Introduction
Abnormalities in tooth number affect approximately
20% of the population (1).
Oligodontia is defined as the congenital absence of six or more
permanent teeth, excluding the third molar. Tooth loss may appear
either as a feature of multi-organ syndromes or as a non-syndromic
isolated trait. Individuals with missing teeth have numerous
problems with aesthetics, phonetics and mastication. The
development of dentition is a notable process that encompasses a
complex series of epithelial-mesenchymal interactions involving
growth factors, transcription factors, signal receptors and other
soluble morphogens (2). Any
disturbance in this tightly balanced process may result in tooth
agenesis or other dental defects. Numerous genes that underlie
dental defects have been identified; however, the occurrence of
non-syndromic cases remains poorly understood. Previous studies
have demonstrated that Msh homeobox 1 (MSX1), paired box
gene 9 (PAX9), ectodysplasin-A (EDA) and axis
inhibition protein 2 (AXIN2) are key regulators of tooth
development. Lammi et al suggested that oligodontia may be
caused by mutations of the AXIN2 gene (3), which is localised on chromosome
17q21-q25. AXIN2 is known as an intracellular antagonist of
Wnt signalling that is expressed in the dental mesenchyme,
odontoblasts and enamel knots (4).
In humans, mutations in AXIN2 cause tooth agenesis affecting
permanent teeth, predominantly including permanent molars, lower
incisors and upper lateral incisors (5,6). The
objective of the present study was to identify the AXIN2
mutations responsible for oligodontia in a family with
non-syndromic oligodontia. In addition, we attempted to explore
genotype-phenotype correlations that could improve our current
understanding of different mutations based on the pattern of tooth
agenesis.
Materials and methods
Patients and controls
The female proband was a patient of the Department
of Stomatology at the First People’s Hospital of Lianyungang City,
Lianyungang, China. A pedigree of this family was constructed by
extended interviews. In this study, all individuals presented with
normal physical development and normal intelligence, and the
clinical examination for other ectodermal abnormalities of the
nails, hair, skin and sweat glands did not reveal any defects.
Thus, we suggested that the family exhibited non-syndromic
oligodontia. Retrospective data were reviewed and the diagnosis of
oligodontia was verified by panoramic dental radiographs for all
available family members. Eight members of the family were studied,
with three members being affected, one of which was deceased, and
five unaffected. Furthermore, 60 unrelated individuals (of
different ages and genders) who were not affected with tooth
agenesis (excluding third molars) were used as controls. The study
was approved by the Institutional Review Board as well as the
Ethics Committee of the First People’s Hospital of Lianyungang
City.
DNA extraction and polymerase chain
reaction (PCR) of candidate genes
Peripheral blood samples from all members of the
family and controls were collected from the Department of
Stomatology at the First People’s Hospital of Lianyungang City.
Genomic DNA was extracted from these samples with the QIAamp DNA
blood mini kit (Qiagen, New York City, NY, USA). To screen for
putative mutations, two exons of MSX1 (GenBank accession
number M97676), four exons of PAX9 (GenBank accession number
AJ238381), ten exons of AXIN2 (GenBank accession number
AE006463) and eight exons of EDA (GenBank accession number
U59228), as well as their exon-intron boundaries, were amplified by
PCR with the use of primers that were previously reported (7) or designed using the Primer 3 online
application (http://frodo.wi.mit.edu/primer3/input.htm). The primer
sequences and optimal annealing temperature for each primer pair
are shown in Tables I–IV. The PCR amplifications used the
GC-rich PCR system (Roche, Nutley, NJ, USA) and the PrimeSTAR PCR
system (Takara Bio, Inc., Otsu, Japan). The amplified fragments of
AXIN2 from individuals of the affected family and controls
were gel-purified with the MinElute gel extraction kit (Qiagen)
according to the manufacturer’s instructions. The sequencing
analyses were performed with an ABI BigDye™ terminator cycle
sequencing ready reaction kit with AmpliTaq DNA polymerase on an
ABI PRISM® 377 XL DNA sequencer (Applied Biosystems Life
Technologies, Carlsbad, CA, USA). Finally, the mutations and
polymorphisms in AXIN2 at the genomic and protein levels
were analysed using MegAlign 5.01 software (DNASTAR, Inc., Madison,
WI, USA), Polyphen-2 software (Harvard University, Cambridge, MA,
USA) and exonic splicing enhancer (ESE) Finder 3.0 software (Cold
Spring Harbor Laboratory, Cold Spring Harbor, NY, USA).
 | Table IMSX1 primer sequences. |
Table I
MSX1 primer sequences.
| Region | Forward primer
(5′-3′) | Reverse primer
(5′-3′) | Size (bp) | Annealing temperature
(°C) |
|---|
| Exon 1 |
GGCTGCTGACATGACTTCTTTGC |
TTGGAACCTTCTTCCTGGGTG | 642 | 65 |
| Exon 2 |
CCAGAAGCAGTACCTGTCCAT |
TCAGGGATCAGACTTCGGAGA | 506 | 65 |
 | Table IVAXIN2 primer sequences. |
Table IV
AXIN2 primer sequences.
| Region | Forward primer
(5′-3′) | Reverse primer | Size (bp) | Annealing
temperature (°C) |
|---|
| Exon 2 |
AGAGAGACCACGCCGATTG |
CATGGCCAGCAGTCCTAAC | 1017 | 64 |
| Exon 3 |
CTTTCTGCCCAGGTGAGC |
CCCACCAAACTGATGTCC | 424 | 61 |
| Exon 4 |
AAGCCAAGTTTCCATAGATGA |
GTTCTTTTCTCTCCCTTTGCC | 471 | 62 |
| Exon 5 |
CGCCTTGCCATCCACTC |
GCACATGCGCACACAGC | 443 | 64 |
| Exon 6 |
ATGGGTTGCGTGTGGGT |
GGGCTGGTGACACGAAAG | 883 | 63 |
| Exon 7 |
CGCATTACAGGCATTTAGTT |
AAGATGGCAGGAGCAAATAG | 499 | 62 |
| Exon 8 |
GACTATTTGCTCCTGCCATC |
GACTTGCCTCACAGATCCTG | 674 | 62 |
| Exon 9 |
CAGGGTCTTGGTTGGGTCT |
CAGGACATGGATGGCAACA | 408 | 64 |
| Exon 10 |
CATGTGGGGTTGGACTGTG |
CGGCTGCTGCTTCGTTATT | 466 | 66 |
| Exon 11 |
AGTCCCAGCTGCCGTCTTA |
CCACTGGCCGATTCTTCC | 431 | 65 |
Results
Clinical examination
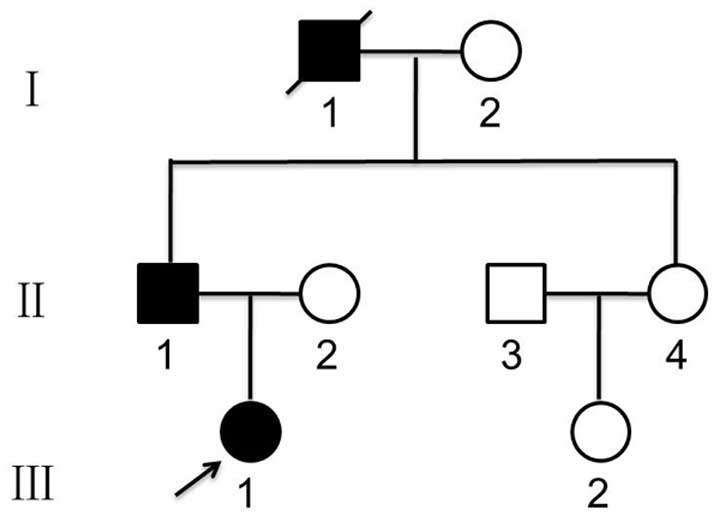
We studied a family in which oligodontia was
segregating in an autosomal-dominant manner. A pedigree of this
family was constructed by extended interviews (Fig. 1). Retrospective data were reviewed,
and the diagnosis of oligodontia was verified based on panoramic
dental radiographs of all available family members. Clinical and
radiographic examinations revealed that the proband’s father (II-1)
was missing 12 permanent teeth: one permanent maxillary lateral
incisor, two permanent maxillary canines, three permanent maxillary
premolars, two permanent mandibular central incisors, two permanent
mandibular lateral incisors and two permanent mandibular premolars
(Fig. 2). The proband (III-1) was
missing 12 permanent teeth: two permanent maxillary lateral
incisors, two permanent maxillary canines, four permanent maxillary
premolars and four permanent mandibular premolars (Fig. 3).
Mutation analysis
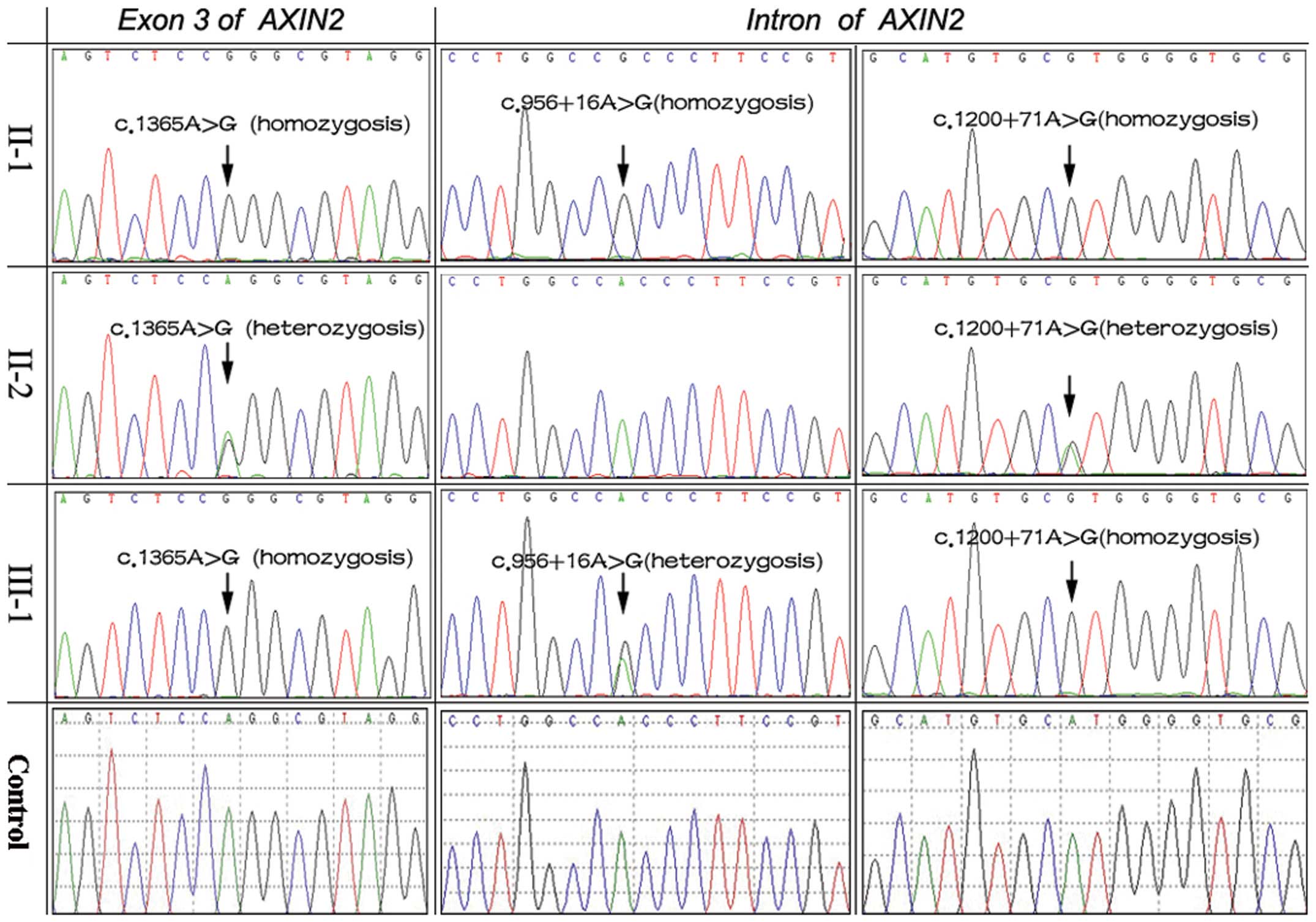
DNA sequencing of the AXIN2 gene revealed
three mutations in the two patients with oligodontia: a homozygotic
silent mutation c.1365A>G (p.Pro455=) in exon 3, two mutations
c.956+16A>G (II-1: homozygosis; III-1: heterozygosis) and
c.1200+71A>G (homozygosis) in the intron. In addition, the
heterozygotic mutations c.1365A>G and c.1200+71A>G were
identified in the proband’s mother (II-2; Fig. 4). The three mutations were thought
to be known polymorphisms (rs9915936, rs35285779 and rs8078753)
according to a bioinformatics analysis. Polyphen-2 software was
used to predict the possible impact of the amino acid substitution
on the structure and function of the AXIN2 protein. The
change was predicted to be benign. We also used ESE Finder 3.0
software to predict whether the change would alter exonic splicing
enhancers. The A to G nucleotide change did not affect any
predicted exonic splicing enhancer site. No mutations were detected
in EDA, PAX9 or MSX1. In addition, the family
members of other descent and the unrelated healthy controls had no
mutations in EDA, MSX1, PAX9 or
AXIN2.
Discussion
Oligodontia appears as a feature of multi-organ
syndromes as well as a non-syndromic isolated character. We
analysed a family in which oligodontia was segregating in an
autosomal-dominant manner in order to define the clinical features
of oligodontia and to localise the gene locus behind this anomaly.
Our clinical examinations and interviews of the kindred revealed
that the affected members were missing more than six teeth but had
no other systemic abnormality. These individuals were diagnosed
with non-syndromic oligodontia.
Numerous genes underlying dental defects have been
identified (8–10); however, the occurrence of
non-syndromic oligodontia remains poorly understood. Based on
results from familial studies, Lammi et al suggested that
oligodontia may be caused by mutations of the AXIN2 gene
(3). The protein product of
AXIN2 is a negative regulator of the canonical Wnt pathway
and suppresses signal transduction by promoting the degradation of
β-catenin (11,12). The inactivating mutations described
in AXIN2 that lead to oligodontia and higher susceptibility
to colon cancer affected incisor development in 11 out of 12 cases.
Mostowska et al suggested that AXIN2 polymorphic
variants may be associated with both hypodontia and oligodontia
(13). This study from Poland
suggested that two AXIN2 variants were in strong linkage
disequilibrium with each other: the silent mutation c.2062C>T
and the intronic variant c.956+16A>G. Linkage disequilibrium
refers to the non-random association of linked genes. This is the
tendency of the alleles of two separate but already linked loci to
be found together more frequently than usual. c.2062C>T was
predicted to disrupt exonic splicing enhancer sequences, and
although a synonymous change, it could contribute to the tooth
agenesis phenotype. The c.956+16A>G mutation was also suggested
to exert a possible effect on splicing as it creates an additional
donor-splicing site within the sequence of exon 2. In our study,
three AXIN2 variations (including the c.956+16A>G
mutation) were identified in the proband and her father, which
confirm the significance of AXIN2 in tooth development.
Previous studies have suggested that different tooth
types are regulated by independent gene expression (14,15).
Numerous Wnt genes are expressed in developing teeth, and changes
in their expression may be one of the factors determining tooth
agenesis (16). There is evidence
that genes associated with the Wnt pathway are differentially
expressed in molars as opposed to incisors. Studies in mice
targeting lymphoid enhancer-binding factor 1 (LEF1) function, a
transcription factor that can be activated by Wnt proteins,
revealed that transgene molar expression of the LEF1 promoter
during molar development was observed only in E12.5 embryos and was
absent in E13.5 embryos. Conversely, LEF1 promoter expression
during incisor tooth development persisted from E12.5 to E17.5
(17). It is reasonable to
hypothesise that AXIN2 alleles affect incisors more often
due to the persistent expression of LEF1 in these developing teeth
compared with molars. In the family studied, II-1 was missing one
permanent maxillary lateral incisor, two permanent maxillary
canines, three permanent maxillary premolars, two permanent
mandibular central incisors, two permanent mandibular lateral
incisors and two permanent mandibular premolars, while III-1 was
missing two permanent maxillary lateral incisors, two permanent
maxillary canines, four permanent maxillary premolars and four
permanent mandibular premolars. Neither of the patients had missing
molars (with the exception of the third molars). These results may
therefore suggest that multiple AXIN2 variants could
contribute to sporadic forms of common incisor agenesis in humans,
which is in agreement with previous studies on the frequency of
tooth loss with AXIN2 mutations (13).
The same type of mutation (c.1365A>G and
c.1200+71A>G) but in heterozygotic form was identified in the
proband’s mother. Possible reasons for this may be gene
polymorphism, multiple gene factors and environmental factors.
First, the rationale for performing the joint effect analysis of
the studied single nucleotide polymorphism (SNP) is that, under a
mixture of various pollutants, the contribution of polymorphisms in
various metabolising enzymes could exert an additive effect in
disease susceptibility that is only possible to identify when
considering the SNPs in combination. This analysis revealed that
the combination of the c.956+16A>G, c.1365A>G and
c.1200+71A>G homozygotic mutations may increase the oligodontia
risk compared with heterozygotic mutations and polymorphisms. The
identification of the causative genetic variants involved in the
phenotypes of interest remain a difficult task. In light of the
non-independence of mammalian dental traits (18), adequate samples of various
pedigrees and references to established databases are required to
determine the association of the genotype and phenotype in
oligodontia. Although the study subjects come from the same
geographical area, notable aspects concerning the source of cases
and controls have to be taken into account. Our findings may imply
that c.956+16A>G (rs35285779, homozygosis), c.1365A>G
(p.Pro455=, rs9915936, homozygosis) and c.1200+71A>G (rs8078753,
homozygosis) mutations are risk factors for oligodontia in the
Chinese population. However, due to the small sample size and the
limited number of studies, our results should be interpreted with
caution.
The second explanation could be the multiple gene
factors. The development of dentition is a notable process that
encompasses a complex series of epithelial-mesenchymal interactions
involving growth factors, transcription factors, signal receptors
and other soluble morphogens. To date, over 300 genes have been
identified as being involved in tooth development (19). This fact highlights the
significance of other presently unknown genes and developmental
factors in tooth development and in the etiology of dental
anomalies. Tooth agenesis may be related to perturbations of the
complex intracellular and intercellular networks that link tissues
and organ systems (20–22). Hence, the pathogenesis of tooth
agenesis may be more heterogeneous than was previously considered.
We conclude that additional genes are responsible for tooth
agenesis, and it is conceivable that a number of these genes also
belong to the large group of genes that are known to be expressed
during tooth morphogenesis.
The third explanation could be environmental
factors. Due to its peculiar properties, during tooth development,
variations in the metabolic status of the individual (23, 24)
may play a role. Our study proved that none of the polymorphisms
(c.956+16A>G, c.1365A>G and c.1200+71A>G) appear to lie on
xenobiotic metabolising genes. At this point, it is essential to
take into account the fact that isolated oligodontia is a
multifactorial process, and genetic-determined suboptimal
xenobiotic metabolising machinery could partly explain not all but
certain oligodontia cases, which have developed under certain
exposure conditions. The human body is a complex open system.
Whether an individual will be affected following contact with
harmful factors not only depends on environmental response genes
and environmental factors, but also on the complex regulatory
processes of various cells, tissues and organs, which cause changes
in gene expression. Hence, tooth anomalies may reflect
environmental and systemic disturbances. A number of environmental
factors (25,26) including irradiation,
chemotherapeutic agents and dioxin are capable of arresting tooth
development. The dental phenotype could be similar for various
genotypes, adverse environments or systemic anomalies, and thus the
search for etiological factors should be extremely accurate. These
factors may be pivotal in the strategy of molecular screening.
Taken together, our results may explain why the polymorphic locus
can exhibit the typical edentulous phenotype, and why the same
mutation but in heterozygotic form was identified in the proband’s
mother, and could serve as a starting point for future studies in
which pollutant activity and genotype influence could be
studied.
Although significant progress has been made in our
understanding of the molecular mechanisms underlying oligodontia
(27–29), a detailed picture of the genetic
background of this syndrome is still lacking. In this study, two
individuals in a family affected by the c.956+16A>G,
c.1365A>G and c.1200+71A>G homozygotic mutations lacked
permanent teeth, which may indicate that these polymorphisms are a
risk factor for oligodontia in the Chinese Han population. We
provide further evidence suggesting a role of AXIN2 in tooth
agenesis, and also confirm the significance of the Wnt pathway in
tooth development. We suggest that other factors, as yet unknown,
may be associated with this common developmental anomaly. Although
only performed on a few cases, this study on the subject of
oligodontia may contribute to our knowledge of the condition.
However, to understand the exact function of AXIN2 in
odontogenesis requires further detailed analysis of each stage of
this process.
Acknowledgements
The present study was supported by the Municipal
Bureau of Science and Technology of Lianyungang city (grant no.
SH1332). The authors are grateful to the patients and their family
members for their kind cooperation and participation.
References
|
1
|
Matalova E, Fleischmannova J, Sharpe PT
and Tucker AS: Tooth agenesis: from molecular genetics to molecular
dentistry. J Dent Res. 87:617–623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cudney SM and Vieira AR: Molecular factors
resulting in tooth agenesis and contemporary approaches for
regeneration: a review. Eur Arch Paediatr Dent. 13:297–304. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lammi L, Arte S, Somer M, et al: Mutations
in AXIN2 cause familial tooth agenesis and predispose to colorectal
cancer. Am J Hum Genet. 74:1043–1050. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Swinnen S, Bailleul-Forestier I, Arte S,
et al: Investigating the etiology of multiple tooth agenesis in
three sisters with severe oligodontia. Orthod Craniofac Res.
11:24–31. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jia S, Zhou J, Gao Y, et al: Roles of Bmp4
during tooth morphogenesis and sequential tooth formation.
Development. 140:423–432. 2013. View Article : Google Scholar :
|
|
6
|
Bailleul-Forestier I, Molla M, Verloes A
and Berdal A: The genetic basis of inherited anomalies of the
teeth: Part 1. Clinical and molecular aspects of non-syndromic
dental disorders. Eur J Med Genet. 51:273–291. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J, Jian F, Chen J, et al: Sequence
analysis of PAX9, MSX1 and AXIN2 genes in a Chinese oligodontia
family. Arch Oral Biol. 56:1027–1034. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bergendal B, Klar J, Stecksén-Blicks C, et
al: Isolated oligodontia associated with mutations in EDARADD,
AXIN2, MSX1, and PAX9 genes. Am J Med Genet A. 155:1616–1622. 2011.
View Article : Google Scholar
|
|
9
|
Gunbay T, Koyuncu BO, Sipahi A, et al:
Multidisciplinary approach to a nonsyndromic oligodontia patient
using advanced surgical techniques. Int J Periodontics Restorative
Dent. 31:297–305. 2011.PubMed/NCBI
|
|
10
|
Nakatomi M, Wang XP, Key D, et al: Genetic
interactions between Pax9 and Msx1 regulate lip development and
several stages of tooth morphogenesis. Dev Biol. 340:438–449. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De Coster PJ, Marks LA, Martens LC and
Huysseune A: Dental agenesis: genetic and clinical perspectives. J
Oral Pathol Med. 38:1–17. 2009. View Article : Google Scholar
|
|
12
|
Letra A, Menezes R, Granjeiro JM, et al:
AXIN2 and CDH1 polymorphisms, tooth agenesis, and oral clefts.
Birth Defects Res A Clin Mol Teratol. 85:169–173. 2009. View Article : Google Scholar :
|
|
13
|
Mostowska A, Biedziak B and Jagodzinski
PP: Axis inhibition protein 2 (AXIN2) polymorphisms may be a risk
factor for selective tooth agenesis. J Hum Genet. 51:262–266. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang J, Song G, Li Q, et al: Novel
missense mutations in PAX9 causing oligodontia. Arch Oral Biol.
57:784–789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Vieira AR: Oral clefts and syndromic forms
of tooth agenesis as models for genetics of isolated tooth
agenesis. J Dent Res. 82:162–165. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang YD, Chen Z, Song YQ, et al: Making a
tooth: growth factors, transcription factors, and stem cells. Cell
Res. 15:301–316. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Amen M, Liu X, Vadlamudi U, et al: PITX2
and β-catenin interactions regulate lef-1 isoform expression. Mol
Cel Biol. 27:7560–7573. 2007. View Article : Google Scholar
|
|
18
|
Kangas AT, Evans AR, Thesleff I, et al:
Nonindependence of mammalian dental characters. Nature.
432:211–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Küchler EC, Lips A, Tannure PN, et al:
Tooth agenesis association with self-reported family history of
cancer. J Dent Res. 92:149–155. 2013. View Article : Google Scholar
|
|
20
|
Paixao-Cortes VR, Braga T, Salzano FM, et
al: PAX9 and MSX1 transcription factor genes in non-syndromic
dental agenesis. Arch Oral Biol. 56:337–344. 2011. View Article : Google Scholar
|
|
21
|
Menezes R, Marazita ML, Goldstein McHenry
T, et al: AXIS inhibition protein 2, orofacial clefts and a family
history of cancer. J Am Dent Assoc. 140:80–84. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ruhin-Poncet B, Ghoul-Mazgar S, Hotton D,
et al: Msx and dlx homeogene expression in epithelial odontogenic
tumors. J Histochem Cytochem. 57:69–78. 2009. View Article : Google Scholar :
|
|
23
|
Kapadia H, Mues G and D’Souza R: Genes
affecting tooth morphogenesis. Orthod Craniofac Res. 10:105–113.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ovári G, Molnár B, Tarján I, et al: Gene
polymorphisms in periodontitis and hypodontia: methodological basis
of investigations. Fogorv Sz. 100:266–272. 259–265. 2007.PubMed/NCBI
|
|
25
|
Townsend G, Bockmann M, Hughes T, et al:
Genetic, environmental and epigenetic influences on variation in
human tooth number, size and shape. Odontology. 100:1–9. 2012.
View Article : Google Scholar
|
|
26
|
Wang J and Abate-Shen C: Transcriptional
repression by the Msx1 homeoprotein is associated with global
redistribution of the H3K27me3 repressive mark to the nuclear
periphery. Nucleus. 3:155–161. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pani SC: The genetic basis of tooth
agenesis: basic concepts and genes involved. J Indian Soc Pedod
Prev Dent. 29:84–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
dos Pinheiro RS, Otero RA, Portela MB and
Castro GF: Severe oligodontia and dental anomalies in a child with
a history of multiple natal teeth: An eight-year retrospective. Gen
Dent. 59:e248–e250. 2011.
|
|
29
|
Bural C, Oztas E, Ozturk S, et al:
Multidisciplinary treatment of non-syndromic oligodontia. Eur J
Dent. 6:218–226. 2012.PubMed/NCBI
|