Introduction
The dead end (Dnd1) gene was first characterized as
a gene required for germ-cell viability in a large-scale screening
of zebrafish (1–3). The mouse Dnd1 gene encodes two
protein isoforms: DND1-isoform α (DND1-α), 352 amino acids in
length, and DND1-isoform β (DND1-β), 340 amino acids in length.
DND1 is found in the fetal testis during the critical period when
testicular germ cell tumors (TGCTs) are hypothesized to develop in
mice (3). In a previous study,
DND1 was demonstrated to be critical for germ cell development
(3–5) and embryonic viability (6) in mice. The likely functional
inactivation of the Dnd1 gene in mice results in severe germ cell
depletion and development of TGCT.
DND1 is a highly conserved RNA-binding protein
important for maintaining the viability of primordial germ cells
(PGCs) in vertebrates, including zebrafish, frogs and mice
(1,3,7).
Knockdown of Dnd1 in zebrafish resulted in loss of PGC motile
behavior and subsequent PGC death (1). Dnd1 mutations have been directly
implicated as a heritable cause of spontaneous tumorigenesis in
mice. In 129 inbred mice, a point mutation in Dnd1 introduced a
stop codon, which resulted in truncated Dnd1, which may give rise
to PGC deficiency, testicular germ cell tumor development and
partial embryonic lethality (3).
Human Dnd1 maps to chromosome 5q31.3. Chromosomal deletions in
human 5q have been identified in germ-cell tumor tissues and
germ-cell tumor cell lines, and Dnd1 mutations have also been found
in TGCTs (8,9).
The mechanism by which the DND1 regulates cell
migration, germ cell survival and TGCT development has yet to be
elucidated. Thus far, studies have found that DND1-α is expressed
in early embryos and in the testis following birth, whereas DND1-β
is expressed in the germ cells of adult testis. DND1-α aids in
maintaining PGC viability and the loss of DND1-α results in
transformation of the PGC cells into embryonal carcinoma cells.
However, the role and mechanism of DND1-β in adult germ cells
remains elusive (1,6). In the present study, proteins that
interact with DND1-β were identified using a yeast two-hybrid
interaction strategy.
Materials and methods
Plasmid construction
The pcDNA3.1+ vector was purchased from Invitrogen
Life Technologies (Carlsbad, CA, USA) in order to construct the
pcDNA3.1-Dnd1-β expression plasmid. The Dnd1-β gene (GenBank ID:
BC034897.1) was amplified by reverse transcription polymerase chain
reaction (RT-PCR) from the mRNA of splenocytes derived from
C57BL/6J mice, obtained from Dr. Matin Angabin ( Texas University
MD Anderson Cancer Center, Houston, TX, USA). The primers were as
follows: Forward 5′-CAGATCTGATGGTCTCCCTCCCATCCCAA-3′ and reverse
5′-GGAATTCCTCACTGCTTAACCATAGTACC-3′ for Dnd1-β. Dnd1-β cDNA was
purified by Shanghai Biological Engineering Company (Shanghai,
China). For yeast two-hybrid screening, full-length Dnd1-β cDNA was
ligated in-frame with the GAL4 DNA-binding domain of the pDBLeu
vector, resulting in pDBLeu/Dnd1-β. For the immunoprecipitation
assay, full-length Dnd1-β cDNA was cloned into the mammalian
expression plasmid p cytomegalovirus (CMV)-Myc vector (Clontech
Laboratories, Mountain View, CA, USA), forming the Myc-tagged
Dnd1-β expression vector, pCMV-Myc-Dnd1-β. Full-length c-Jun cDNA
was inserted into a pCMV-human influenza haemagluttinin (HA) vector
(Clontech Laboratories), forming the HA-tagged c-Jun expression
vector, pCMV-HA-c-Jun. For the co-localization assay, full-length
Dnd1-β cDNA was cloned into a p enhanced green fluorescent protein
(EGFP)-c3 vector, forming the pEGFP-c3-Dnd1-β vector, and
full-length c-Jun cDNA was inserted into a pm red fluorescent
protein (RFP) vector Clontech Laboratories), forming the
pmRFP-c-Jun vector. The pGEX-4T-2 vector (GE Healthcare Life
Sciences Corp, Shanghai, China) was used to construct vectors
expressing glutathione S-transferase (GST)-DND1-β fusion proteins.
The cDNA fragment encoding full-length Dnd1-β was cloned in-frame
with respect to GST into the pGEX-4T-2 vector. The pQE-N3 Plasmid
(Qiagen, Hilden, Germany) was used to generate vectors expressing
polyhistidine (His)-tagged c-Jun fusion proteins. AP1-luc
(Stratagene, La Jolla, CA, USA) is a reporter plasmid encoding the
firefly luciferase gene driven by several copies of an AP-1
enhancer element. The pCMV-LacZ vector was constructed by fusing
the LacZ gene to pCMV-Myc (Qiagen, Hilden, Germany).
Cell culture and transient
transfection
The GC-1 cell line (CRL-2053; American Type Culture
Collection, Manassas, VA, USA). was cultured in Dulbecco’s modified
Eagle’s medium (DMEM; Sigma-Aldrich, St. Louis, MO, USA)
supplemented with 10% fetal bovine serum (Invitrogen Life
Technologies), penicillin (100 U/ml) and streptomycin (100 mg/ml)
in a 5% CO2 atmosphere at 37°C. The GC-1 cells were
plated in collagen-coated dishes and were harvested at 100%
confluency for co-immunoprecipitation analysis and
immunofluorescence. The GC-1 cells were transiently transfected
with plasmid constructs at 70% confluence using Lipofectamine 2000
(Invitrogen Life Technologies) according to the manufacturer’s
instructions. After 48 h transfection time, the cells were
harvested for the co-immunoprecipitation assay.
Yeast two-hybrid analysis
The ProQuest™ yeast two-hybrid system was obtained
from Invitrogen Life Technologies. A 10.5-day old mouse embryo cDNA
library (Invitrogen Life Technologies) cloned in-frame with the
GAL4 activation domain in the pPC86 vector (Invitrogen Life
Technologies) was used to screen for Dnd1-β-interacting clones. The
MaV203 yeast strain (Gibco-BRL, Carlsbad, CA, USA) was transformed
with pDBLeu-Dnd1-β and analyzed for basal expression activity, as
described in the manufacturer’s instructions. The bait-containing
MaV203 cells were subsequently transformed with the 10.5-day old
mouse embryo cDNA library and transformants were selected by
growing the cells in medium lacking leucine, tryptophan, uracil and
histidine (SD-leu-, Trp-, Ura-, His-) supplemented with 30 mM
3-amino-1,2,4-triazole (3-AT; Invitrogen Life Technologies). False
positive clones were eliminated by retransforming the prey DNA to
the original bait strain and positive clones were further verified
using an X-gal filter assay. The plasmids from positive clones were
sequenced and characterized as previously described (10).
GST pull-down assay
The GST pull-down assay was performed as previously
described (11). The BL21 (DE3)
cells were transformed with plasmids encoding GST, pGEX-4T-2/Dnd1-β
and pQE3/c-Jun fusion proteins. Overnight cultures were induced for
4 h with 0.1 mM isopropyl-β-d-thiogalactopyranoside (GE Healthcare
Life Sciences Corp), subsequent to which the bacteria were
harvested, resuspended in phosphate-buffered saline (PBS)
containing 1% (vol/vol) Triton X-100 and lysozyme (0.1 mg/ml), and
lysed by sonication. The bacterial extracts were clarified and
mixed with 50% (vol/vol) glutathione-Sepharose 4B ((GE Healthcare
Life Sciences Corp) slurry in PBS (50 μl per 1 ml of lysates).
After 15 min on ice, the beads were pelleted, washed and
resuspended in 0.4 ml portions of either cytoplasmic supernatant
from c-jun-infected cell lysates containing in
vitro-translated c-jun. After 30 min, the beads were pelleted
and washed. Complexes of GST-DND1-β bound to c-jun were eluted with
a small volume of 10 mM glutathione in 50 mM Tris (pH 8.0) and
analyzed by SDS-PAGE. [35S]methionine-labeled c-Jun from
the in vitro translation reactions was detected by
autoradiography (x-ray film) of the dried gel, while unlabeled
c-Jun from the lysates of infected cells was detected by Western
blot analysis with mouse monoclonal anti-c-Jun antibody (Abcam,
Cambridge, MA, USA).
Co-immunoprecipitation
GC-1 cells were co-transfected with pCMV-Myc-Dnd1-β
and pCMV-HA-c-jun. At 24 h after transfection, the GC-1 cells were
washed with PBS and homogenized in co-immunoprecipitation lysis
buffer [20 mM hydroxyethyl piperazineethanesulfonic acid, pH 7.4,
125 mM NaCl, 1% Triton X-100, 10 mM ethylene glycol tetraacetic
acid (EGTA), 2 mM Na3VO4, 50 mM NaF, 20 mM
ZnCl2, 10 mM sodium pyrophosphate, 1 mM dithiothreitol
and 1 mM phenylmethylsulfonyl fluoride]. Subsequently, 1X Complete
Protease Inhibitor mixture (Sigma-Aldrich, Shanghai, China) was
added prior to use. The supernatant fractions were collected and
incubated with mouse monoclonal anti-Myc antibody or rabbit
polyclonal anti-HA antibody (Abcam; 1:200) and GammaBind
Plus-Sepharose beads (Amersham Pharmacia Biotech) with agitation
overnight at 4°C as previously described (12). Co-precipitated proteins were
subjected to electrophoresis by 13% SDS-PAGE, and then analyzed by
western blotting using rabbit polyclonal anti-HA antibody,
monoclonal anti-Myc antibody or rabbit polyclonal anti-Dnd1-β
antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA).
Western blot analysis
Proteins were analyzed by SDS-PAGE and then
transferred to a nitrocellulose membrane. After >4 h blocking in
5% milk with TBST buffer (20 mM Tris-HCl, pH 7.6, 137 mM NaCl and
0.5% Tween 20), the blots were incubated with appropriate primary
antibodies at 4°C overnight. Subsequent to washing with TBST for 30
min, the membranes were incubated with 1:2,000 corresponding
alkaline phosphatase-conjugated secondary antibodies for 2 h.
Following another wash with TBST for 30 min, immunoreactivity was
visualized by enhanced chemiluminescence (ECL; Pierce
Biotechnology, Inc., Rockford, IL, USA). The images were captured
on X-ray film (Kodak, Rochester, NY, USA) and protein
quantification was determined using Gel Pro Analyzer 4 image
analysis software (Media Cybernetics, Inc., Rockville, MD,
USA).
Confocal microscopy
GC-1 cells were transiently co-transfected with
pEGFP-c3-Dnd1-β and pmRFP-c-Jun on glass slide chambers. Adherent
cells were washed three times with PBS containing 2 mM EGTA and
then fixed for 30 min in 3% paraformaldehyde in PBS with 2 mM EGTA.
Subsequent to two rinses with PBS, the cells were permeabilized for
2 min in 0.05% Triton X-100, followed by two additional rinses with
PBS. The chamber slides were observed with a Zeiss LSM 510 confocal
laser scanning microscope (LSM; Carl Zeiss AG, Oberkochen, Germany)
with a 100x oil (1.4-numerical aperture) immersion objective.
Briefly, GFP-DND1-β and RFP-c-Jun were observed in GFP and RFP
channels, respectively. The obtained images were merged to compare
the two signal patterns and the overlapping areas of the images
were measured using the overlay tool of LSM Image Browser software
(Ver. 4.2.0.121; Carl Zeiss AG). The images were converted to 8-bit
gray scale using Image J software (Ver. 1.43b) and the analysis was
performed with the co-localization-finder plug-in. Merged
compartments with >95% overlap were determined to be
co-localized.
Dual-luciferase reporter assay
At 24 h prior to transfection, 1×105
cells/well were plated in a 24-well plate at a density of 60–80%
confluence and transfected in the same medium using DMEM. The
luciferase reporter PCMV-HA-DND1-β containing full-length Dnd1-β
promoter and AP-1 luciferase reporter pAP-1-luc containing seven
AP-1 binding sites were used for Dnd1-β promoter and AP-1
transcriptional activity assays, respectively, in cells under
various conditions. The pRL-TK vector (Promega Corporation,
Madison, WI, USA) served as an internal control. In each
transfection, the cells were transfected with various combinations
of a luciferase reporter plasmid and the desired expression
vectors. The quantity of each expression vector was identical
unless specified otherwise. Total DNA quantity was maintained at a
constant level with pCMV or the relevant empty vectors. Transient
cell transfections with AP1-Luc, pCMV-LacZ and the indicated
expression vectors were performed using Lipofectamine 2000. At 24 h
after transfection, the cells were lysed and dual luciferase assays
were performed (Promega Corporation). Luciferase activity was
normalized to renilla luciferase activity. The results are
presented as the fold induction, which is the relative luciferase
activity of the treated cells over that of control cells. All
transfection experiments were conducted in triplicate wells and
repeated separately at least three times.
Statistical analysis
The significance of the differences among values was
determined by one-way analysis of variance followed by Dunnett’s
multiple comparison tests. An unpaired Student’s t-test was also
applied. P<0.05 was considered to indicate a statistically
significant difference. All data are presented as mean ± standard
error of the mean.
Results
Identification of c-Jun as a
DND1-β-binding protein using yeast two-hybrid assay
To identify the proteins that associate with DND1-β,
a yeast two-hybrid screening was performed with a 10.5-day old
mouse embryo cDNA library and the full-length Dnd1-β as the bait.
The transactivational activity of the GAL4-DND1-β fusion protein in
yeast was inhibited by 30 mM 3-AT. A total of 172 positive clones
were obtained in the first round of screening the mouse embryo cDNA
library, 10 positive clones were obtained in the second round and 8
positive clones were confirmed as positive in the third round
(Fig. 1). These eight sequences
were sequenced and compared with the sequences in GenBank; four of
the corresponding proteins were found to interact with DND1-β.
Through bioinformatic analysis, these four proteins were identified
as c-Jun, actinin alpha 1, nuclear receptor binding protein 1 and
chloride channels 2. As the association between c-Jun expression
and TGCT has been widely investigated, c-jun was selected for
further experiments.
 | Figure 1(A and B) Positive colonies from 10.5
day-old mouse embryo cDNA library screening. (C and D) Positive
control colonies pD+pP, MaV203 yeast cell colonies transformed with
bait vector pDBLeu and target vector pPC86; pD-Dnd1-β+pP, MaV203
cells transformed with bait plasmid pDBLeu-Dnd1-β and target vector
pPC86, serving as negative controls; (A) 2-1, 3-1, 4-1, 7-2, 12-1,
U2, U3 and (B) 11-4, positive colonies obtained from
MaV203-pDBLeu-Dnd1-β transformed with mouse embryo cDNA library and
subsequently screening of the transformants. (C and D) Assessment
of self-activation of the eight positive library plasmids and
confirmation of the specificity of the interaction of the putative
eight targets with pDBLeu-Dnd1-β. (C) Analysis of self-activation
of the eight positive library plasmids: C and D, positive control
colonies; pD+pP and pD-Dnd1-β+pP, negative controls as previously
described; 1–8, MaV203 cells cotransformed with the eight positive
library plasmids with pDBLeu, respectively. (D) Verification of the
interaction of the eight positive targets with pDBLeu-Dnd1-β in
yeast: C and D, positive control colonies; pD+pP and pD-Dnd1-β+pP,
negative controls; 2-1, 3-1, 4-1, 7-2, 11-4, 12-1, U2 and U3,
positive colonies. (E) Identification of the positive colonies by
polymerase chain reaction. Lane M, DNA marker (DL2000); Lane 1,
pPC86-2-1; Lane 2, pPC86-3-1; Lane 3, pPC86-4-1; Lane 4, pPC86-7-2;
Lane 5, pPC86-11-4; Lane 6, pPC86-12-1; Lane 7, pPC86-U3-1; Lane 8,
pPC86-U3-2. Dnd, dead end. |
DND1-β interacts with c-Jun in GST
pull-down assay
The DND1-β-c-Jun interaction is possibly indirect,
as other protein factors in the whole cell extract may mediate the
interaction, for example, by acting as ‘bridging’ factors. To
determine whether DND1-β and c-Jun interact in mammalian cells, GST
pull-down assays were performed using lysates from GC-1 cells. GST,
GST fusion proteins and His fusion proteins were expressed and
purified. The results revealed that c-Jun was pulled down by
GST-fused DND1-β but not by GST alone, indicating that DND1-β and
c-Jun specifically and directly interact in vitro (Fig. 2A).
 | Figure 2DND1-β interacted with full-length
c-Jun (A) in vitro and (B and C) in vivo. (A) GST
pull-down. GST-DND1-β: GST-DND1-β incubated with purified
His-c-Jun. Input, purified His-c-Jun; GST, GST incubated with
purified His-c-Jun. (B and C) Co-immunoprecipitation assay.
Transfected cell lysates were prepared and immunoprecipitated with
(B) monoclonal anti-DND1-β antibodies and (C) anti-c-Jun antibodies
followed by immunoblotting with an anti-c-Jun and anti-DND1-β
antibody, respectively. DND, dead end; GST, glutathione
S-transferase His, histidine; IgG, immunoglobulin G; IP,
immunoprecipitation; IB, immunoblot; HC, heavy chain. |
DND1-β and c-Jun are
co-immunoprecipitated in GC-1 cells
To determine whether endogenous DND1-β interacts
with c-Jun in mammalian cells, a co-immunoprecipitation experiment
was conducted in GC-1 cells with anti-DND1-β, anti-c-Jun and
non-specific rabbit immunoglobulin G antibodies (Fig. 2B and C). The lysates of GC-1 cells
were immunoprecipitated with an anti-DND1-β antibody followed by
immunoblotting with an anti-c-Jun antibody. The result demonstrated
that c-Jun was precipitated by DND1-β. When the cell lysates were
subjected to immunoprecipitation with an anti-c-Jun antibody,
DND1-β was consistently detected in the precipitant. These data
suggested that DND1-β interacted with c-Jun in GC-1 cells.
Dnd1-β co-localizes with c-Jun in GC-1
cells nuclei
As a tight association between DND1-β and c-Jun had
been detected in immunoprecipitation and GST pull-down experiments,
the next aim was to investigate whether these proteins were present
in the same region in cells. Images captured by confocal laser
scanning microscopy revealed that GFP-tagged DND1-β and RFP-tagged
c-Jun were mainly localized in the nuclei of cells (Fig. 3). Following overlay, yellow
co-localized signals were clearly observed (Fig. 3). White areas in Fig. 3F indicate the co-localization
between DND1-β and c-Jun with the following correlation
coefficients: Pearson’s Rr=0.0193; overlap R=0.997948.
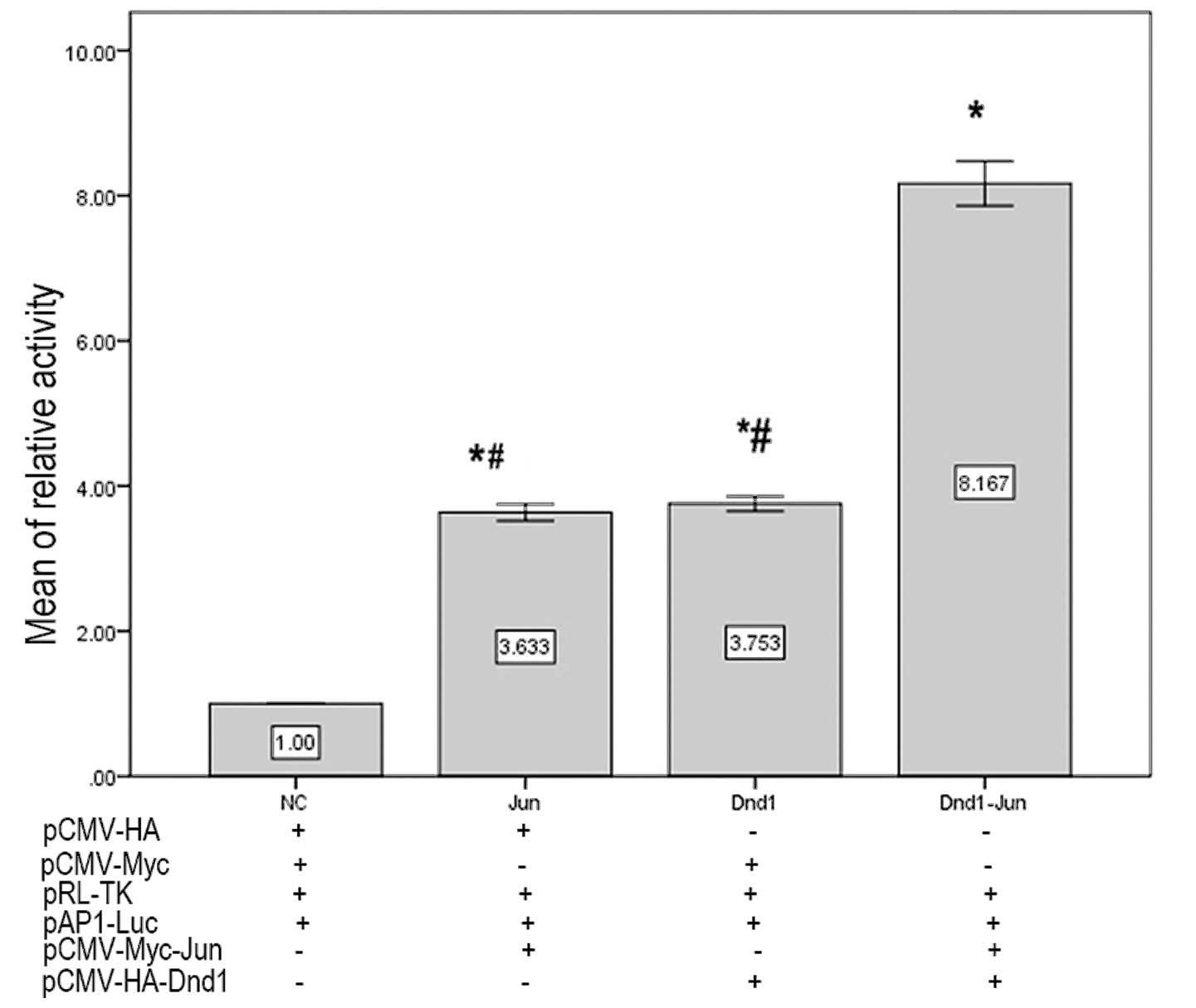
Interaction of DND1-β and c-Jun
stimulates activation of AP-1 transcription
To investigate the physiological relevance of the
DND1-β-c-Jun interaction, the effect of DND1-β on AP-1
transcriptional activity was analyzed. Either DND1-β or c-Jun alone
significantly stimulated luciferase expression (Fig. 4) in GC-1 cells compared with the
negative control, but co-transfection with pCMV-Myc-Dnd1-β and
pCMV-HA-c-Jun significantly stimulated the luciferase activity
further than either individual protein (P<0.05). These results
suggested that the interaction between DND1-β and c-Jun activated
AP-1 transcription.
Discussion
DND1 has been reported to be the first protein with
an RNA recognition motif (RRM) and has been directly implicated as
a heritable cause of spontaneous tumorigenesis (3). To understand in greater detail the
mechanism by which DND1-β influences protein expression levels in
mice, DND1-β was identified as a c-jun-binding protein by yeast
two-hybrid screening. The data from in vitro and in
vivo experiments revealed that DND1-β and c-jun directly
interact, and the biological relevance of this interaction was
investigated.
AP-1 is a homo- or heterodimeric transcription
factor protein composed of other proteins belonging to the Jun,
Fos, activating transcription factor and Jun dimerization protein
subfamilies (13,14). AP-1 was found at the receiving end
of multiple signaling pathways and regulates gene expression in
response to a variety of stimuli, including cytokines, growth
factors, stress, as well as bacterial and viral infections. AP-1
controls a broad range of biological processes, including
proliferation, transformation, cell differentiation, cell migration
and apoptosis (15–18). AP-1 activity is induced by multiple
environmental insults and physiological stimuli, which activate
mitogen-activated protein kinase cascades (19,20).
AP-1 activation may result in either induction or prevention of
apoptosis, depending on the tissue involved and on the
developmental stage of the animal (21–25).
c-Jun is the most extensively investigated AP-1
protein, and is involved in numerous cell functions, including
proliferation, apoptosis, survival, tumorigenesis and tissue
morphogenesis. The majority of these activities rely on the
function of c-Jun as a transcription factor. c-Jun is involved in
spermatogenesis, but is also a proto-oncogene. c-Jun is
stage-specific with expression levels increasing during active
spermatogenesis (26). Following
verification of the changes in c-Jun mRNA and protein expression
levels at different stages of spermatogenesis in mice, scientists
have demonstrated that c-Jun was involved in the transcriptional
events regulating the proliferation and differentiation of
spermatogenic cells during different stages in the seminiferous
epithelium cycle (27–29). c-Jun does not usually act alone;
rather, c-Jun physically interacts with other transcription
factors, allowing signal integration on promoter DNA for
combinatorial transcriptional regulation of gene expression. This
type of interaction expands the diversity of c-Jun effects on gene
regulation. For instance, interaction with other proteins may
promote c-Jun localization to a non-AP-1 site to regulate gene
expression.
In the present study, c-Jun was identified as a
novel binding partner of DND1-β. In vivo binding of DND1-β
with c-Jun was identified by immunoprecipitation and
immunofluorescence, and in vitro interaction between DND1-β
and c-Jun was demonstrated by GST pull-down assays.
The dual luciferase reporter assay is a system that
employs two luciferase proteins, firefly and renilla, which are
assayed independently in one reaction mixture (30,31).
AP-1 activity was measured with the dual luciferase reporter gene
assay. DND1 or c-Jun alone were found to activate the luciferase
reporter to similar extents; however, the two proteins together
exerted an effect marginally greater than the sum of the individual
effects. This suggested that the interaction between DND1-β and
c-Jun subsequently resulted in the activation of AP-1.
In conclusion, DND1-β, which was previously
described as an RNA-binding protein and involved in germ cell
development, embryonic viability and tumorigenesis, was
demonstrated to also interact specifically with the c-Jun
transcription factor. The consequence of this interaction is AP-1
activation, which may be important in germ cell development,
embryonic viability and tumorigenesis, including that which occurs
in TGCT.
Acknowledgements
This study was supported by grants from the Natural
Science Foundation of China (nos. 81071628 and 30971270) and the
Hunan Provincial Natural Science Foundation of China (no.
11jj2013).
References
|
1
|
Weidinger G, Stebler J, Slanchev K, et al:
Dead end, a novel vertebrate germ plasm component, is required for
zebrafish primordial germ cell migration and survival. Curr Biol.
13:1429–1434. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sakurai T, Iguchi T, Moriwaki K and
Noguchi M: The ter mutation first causes primordial germ cell
deficiency in ter/ter mouse embryos at 8 days of gestation. Dev
Growth Differ. 37:293–302. 1995. View Article : Google Scholar
|
|
3
|
Youngren KK, Coveney D, Peng X, et al: The
Ter mutation in the dead end gene causes germ cell loss and
testicular germ cell tumours. Nature. 435:360–364. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu R, Bhattacharya C and Matin A: The
role of dead-end in germ-cell tumor development. Ann N Y Acad Sci.
1120:181–186. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cook MS, Munger SC, Nadeau JH and Capel B:
Regulation of male germ cell cycle arrest and differentiation by
DND1 is modulated by genetic background. Development. 138:23–32.
2011. View Article : Google Scholar :
|
|
6
|
Bhattacharya C, Aggarwal S, Zhu R, et al:
The mouse dead-end gene isoform alpha is necessary for germ cell
and embryonic viability. Biochem Biophys Res Commun. 355:194–199.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Horvay K, Claussen M, Katzer M, Landgrebe
J and Pieler T: Xenopus dead end mRNA is a localized maternal
determinant that serves a conserved function in germ cell
development. Dev Biol. 291:1–11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng HQ, Bailey D, Bronson D, Goss PE and
Hogg D: Loss of heterozygosity of tumor suppressor genes in testis
cancer. Cancer Res. 55:2871–2875. 1995.PubMed/NCBI
|
|
9
|
McIntyre A, Summersgill B, Jafer O, et al:
Defining minimum genomic regions of imbalance involved in
testicular germ cell tumors of adolescents and adults through
genome wide microarray analysis of cDNA clones. Oncogene.
23:9142–9147. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Muller EE, Fayemiwo SA and Lewis DA:
Characterization of a novel beta-lactamase-producing plasmid in
Neisseria Gonorrhoeae: sequence analysis and molecular typing of
host Gonococci. J Antimicrob Chemother. 66:1514–1517. 2011.
View Article : Google Scholar
|
|
11
|
Tian Y, Ke S, Chen M and Sheng T:
Interactions between the aryl hydrocarbon receptor and P-TEFb.
Sequential recruitment of transcription factors and differential
phosphorylation of C-terminal domain of RNA polymerase II at cyp1a1
promoter. J Biol Chem. 278:44041–44048. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lauffart B, Howell SJ, Tasch JE, Cowell JK
and Still IH: Interaction of the transforming acidic coiled-coil 1
(TACC1) protein with ch-TOG and GAS41/NuBI1 suggests multiple
TACC1-containing protein complexes in human cells. Biochem J.
363(Pt 1): 195–200. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wagner EF: AP-1 - Introductory remarks.
Oncogene. 20:2334–2335. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shaulian E and Karin M: AP-1 in cell
proliferation and survival. Oncogene. 20:2390–2400. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Leppä S and Bohmann D: Diverse functions
of JNK signaling and c-Jun in stress response and apoptosis.
Oncogene. 18:6158–6162. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnson GL and Lapadat R:
Mitogen-activated protein kinase pathways mediated by ERK, JNK, and
p38 protein kinases. Science. 298:1911–1912. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shaulian E and Karin M: AP-1 as a
regulator of cell life and death. Nat Cell Biol. 4:E131–E136. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ameyar M, Wisniewska M and Weitzman JB: A
role for AP-1 in apoptosis: the case for and against. Biochimie.
85:747–752. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maeda S and Karin M: Oncogene at last -
c-Jun promotes liver cancer in mice. Cancer Cell. 3:102–104. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jochum W, Passegué E and Wagner EF: AP-1
in mouse development and tumorigenesis. Oncogene. 20:2401–2412.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Colotta F, Polentarutti N, Sironi M and
Mantovani A: Expression and involvement of c-fos and c-jun
protooncogenes in programmed cell death induced by growth factor
deprivation in lymphoid cell lines. J Biol Chem. 267:18278–18283.
1992.PubMed/NCBI
|
|
22
|
Hilberg F, Aguzzi A, Howells N and Wagner
EF: c-jun is essential for normal mouse development and
hepatogenesis. Nature. 365:179–181. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Smeyne RJ, Vendrell M, Hayward M, et al:
Continuous c-fos expression precedes programmed cell death in vivo.
Nature. 363:166–169. 1993. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Estus S, Zaks WJ, Freeman RS, Gruda M,
Bravo R and Johnson EM Jr: Altered gene expression in neurons
during programmed cell death: identification of c-jun as necessary
for neuronal apoptosis. J Cell Biol. 127(6 Pt 1): 1717–1727. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hafezi F, Steinbach JP, Marti A, et al:
The absence of c-fos prevents light-induced apoptotic cell death of
photoreceptors in retinal degeneration in vivo. Nat Med. 3:346–349.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cohen DR, Vandermark SE, McGovern JD and
Bradley MP: Transcriptional regulation in the testis: a role for
transcription factor AP-1 complexes at various stages of
spermatogenesis. Oncogene. 8:443–455. 1993.PubMed/NCBI
|
|
27
|
Shalini S and Bansal MP: Role of selenium
in spermatogenesis: differential expression of cjun and cfos in
tubular cells of mice testis. Mol Cell Biochem. 292:27–38. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wolfes H, Kogawa K, Millette CF and Cooper
GM: Specific expression of nuclear proto-oncogenes before entry
into meiotic prophase of spermatogenesis. Science. 245:740–743.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chieffi P, Angelini F and Pierantoni R:
Proto-oncogene activity in the testis of the lizard, Podarcis s.
sicula, during the annual reproductive cycle. Gen Comp Endocrinol.
108:173–181. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Grentzmann G, Ingram JA, Kelly PJ,
Gesteland RF and Atkins JF: A dual-luciferase reporter system for
studying recoding signals. RNA. 4:479–486. 1998.PubMed/NCBI
|
|
31
|
Lee JY, Kim S, Hwang do W, et al:
Development of a dual-luciferase reporter system for in vivo
visualization of MicroRNA biogenesis and posttranscriptional
regulation. J Nucl Med. 49:285–294. 2008. View Article : Google Scholar : PubMed/NCBI
|