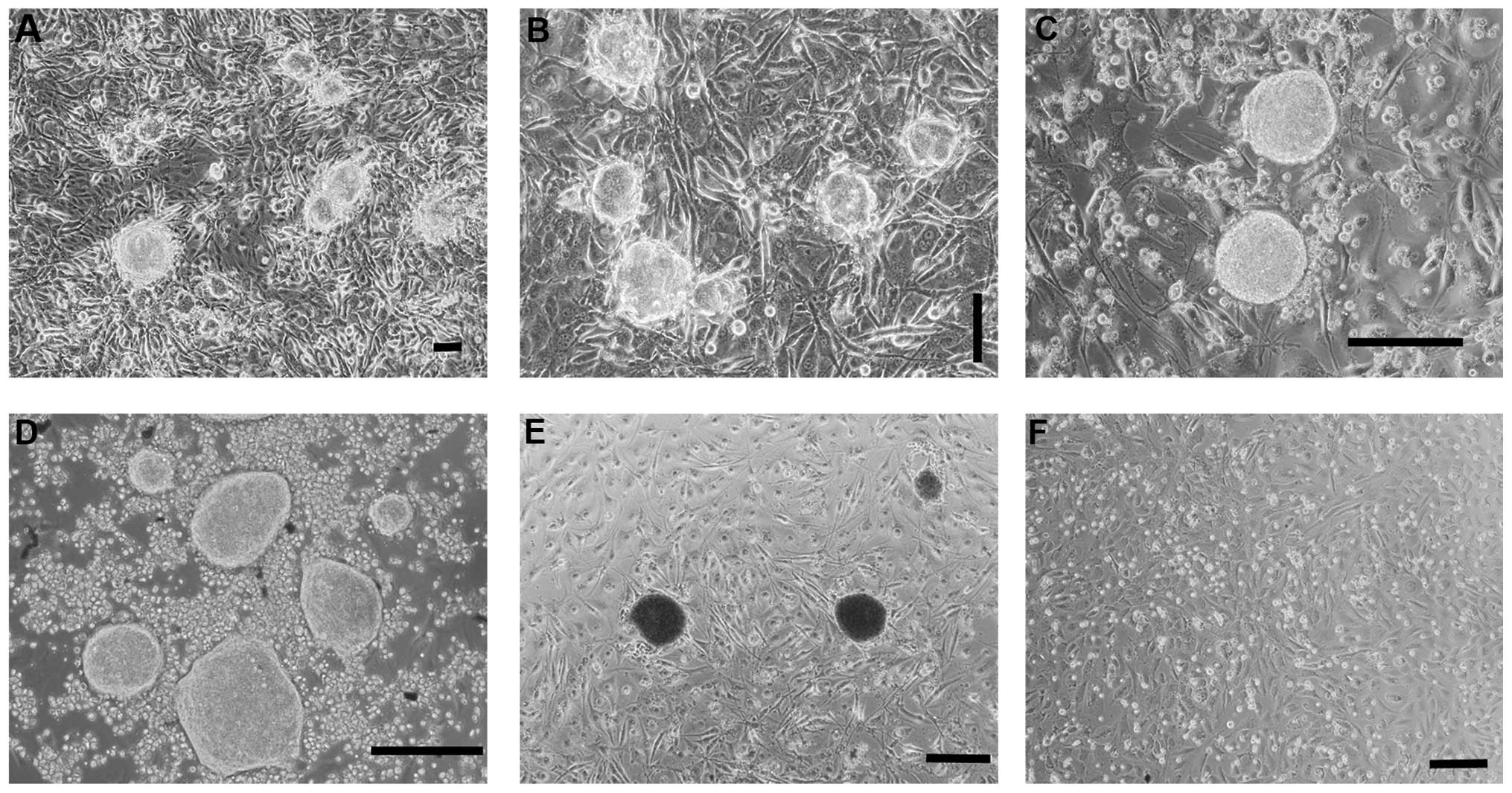
Following 14 days, colonies with an embryonic stem
(ES)-like morphology were observed in REFs transfected with PB-OSKM
(Fig. 1A), which also stained
positive with alkaline phosphatase (AP) (Fig. 1E). Following being supplemented
with 1 μM PD0325901, the colonies remained intact at 21 days
(Fig. 1B). At 35 days, its shape
was large and round with clear boundaries (Fig. 1C), which was very similar to the
shape of rat ES (Fig. 1D).
However, the empty vehicle control group (Fig. 1F) exhibited no clone formation at
any time.
iPSC clone identification
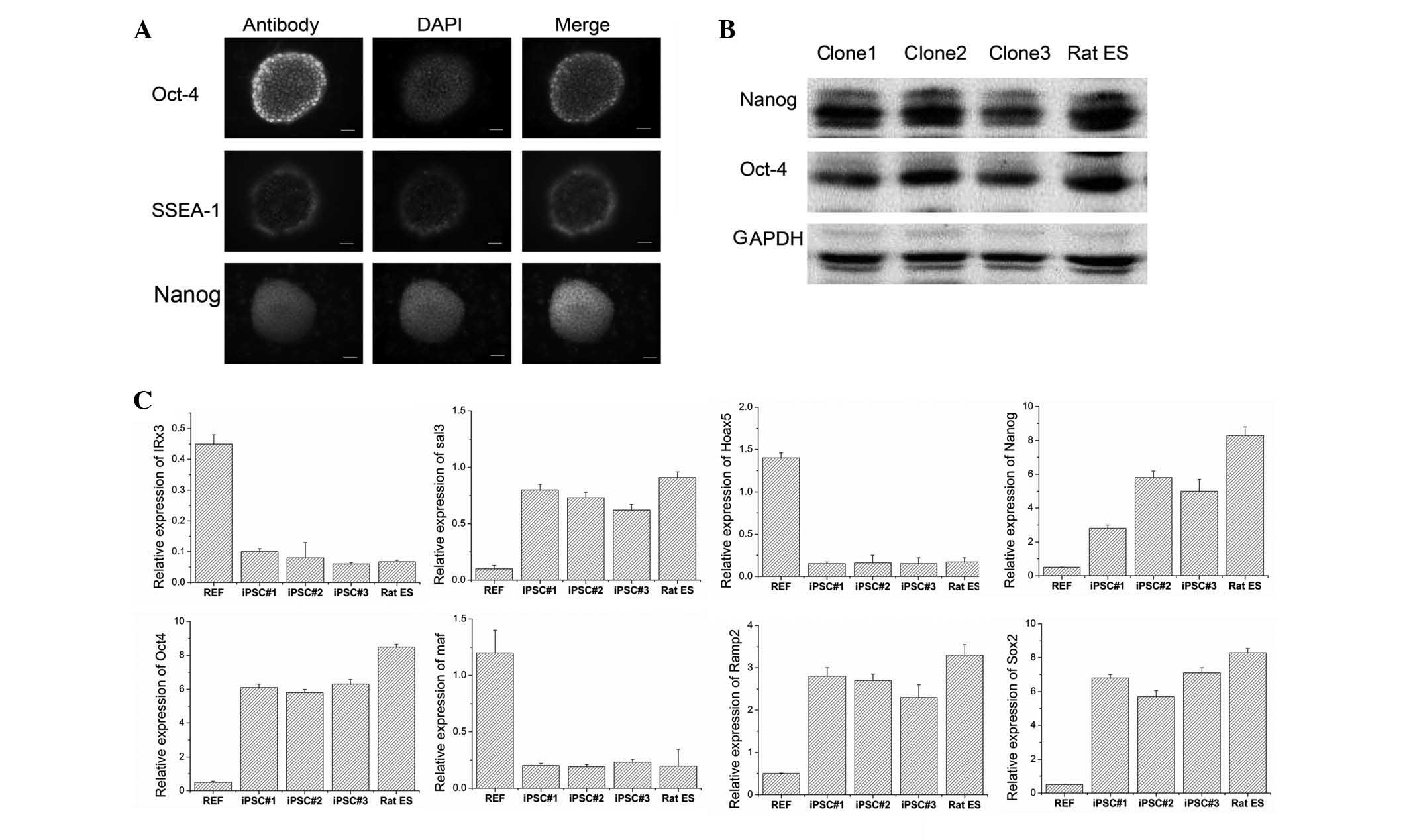
In the present study, three clones were selected for
examining the associated genes. The immunostaining (Fig. 2A) and western blotting results
(Fig. 2B) revealed that the cells
were positive for surface marker genes, including Oct4, Nanog and
SSEA1. These findings indicated that the iPSCs were similar to rat
ES regarding their biological characteristics. qPCR analysis
(Fig. 2C) indicated that the
clones exhibited lower expression levels of IRX3, Maf and Hoax5
compared with REF, while Oct4, nanog, Ramp2, Sox2 and Sal3 were
expressed at higher levels in the clones compared with those of
REF. These findings indicated that iPSCs expressed pluripotent
markers and exhibited a similar gene expression to that of rat
ES.
Transposon removal from primary
iPSCs
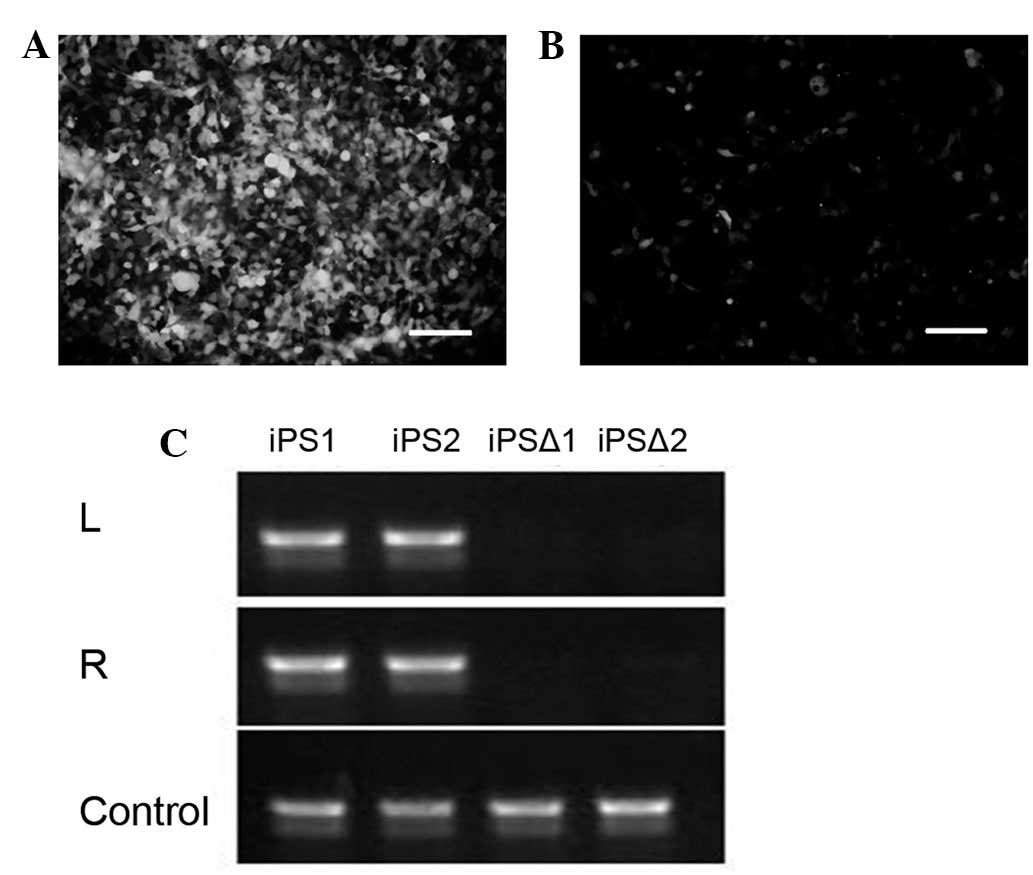
In order to further examine whether the tranposase
mPB system may enter into the cells to remove piggyBac-carried
genes, the piggyBac-green fluorescent protein (GFP) plasmid was
transduced into 293T cells (Fig.
3A). Following 48 h, 0.2 μg/ml of the mPB protein was added for
8 h. Approximately 70% of the GFP was removed following 48 h
(Fig. 3B), indicating the mPB
protein had the capability to remove piggyBac transposon-carrying
genes efficiently. Following adding FIAU, the cells without gene
removal gradually died, while the cells with gene removal were able
to survive. Finally, the iPSC clones without genetic modification
were obtained (Fig. 3C).
Chondrogenic differentiation of
reprogrammed iPSCs
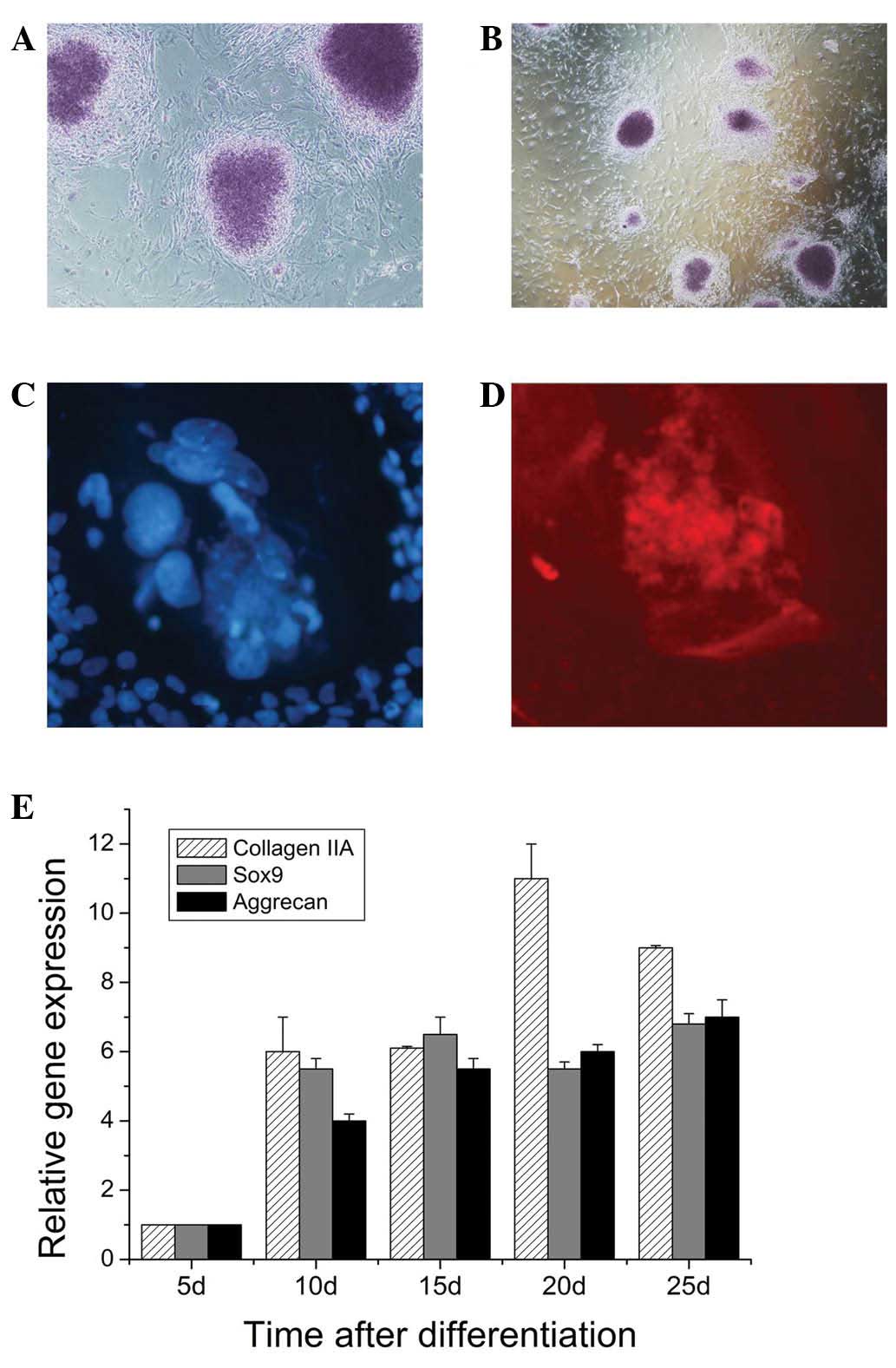
In EB outgrowths derived from iPSCs, numerous Alcian
blue-stained regions were found, indicating the presence of acidic
proteoglycans (Fig. 4A and B).
Acidic proteoglycans were suggestive for cartilaginous tissue. The
cells in these areas produced collagen II as demonstrated by
immunostaining (Fig. 4C and D).
qPCR analysis demonstrated that genes encoding transcription
factors involved in mesenchymal differentiation, including
extracellular matrix proteins of cartilage tissue (Type II
collagen, aggrecan) were found to be expressed in EBs during
culture (Fig. 4E). On the 20th day
of culture time, the expression levels of Type II were highest.
Discussion
In the present study, piggyBac plasmid (carrying
four or five genes) and transposase mPB system were applied to
obtain rat iPSCs. Particularly, the transposase mPB system was used
to successfully remove the transposons-carried genes to obtain the
iPSCs without genetic modification. The preliminary study also
revealed that iPSCs differentiate into chondrocytes in
vitro.
Previously, the formation of iPSCs mostly depended
on the mediation of virus (retrovirus, lentivirus and adenovirus).
In 2006, Takahashi and Yamanaka demonstrated that
retrovirus-mediated introduction of four transcription factors
(Oct-3/4, Sox2, c-Myc and KLF4) into mouse embryonic or adult
fibroblasts and selection for the expression of Fbx15, a target of
Oct-3/4 and Sox2, resulted in the generation of iPSCs, which
exhibited the morphology and growth properties of ES cells and
expressed ES cell marker genes (16,17).
In 2009, Sommer et al (18)
described the application of a single lentiviral vector expressing
a ‘stem cell cassette’ composed of the four transcription factors
and a combination of 2A peptide and internal ribosome entry site
technology, generating iPSCs from postnatal fibroblasts. Using a
doxycycline-inducible lentiviral system, Maherali et al
(19) developed a strategy to
induce human iPSCs formation at high efficiency, and they obtained
‘secondary’ human iPSCs at a frequency of at least 100-fold greater
than the initial conversion upon addition of doxycycline to human
iPSCs-derived differentiated cells. However, a major limitation of
this technology by viral expression of the transcription factors is
the use of potentially harmful genome-integrating viruses.
Stadtfeld et al (20)
obtained mouse iPSCs from fibroblasts and liver cells by using
non-integrating adenoviruses transiently expressing Oct4, Sox2,
Klf4 and c-Myc. These adenoviral iPSCs revealed DNA demethylation
characteristic of reprogrammed cells, expressed endogenous
pluripotency genes, formed teratomas and contributed to multiple
tissues. However, viral integration into the host genome increases
the risk of tumorigenicity, hindering its clinical applications
(21). In order to avoid the use
of a viral vectors, Okita et al (12,22)
described an alternative method to generate iPSCs from MEF by
continual transfection of plasmid vectors. However, the
reprogramming efficiency of this protocol was very low. Previously,
Yusa et al (13) reported
an efficient piggyBac transposon-based approach to generate
integration-free iPSCs in MEF. In the present study, piggyBac
transposon-based reprogramming was successfully used to generate
iPSCs in REF. Furthermore, PCDH-cPP-mPB-HIS was applied to
successfully remove the piggyBac transposon, to obtain iPSCs clones
without any genetic alteration.
Pluripotent mouse ES cells have been derived and
maintained by using various empirical combinations of feeder cells,
conditioned media, cytokines, growth factors, hormones, fetal calf
serum and serum extracts. It has been reported that ES cells have
an innate program for self-replication, which does not require
extrinsic instruction (23). The
self-renewal factors present in embryonic carcinoma cell-derived
conditioned medium may be responsible for the self-renewal capacity
of embryonic carcinoma and ES cells independently of leukemia
inhibitory factor signaling (24).
In the present study, the L-wnt3a feeding layer was applied to
enhance ES cell adherence. In the culture of iPSCs, L-wnt3a with
PD0325901 was able to maintain its pluripotent state. These
findings indicated that Wnt3a activated Wnt in the conventional way
to maintain its self-renewal.
Generally, chondrogenic differentiation of iPSCs may
be achieved in EBs by co-culturing with chondrocytes or adding
selected growth factors to the medium. Qu et al (25) studied whether co-culture with
primary chondrocytes induces human ES cells or iPSCs to
differentiate into chondrocyte lineages, and their results
demonstrated that co-culture of human ES cells or iPSCs with
primary chondrocytes induced specific chondrogenic differentiation.
Wei et al (26) used a
lentivirus to transduce iPSCs seeded in an alginate matrix with
TGF-β1 and then co-cultured these iPSCs with chondrocytes in
vitro. The authors observed an increased expression of
cartilage-associated genes, including collagen II, aggrecan and
cartilage oligomeric matrix protein, and decreased gene expression
of the degenerative cartilage marker, vascular endothelial growth
factor and the histological results also revealed a dense sulphated
extracellular matrix in the co-culture of TGF-β1-transfected iPSCs
with chondrocytes in the alginate matrix. Kuboth et al
(27) demonstrated the induction
of chondrogenic iPSCs differentiation by certain members of the
TGF-β family (BMP-2, TGF-β1). Their results demonstrated that the
number of Alcian blue-positive nodules and the expression of
cartilage marker molecule collagen type II increased following
application of BMP-2. In the present study, the combination of
BMP-2 and TGF-β1 was used to enhance chondrogenic differentiation
of iPSCs. The results revealed that extracellular matrix proteins
of cartilage tissue (Type II collagen) were found to be highly
expressed in EBs during culture. The gene expression analysis
demonstrated that the established iPSCs differentiated into
chondrocytes with increased expression of cartilage-associated
genes, including collagen II and aggrecan.
Koyama et al (28) examined the chondrogenic
differentiation capability of human iPSCs using a multistep culture
method consisting of EB formation, cell outgrowth from EBs,
monolayer culture of sprouted cells from EBs and 3-dimensional
pellet culture. The authors identified that the cells in pellets
exhibited a spherical morphology typical of chondrocytes and were
surrounded by extracellular matrix that contained acidic
proteoglycans following 2–3 weeks of pellet culture, and the
expression of type II collagen and aggrecan in pellets
progressively increased. Furthermore, BMP-2 treatment of
iPSC-derived MSC-like micromass cultures resulted in a phenotype
more typical of articular chondrocytes compared with the pellet
culture differentiation, characterized by the enrichment of
cartilage-specific type II collagen, decreased expression of type I
collagen as well as lack of chondrocyte hypertrophy (29). In the present study, cells were
cultivated in ‘hanging drops’ for two days and subsequently in
suspension in petri dishes for an additional three days. In the EB
outgrowths derived from iPSCs, numerous Alcian blue-stained regions
were found, indicating the presence of cartilaginous tissue.
In conclusion, the present study indicated that
iPSCs may be generated by applying the piggyBac plasmid (carrying
four or five genes) and the transposase mPB system. Particularly,
the transposase mPB system was used to successfully remove the
transposons-carried genes to obtain rat iPSCs without genetic
modification. Additionally, the present study also revealed that
the iPSCs were able to differentiate into chondrocytes in
vitro.
References
|
1
|
Roelofs AJ, Rocke JP and De Bari C:
Cell-based approaches to joint surface repair: a research
perspective. Osteoarthritis Cartilage. 21:892–900. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Teramura T, Onodera Y, Mihara T, et al:
Induction of mesenchymal progenitor cells with chondrogenic
property from mouse-induced pluripotent stem cells. Cell Reprogram.
12:249–261. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Takahashi K, Tanabe K, Ohnuki M, et al:
Induction of pluripotent stem cells from adult human fibroblasts by
defined factors. Cell. 131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Szabo E, Rampalli S, Risueno RM, et al:
Direct conversion of human fibroblasts to multilineage blood
progenitors. Nature. 468:521–526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Efe JA, Hilcove S, Kim J, et al:
Conversion of mouse fibroblasts into cardiomyocytes using a direct
reprogramming strategy. Nat Cell Biol. 13:215–222. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim J, Efe JA, Zhu S, et al: Direct
reprogramming of mouse fibroblasts to neural progenitors. Proc Natl
Acad Sci USA. 108:7838–7843. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jang YY, Collector MI, Baylin SB, Diehl AM
and Sharkis SJ: Hematopoietic stem cells convert into liver cells
within days without fusion. Nat Cell Biol. 6:532–539. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yu B, He ZY, You P, et al: Reprogramming
fibroblasts into bipotential hepatic stem cells by defined factors.
Cell Stem Cell. 13:328–340. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hanna J, Wernig M, Markoulaki S, et al:
Treatment of sickle cell anemia mouse model with iPS cells
generated from autologous skin. Science. 318:1920–1923. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wernig M, Zhao JP, Pruszak J, et al:
Neurons derived from reprogrammed fibroblasts functionally
integrate into the fetal brain and improve symptoms of rats with
Parkinson’s disease. Proc Natl Acad Sci USA. 105:5856–5861. 2008.
View Article : Google Scholar
|
|
11
|
Nishimura K, Nakagawa T, Ono K, et al:
Transplantation of mouse induced pluripotent stem cells into the
cochlea. Neuroreport. 20:1250–1254. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Okita K, Nakagawa M, Hyenjong H, Ichisaka
T and Yamanaka S: Generation of mouse induced pluripotent stem
cells without viral vectors. Science. 322:949–953. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yusa K, Rad R, Takeda J and Bradley A:
Generation of transgene-free induced pluripotent mouse stem cells
by the piggyBac transposon. Nat Methods. 6:363–369. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Peitz M, Pfannkuche K, Rajewsky K and
Edenhofer F: Ability of the hydrophobic FGF and basic TAT peptides
to promote cellular uptake of recombinant Cre recombinase: a tool
for efficient genetic engineering of mammalian genomes. Proc Natl
Acad Sci USA. 99:4489–4494. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kramer J, Hegert C, Guan K, et al:
Embryonic stem cell-derived chondrogenic differentiation in vitro:
activation by BMP-2 and BMP-4. Mech Dev. 92:193–205. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamanaka S: Strategies and new
developments in the generation of patient-specific pluripotent stem
cells. Cell Stem Cell. 1:39–49. 2007. View Article : Google Scholar
|
|
18
|
Sommer CA, Stadtfeld M, Murphy GJ, et al:
Induced pluripotent stem cell generation using a single lentiviral
stem cell cassette. Stem Cells. 27:543–549. 2009. View Article : Google Scholar
|
|
19
|
Maherali N, Ahfeldt T, Rigamonti A, et al:
A high-efficiency system for the generation and study of human
induced pluripotent stem cells. Cell Stem Cell. 3:340–345. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stadtfeld M, Nagaya M, Utikal J, Weir G
and Hochedlinger K: Induced pluripotent stem cells generated
without viral integration. Science. 322:945–949. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakagawa M, Koyanagi M, Tanabe K, et al:
Generation of induced pluripotent stem cells without Myc from mouse
and human fibroblasts. Nat Biotechnol. 26:101–106. 2008. View Article : Google Scholar
|
|
22
|
Okita K, Hong H, Takahashi K and Yamanaka
S: Generation of mouse-induced pluripotent stem cells with plasmid
vectors. Nat Protoc. 5:418–428. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ying QL, Wray J, Nichols J, et al: The
ground state of embryonic stem cell self-renewal. Nature.
453:519–523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawazoe S, Ikeda N, Miki K, et al:
Extrinsic factors derived from mouse embryonal carcinoma cell lines
maintain pluripotency of mouse embryonic stem cells through a novel
signal pathway. Dev Growth Differ. 51:81–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qu C, Puttonen KA, Lindeberg H, et al:
Chondrogenic differentiation of human pluripotent stem cells in
chondrocyte co-culture. Int J Biochem Cell Biol. 45:1802–1812.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wei Y, Zeng W, Wan R, et al: Chondrogenic
differentiation of induced pluripotent stem cells from
osteoarthritic chondrocytes in alginate matrix. Eur Cell Mater.
23:1–12. 2012.PubMed/NCBI
|
|
27
|
Kuboth S, Kramer J and Rohwedel J:
Chondrogenic differentiation in vitro of murine two-factor induced
pluripotent stem cells is comparable to murine embryonic stem
cells. Cells Tissues Organs. 196:481–489. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Koyama N, Miura M, Nakao K, et al: Human
induced pluripotent stem cells differentiated into chondrogenic
lineage via generation of mesenchymal progenitor cells. Stem Cells
Dev. 22:102–113. 2013. View Article : Google Scholar
|
|
29
|
Guzzo RM, Gibson J, Xu RH, Lee FY and
Drissi H: Efficient differentiation of human iPSC-derived
mesenchymal stem cells to chondroprogenitor cells. J Cell Biochem.
114:480–490. 2013. View Article : Google Scholar
|